ERCC1 and MGMT Methylation as a Predictive Marker of Relapse and FOLFOX Response in Colorectal Cancer Patients from South Tunisia
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Samples
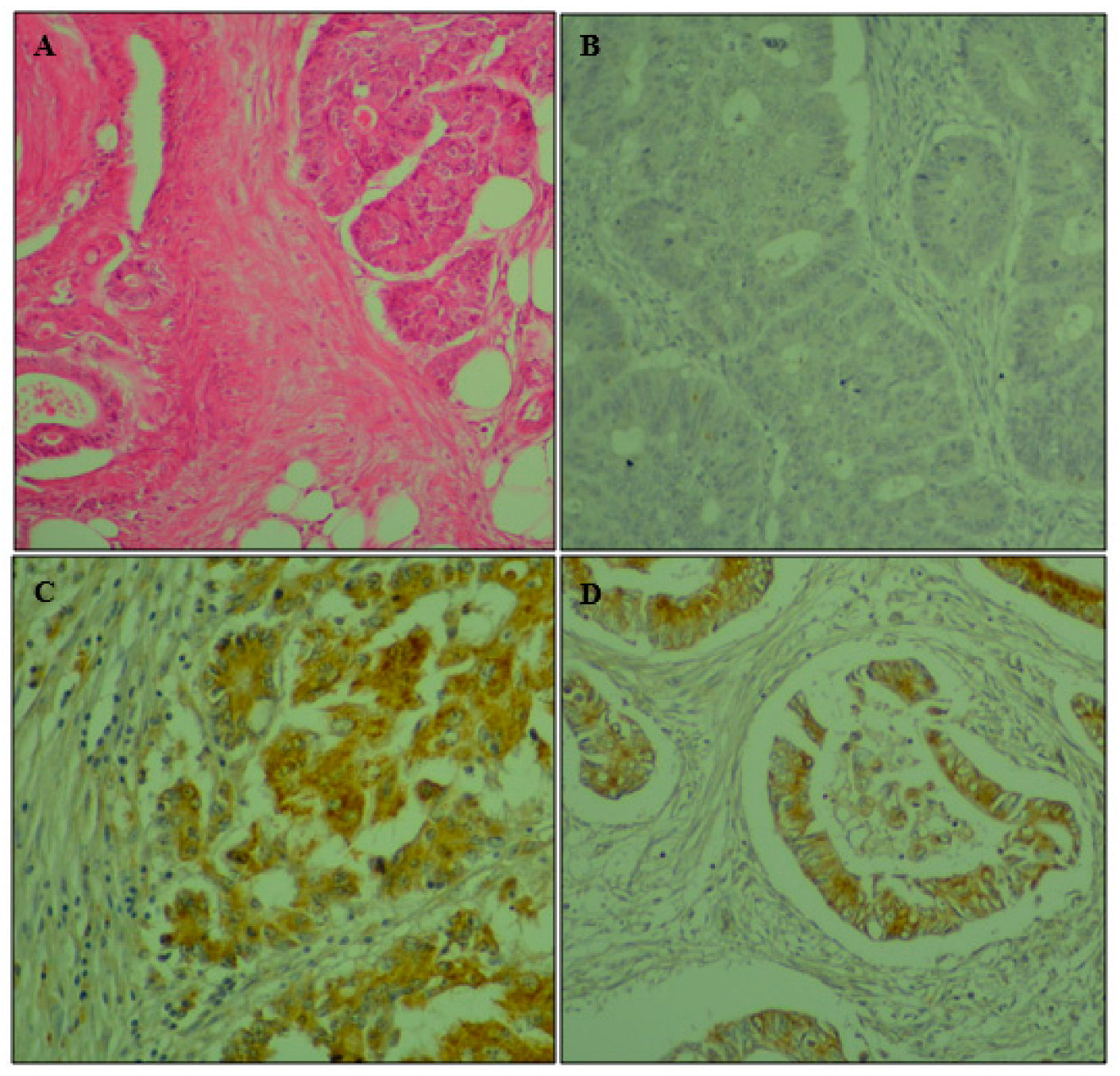
2.2. Immunohistochemistry
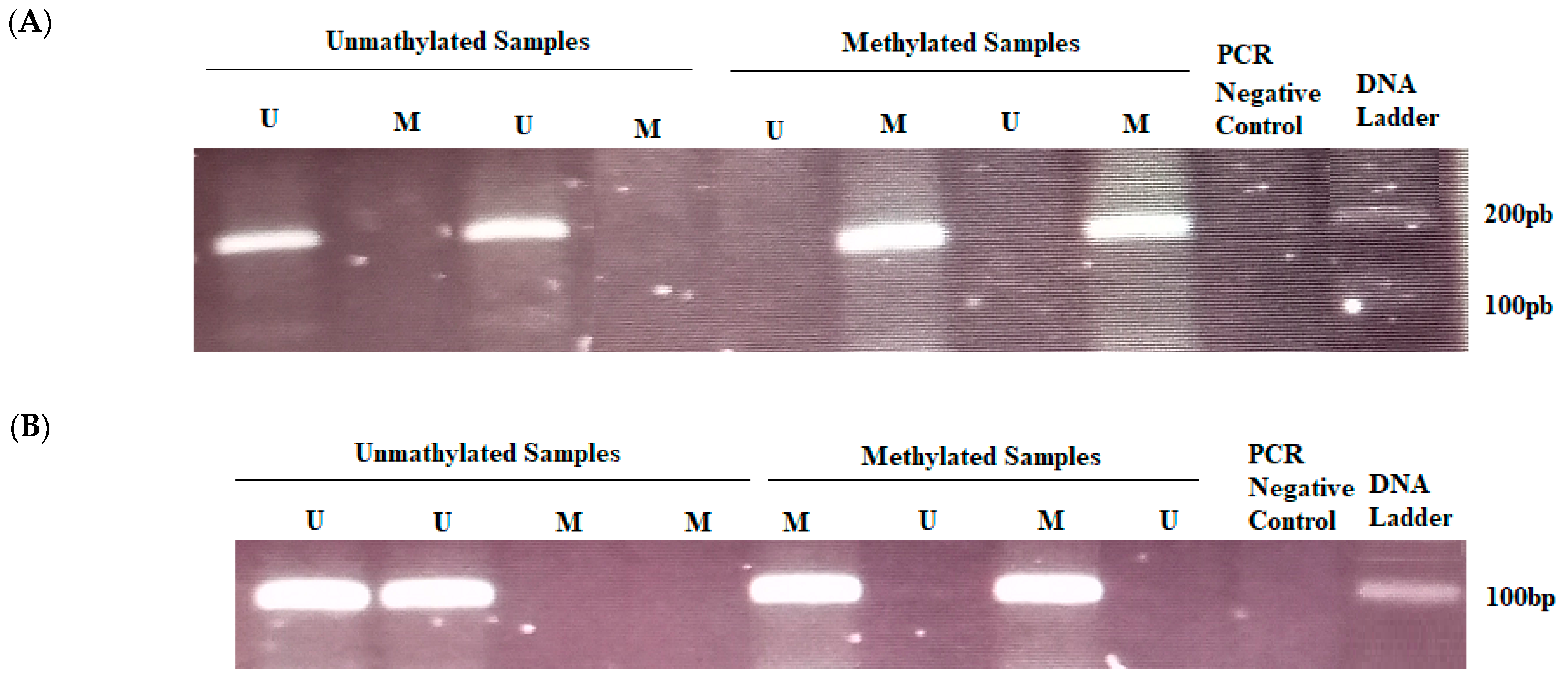
2.3. DNA Extraction, Bisulfite Treatment, and Methylation-Specific PCR
2.4. ERCC1 Polymorphism Study by PCR-RFLP
2.5. Statistical Analysis
3. Results
3.1. Patients
3.2. Immunohistochemistry
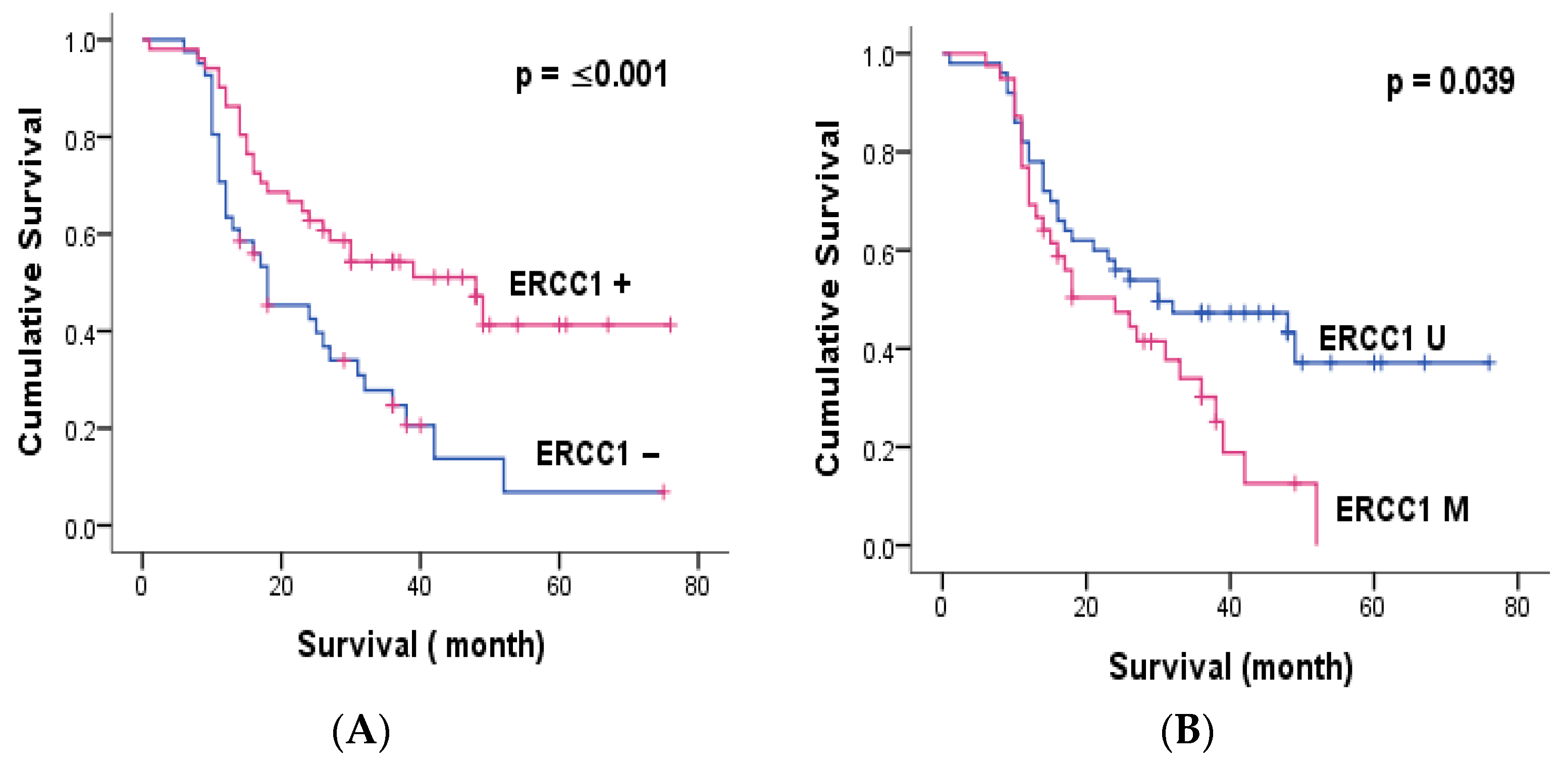
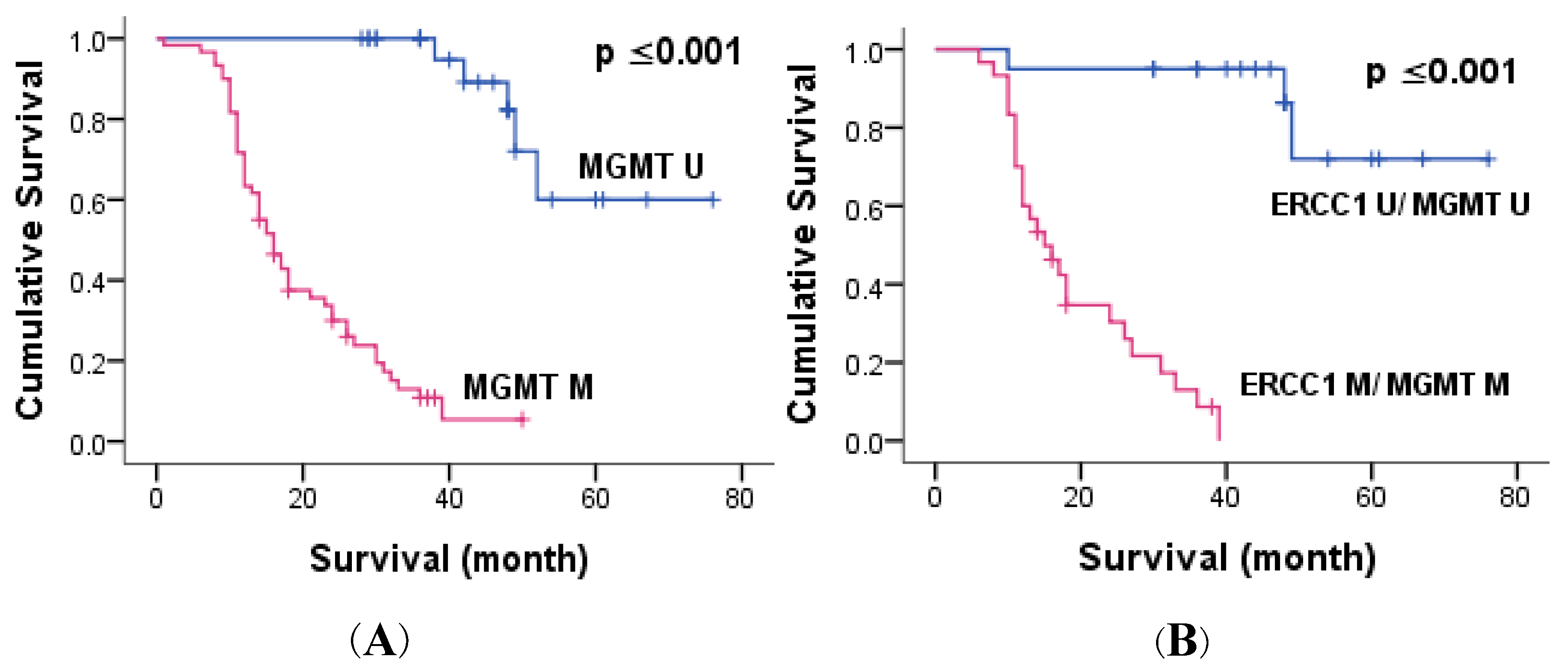
3.3. ERCC1 and MGMT Methylation
3.4. Correlation of ERCC1 Polymorphism with Clinicopathological Parameters, ERCC1 Expression and Methylation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Torgovnick, A.; Schumacher, B. DNA Repair Mechanisms in Cancer Development and Therapy. Front. Genet. 2015, 6, 157. [Google Scholar] [CrossRef] [PubMed]
- Martin, L.P.; Hamilton, T.C.; Schilder, R.J. Platinum Resistance: The Role of DNA Repair Pathways. Clin. Cancer Res. 2008, 14, 1291–1295. [Google Scholar] [CrossRef]
