A Novel Homozygous GPAA1 Variant in a Patient with a Glycosylphosphatidylinositol Biosynthesis Defect
Abstract
:1. Introduction
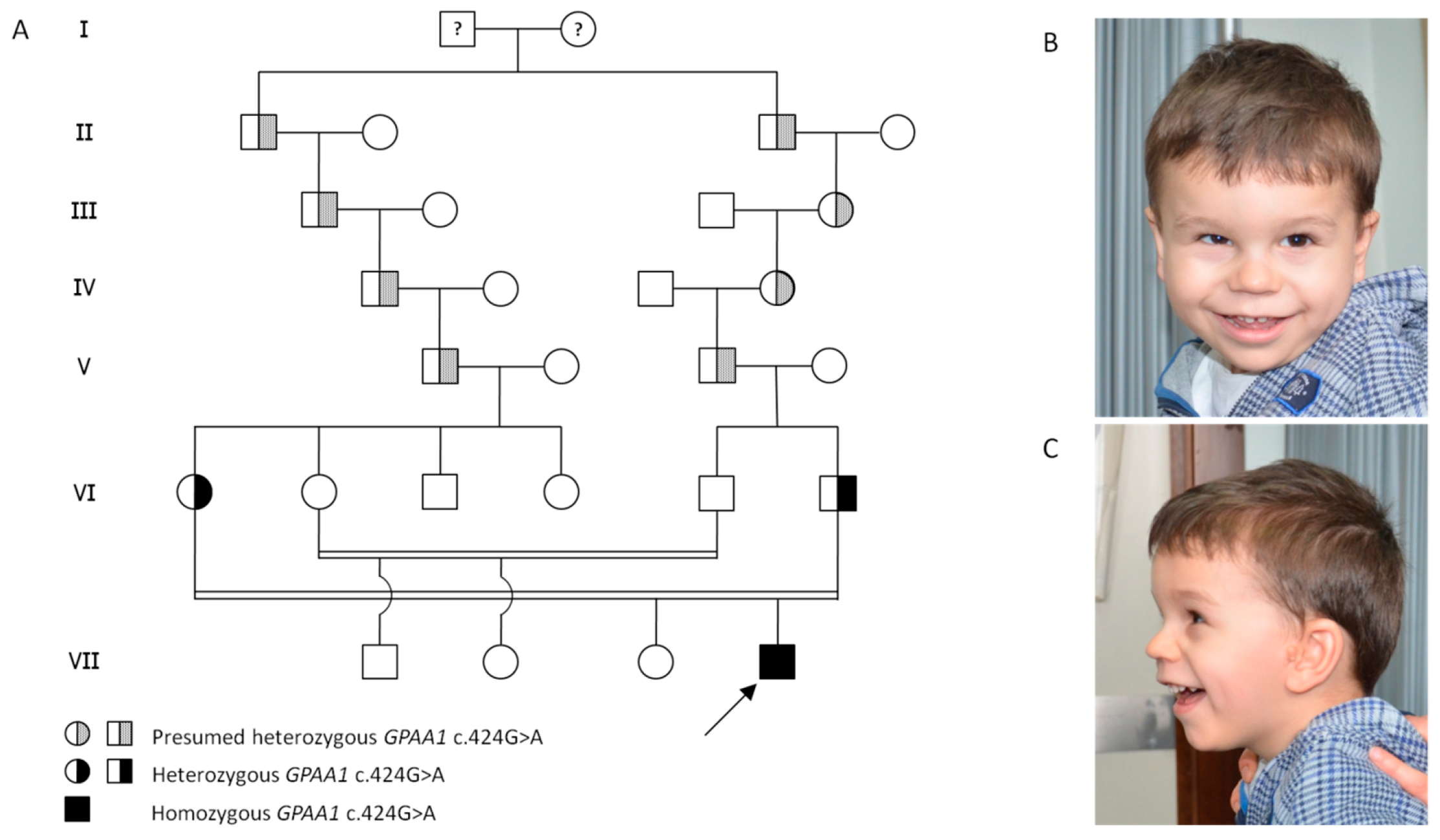
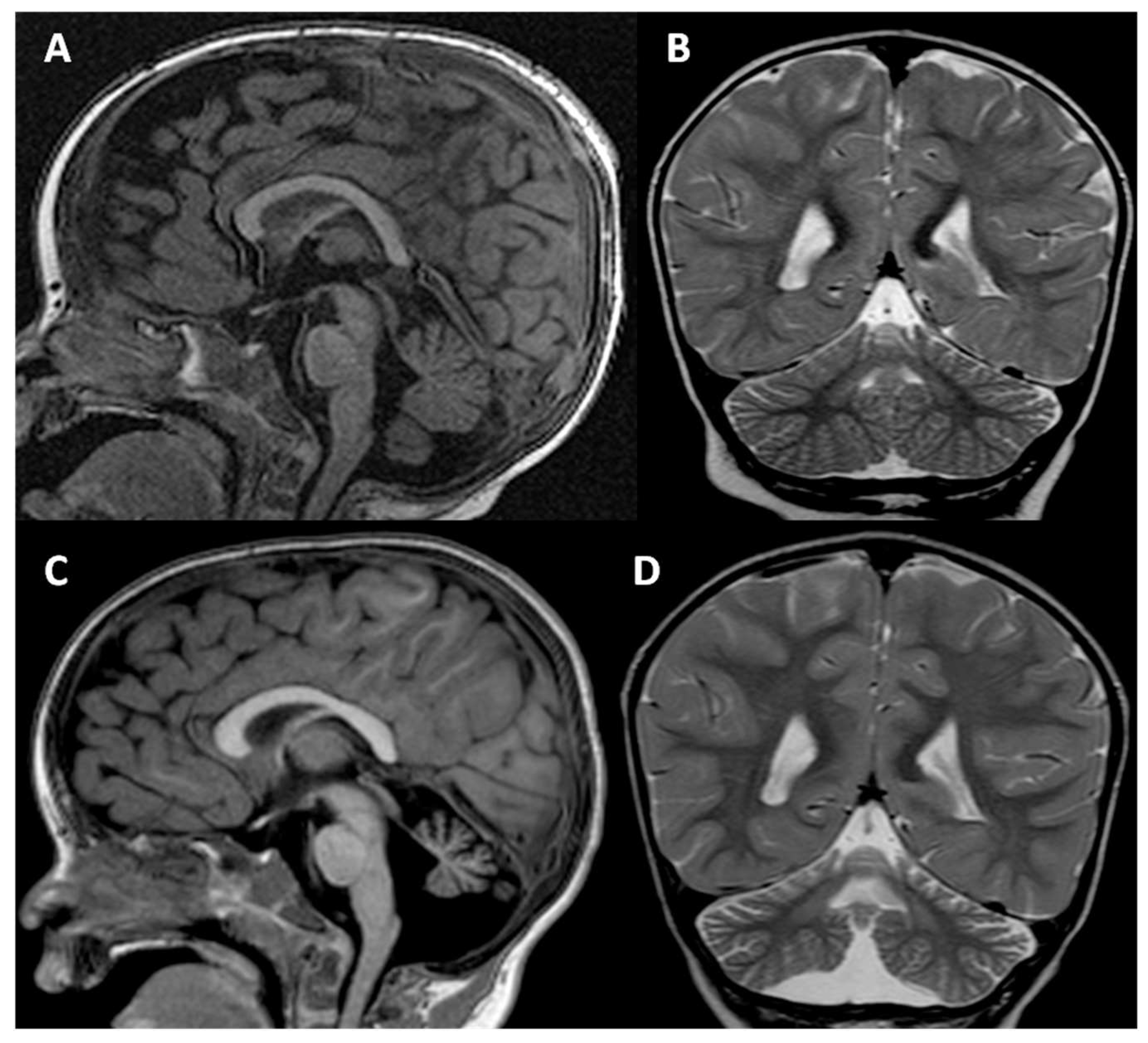
2. Clinical Report
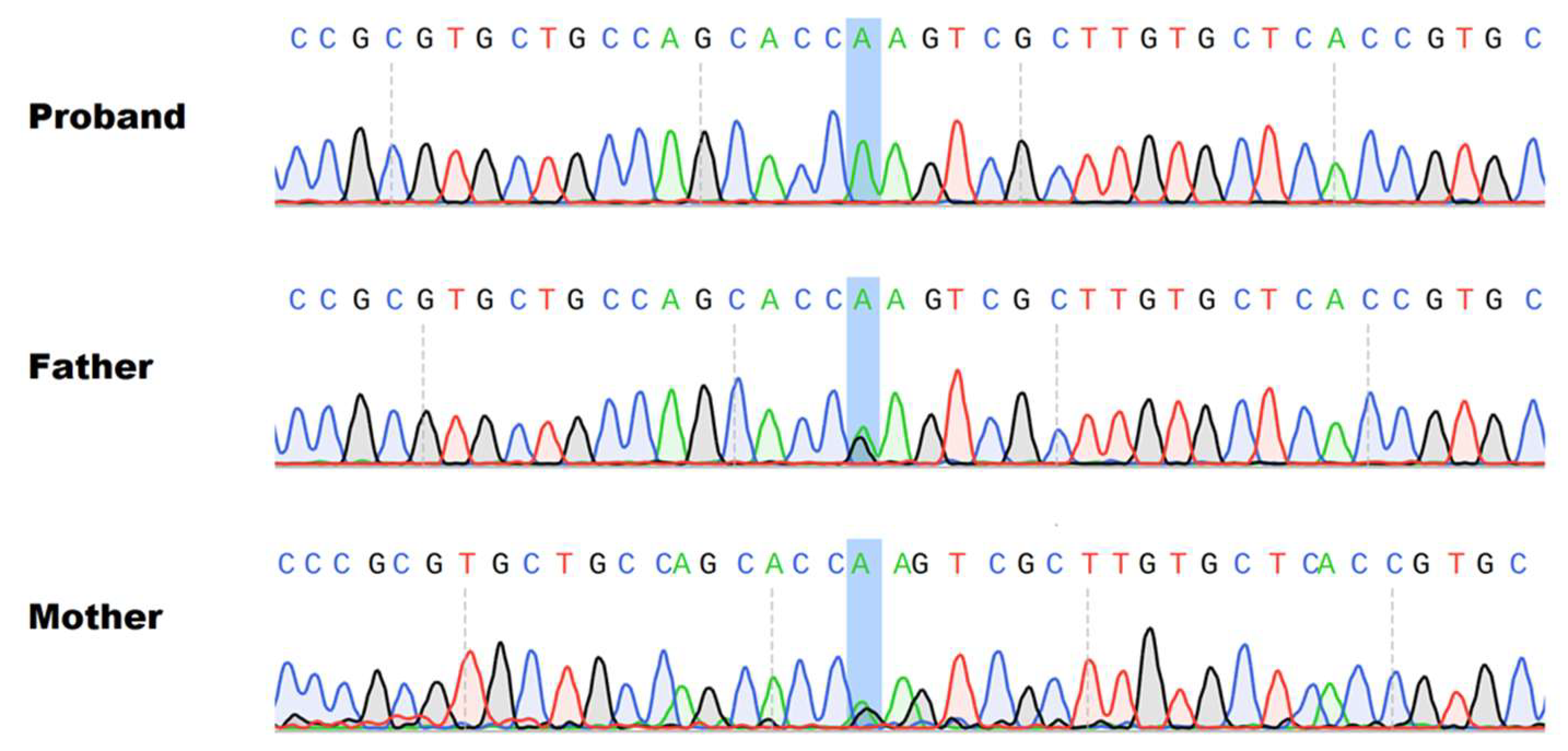
3. Materials and Methods
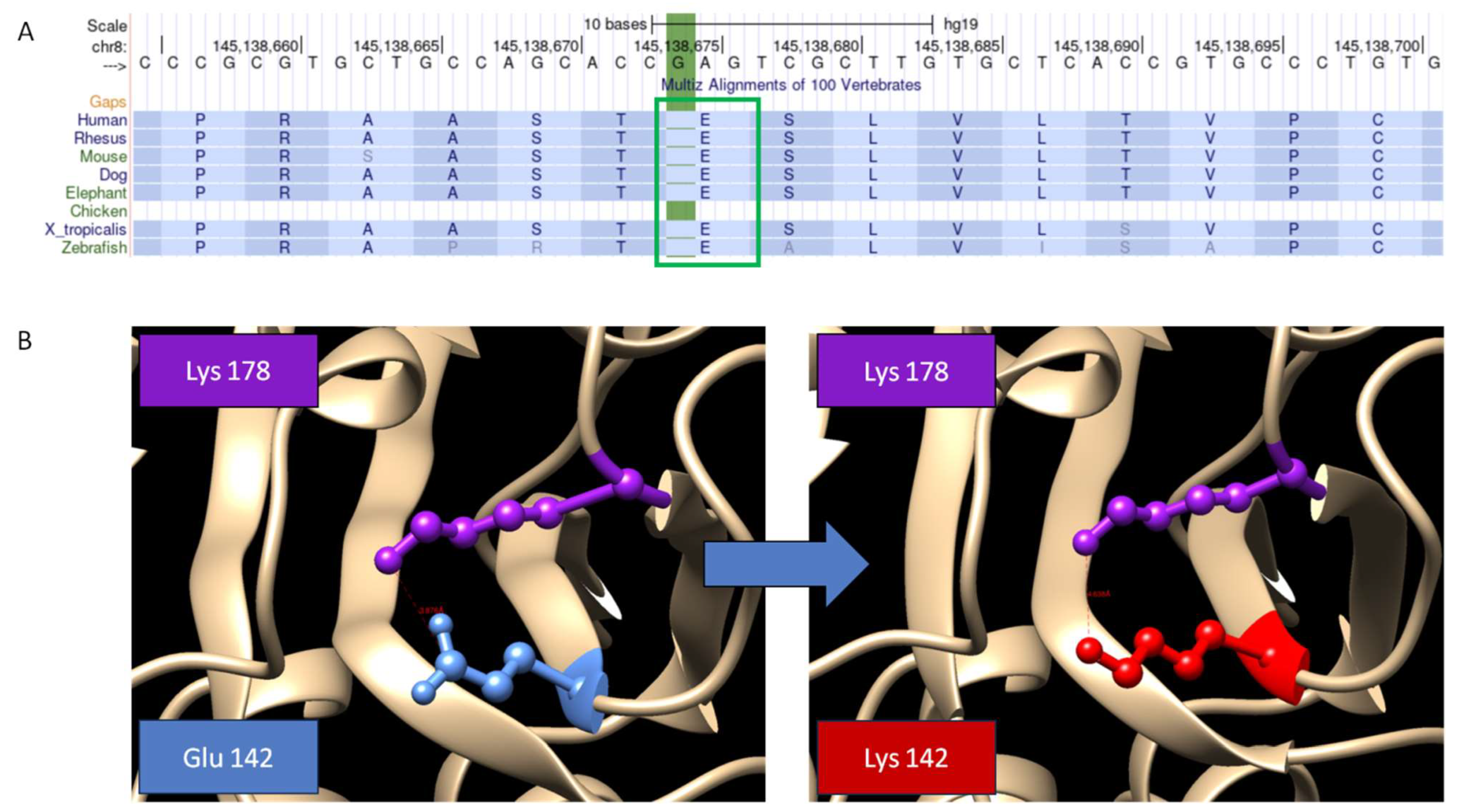
4. Results
5. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Chang, I.J.; He, M.; Lam, C.T. Congenital disorders of glycosylation. Ann. Transl. Med. 2018, 6, 477. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Yin, F.; Guang, S.; He, F.; Yang, L.; Peng, J. The Glycosylphosphatidylinositol biosynthesis pathway in human diseases. Orphanet J. Rare Dis. 2020, 15, 129. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, T. Biosynthesis and biology of mammalian GPI-anchored proteins. Open Biol. 2020, 10, 190290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kinoshita, T. Glycosylphosphatidylinositol (GPI) Anchors: Biochemistry and Cell Biology: Introduction to a Thematic Review Series. J. Lipid Res. 2016, 57, 4–5. [Google Scholar] [CrossRef] [Green Version]
- Vainauskas, S.; Maeda, Y.; Kurniawan, H.; Kinoshita, T.; Menon, A.K. Structural requirements for the recruitment of Gaa1 into a functional glycosylphosphatidylinositol transamidase complex. J. Biol. Chem. 2002, 277, 30535–30542. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, T.T.M.; Murakami, Y.; Sheridan, E.; Ehresmann, S.; Rousseau, J.; St-Denis, A.; Chai, G.; Ajeawung, N.F.; Fairbrother, L.; Reimschisel, T.; et al. Mutations in GPAA1, Encoding a GPI Transamidase Complex Protein, Cause Developmental Delay, Epilepsy, Cerebellar Atrophy, and Osteopenia. Am. J. Hum. Genet. 2017, 101, 856–865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castle, A.M.; Salian, S.; Bassan, H.; Sofrin-Drucker, E.; Cusmai, R.; Herman, K.C.; Heron, D.; Keren, B.; Johnstone, D.L.; Mears, W.; et al. Expanding the Phenotypic Spectrum of GPI Anchoring Deficiency Due to Biallelic Variants in GPAA1. Neurol. Genet. 2021, 7, e631. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yang, L.; Yang, J.; Shi, J.; Chai, P.; Ge, S.; Wang, Y.; Fan, X.; Jia, R. A novel variant in GPAA1, encoding a GPI transamidase complex protein, causes inherited vascular anomalies with various phenotypes. Hum. Genet. 2020, 139, 1499–1511. [Google Scholar] [CrossRef] [PubMed]
- Musacchia, F.; Tudp; Ciolfi, A.; Mutarelli, M.; Bruselles, A.; Castello, R.; Pinelli, M.; Basu, S.; Banfi, S.; Casari, G.; et al. VarGenius executes cohort-level DNA-seq variant calling and annotation and allows to manage the resulting data through a PostgreSQL database. BMC Bioinform. 2018, 19, 477. [Google Scholar] [CrossRef] [PubMed]