- Sharma, S.; Salehi, F.; Scheithauer, B.W.; Rotondo, F.; Syro, L.V.; Kovacs, K. Role of MGMT in Tumor Development, Progression, Diagnosis, Treatment and Prognosis. Anticancer Res. 2009, 29, 3759–3768. [Google Scholar] [PubMed]
- Shuck, S.C.; Short, E.A.; Turchi, J.J. Eukaryotic Nucleotide Excision Repair: From Understanding Mechanisms to Influencing Biology. Cell Res. 2008, 18, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Sancar, A. DNA Excision Repair. Annu. Rev. Biochem. 1996, 65, 43–81. [Google Scholar] [CrossRef]
- Petit, C.; Sancar, A. Nucleotide Excision Repair: From E. coli to Man. Biochimie 1999, 81, 15–25. [Google Scholar] [CrossRef]
- Spivak, G. Nucleotide Excision Repair in Humans. DNA Repair 2015, 36, 13–18. [Google Scholar] [CrossRef]
- Jiang, H.; Li, B.; Wang, F.; Ma, C.; Hao, T. Expression of ERCC1 and TYMS in Colorectal Cancer Patients and the Predictive Value of Chemotherapy Efficacy. Oncol. Lett. 2019, 18, 1157–1162. [Google Scholar] [CrossRef]