- Lukacs, M.; E Blizzard, L.; Stottmann, R.W. CNS glycosylphosphatidylinositol deficiency results in delayed white matter development, ataxia and premature death in a novel mouse model. Hum. Mol. Genet. 2020, 29, 1205–1217. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.T.M.; Murakami, Y.; Wigby, K.M.; Baratang, N.V.; Rousseau, J.; St-Denis, A.; Rosenfeld, J.A.; Laniewski, S.C.; Jones, J.; Iglesias, A.D.; et al. Mutations in PIGS, Encoding a GPI Transamidase, Cause a Neurological Syndrome Ranging from Fetal Akinesia to Epileptic Encephalopathy. Am. J. Hum. Genet. 2018, 103, 602–611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kvarnung, M.; Nilsson, D.; Lindstrand, A.; Korenke, G.C.; Chiang, S.C.C.; Blennow, E.; Bergmann, M.; Stödberg, T.; Mäkitie, O.; Anderlid, B.-M.; et al. A novel intellectual disability syndrome caused by GPI anchor deficiency due to homozygous mutations inPIGT. J. Med. Genet. 2013, 50, 521–528. [Google Scholar] [CrossRef] [PubMed]
- Knaus, A.; Kortüm, F.; Kleefstra, T.; Stray-Pedersen, A.; Đukić, D.; Murakami, Y.; Gerstner, T.; van Bokhoven, H.; Iqbal, Z.; Horn, D.; et al. Mutations in PIGU Impair the Function of the GPI Transamidase Complex, Causing Severe Intellectual Disability, Epilepsy, and Brain Anomalies. Am. J. Hum. Genet. 