- Gajjar, K.K.; Yadav, D.K.; Kobawala, T.P.; Trivedi, T.I.; Vora, H.H.; Ghosh, N.R. ERCC1 Expression in Patients with Colorectal Cancer: A Pilot Study. J. Cancer Metastasis Treat. 2016, 2, 471. [Google Scholar] [CrossRef]
- Rao, D.; Mallick, A.B.; Augustine, T.; Daroqui, C.; Jiffry, J.; Merla, A.; Chaudhary, I.; Seetharam, R.; Sood, A.; Gajavelli, S.; et al. Excision Repair Cross-Complementing Group-1 (ERCC1) Induction Kinetics and Polymorphism Are Markers of Inferior Outcome in Patients with Colorectal Cancer Treated with Oxaliplatin. Oncotarget 2019, 10, 5510–5522. [Google Scholar] [CrossRef]
- Zaanan, A.; Dalban, C.; Emile, J.F.J.F.; Blons, H.; Fléjou, J.F.; Goumard, C.; Istanbullu, M.; Calmel, C.; Alhazmi, K.; Validire, P.; et al. ERCC1, XRCC1 and GSTP1 Single Nucleotide Polymorphisms and Survival of Patients with Colon Cancer Receiving Oxaliplatin-Based Adjuvant Chemotherapy. J. Cancer 2014, 5, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Salimzadeh, H.; Lindskog, E.B.; Gustavsson, B.; Wettergren, Y.; Ljungman, D. Association