2019, 105, 395–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, T.T.M.; Murakami, Y.; Mobilio, S.; Niceta, M.; Zampino, G.; Philippe, C.; Moutton, S.; Zaki, M.S.; James, K.N.; Musaev, D.; et al. Bi-allelic Variants in the GPI Transamidase Subunit PIGK Cause a Neurodevelopmental Syndrome with Hypotonia, Cerebellar Atrophy, and Epilepsy. Am. J. Hum. Genet. 2020, 106, 484–495. [Google Scholar] [CrossRef] [PubMed]
- Efthymiou, S.; Dutra-Clarke, M.; Maroofian, R.; Kaiyrzhanov, R.; Scala, M.; Alvi, J.R.; Sultan, T.; Christoforou, M.; Nguyen, T.T.M.; Mankad, K.; et al. Expanding the phenotype of PIGS -associated early onset epileptic developmental encephalopathy. Epilepsia 2021, 62, e35–e41. [Google Scholar] [CrossRef] [PubMed]
- Beleza-Meireles, A.; Hart, R.; Clayton-Smith, J.; Oliveira, R.; Reis, C.F.; Venâncio, M.; Ramos, F.; Sá, J.; Ramos, L.; Cunha, E.; et al. Oculo-auriculo-vertebral spectrum: Clinical and molecular analysis of 51 patients. Eur. J. Med Genet. 2015, 58, 455–465. [Google Scholar] [CrossRef] [PubMed]
| Variants | Inheritance | Age Years | Height % | Weight % | OFC % | DD ID | Hypotonia | Cerebellar Atrophy | Epilepsy | Nystagmus | Ataxia | Osteo-Penia | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| This Case | c.424G > A | homoz | 4 | 10 | 5 | 55 | + | + | + | + | + | + | - |
| N. 1a | c.872T > C c.981_993del | compound heteroz | 15 | 8 | 58 | 14 | + | + | + | + | + | + | + |
| N. 1b | c.872T > C c.981_993del | compound heteroz | 10 | 4 | 16 | 27 | + | + | + | + | + | + | + |
| N. 2 | c.152C > T c.1164 + 5C > T | compound heteroz | 6 | 59 | 30 | 87 | + | + | - | + | - | NA | + |
| N. 3a | c.920delG c.1165G > C | compound heteroz | 10 | 1 | 49 | 1 | + | + | + | + | + | NA | + |
| N. 3b | c.920delG c.1165G > C | compound heteroz | 3 | 15 | 19 | 19 | + | + | + | + | + | NA | + |
| N. 4a | c.527G > C | homoz | 8 | 1 | 27 | 31 | + | + | + | - | + | + | + |
| N. 4b | c.527G > C | homoz | 5 | 1 | 23 | 4 | + | + | + | - | - | + | + |
| N. 4c | c.527G > C | homoz | 4 | 1 | 31 | 18 | + | + | + | - | + | + | + |
| N. 5a | c.160_161delinsAA c.869T > C | compound heteroz | 30 | −2 SD | 48 | 50 | + | + | + | + | + | + | NA |
| N. 5b | c.160_161delinsAA c.869T > C | compound heteroz | 25 | −1.8 SD | 31 | 50 | + | + | + | + | + | + | NA |
| C. I | c.164T > C | homoz | 38 | 17 | 72 | 93 | + | + | + | + | - | + | + |
| C. II | c.1049T > G | homoz | 1 | <1 | 23 | 13 | + | - | - | - | - | - | NA |
| C. III | c.917A > G c.1559T > G | compound heteroz | 3 | <1 | 3 | 1 | + | + | + | + | - | - | NA |
| C. IV | c.947C > T | homoz | 5 | 33 | 96 | <1 | + | + | - | + | + | + | NA |
| C. V | c.947C > T c.1233_1239del | compound heteroz | 3 | 1 | <1 | 2 | + | + | + | + | + | - | NA |
| C. VI | c.1477_1478del c.1831T > C | compound heteroz | 5 | 7 | 31 | 69 | + | + | NA | + | - | - | NA |
| C. VII | c.619delA c.149T > A | compound heteroz | 3 | 59 | 72 | 34 | + | + | - | + | - | + | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fontana, P.; Budillon, A.; Simeone, D.; Del Vecchio Blanco, F.; Caiazza, M.; D’Amico, A.; Lonardo, F.; Nigro, V.; Limongelli, G.; Scarano, G. A Novel Homozygous GPAA1 Variant in a Patient with a Glycosylphosphatidylinositol Biosynthesis Defect. Genes 2023, 14, 1444. https://doi.org/10.3390/genes14071444
Fontana P, Budillon A, Simeone D, Del Vecchio Blanco F, Caiazza M, D’Amico A, Lonardo F, Nigro V, Limongelli G, Scarano G. A Novel Homozygous GPAA1 Variant in a Patient with a Glycosylphosphatidylinositol Biosynthesis Defect. Genes. 2023; 14(7):1444. https://doi.org/10.3390/genes14071444
Chicago/Turabian StyleFontana, Paolo, Alberto Budillon, Domenico Simeone, Francesca Del Vecchio Blanco, Martina Caiazza, Alessandra D’Amico, Fortunato Lonardo, Vincenzo Nigro, Giuseppe Limongelli, and Gioacchino Scarano. 2023. "A Novel Homozygous GPAA1 Variant in a Patient with a Glycosylphosphatidylinositol Biosynthesis Defect" Genes 14, no. 7: 1444. https://doi.org/10.3390/genes14071444
APA StyleFontana, P., Budillon, A., Simeone, D., Del Vecchio Blanco, F., Caiazza, M., D’Amico, A., Lonardo, F., Nigro, V., Limongelli, G., & Scarano, G. (2023). A Novel Homozygous GPAA1 Variant in a Patient with a Glycosylphosphatidylinositol Biosynthesis Defect. Genes, 14(7), 1444. https://doi.org/10.3390/genes14071444