of DNA Repair Gene Variants with Colorectal Cancer: Risk, Toxicity, and Survival. BMC Cancer 2020, 20, 409. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Sun, N.; Hu, G.; Chen, X.; Jiang, J.; Wu, H.; Luo, G. Association of ERCC1 Polymorphisms with the Risk of Colorectal Cancer: A Meta-Analysis. Crit. Rev. Eukaryot. Gene Expr. 2017, 27, 267–275. [Google Scholar] [CrossRef]
- Shalaby, S.M.; El-Shal, A.S.; Abdelaziz, L.A.; Abd-Elbary, E.; Khairy, M.M. Promoter Methylation and Expression of DNA Repair Genes MGMT and ERCC1 in Tissue and Blood of Rectal Cancer Patients. Gene 2018, 644, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Tubbs, J.L.; Pegg, A.E.; Tainer, J.A. DNA Binding, Nucleotide Flipping, and the Helix-Turn-Helix Motif in Base Repair by O6-Alkylguanine-DNA Alkyltransferase and Its Implications for Cancer Chemotherapy. DNA Repair 2007, 6, 1100–1115. [Google Scholar] [CrossRef]
- Ahmed, M.; Bayoumi, B.; Abdallah, S.; Elserafy, M. Mgmt Immunohistochemical Expression in Colorectal Carcinoma and Its Correlation with Tumor Progression. Open Access Maced. J. Med. Sci. 2021, 9, 244–251. [Google Scholar] [CrossRef]
- Li, Y.; Lyu, Z.; Zhao, L.; Cheng, H.; Zhu, D.; Gao, Y.; Shang, X.; Shi, H. Prognostic Value of MGMT Methylation in Colorectal Cancer: A Meta-Analysis and Literature Review. Tumor Biol. 2015, 36, 1595–1601. [Google Scholar] [CrossRef]
- Kassem, A.B.; Salem, S.E.; Abdelrahim, M.E.; Said, A.S.A.; Salahuddin, A.; Hussein, M.M.; Bahnassy, A.A. ERCC1 and ERCC2 as Predictive Biomarkers to Oxaliplatin-Based Chemotherapy in Colorectal Cancer Patients from Egypt. Exp. Mol. Pathol. 2017, 102, 78–85. [Google Scholar] [CrossRef]
- Park, S.M.; Choi, S.B.; Lee, Y.S.; Lee, I.K. Predictive Value of KRAS Mutation and Excision Repair Cross-Complementing 1 (ERCC1) Protein Overexpression in Patients with Colorectal Cancer Administered FOLFOX Regimen. Asian J. Surg. 2021, 44, 715–722. [Google Scholar] [CrossRef]
- Gabr, A.; Elsaba, T.M.; Razek, K.; Tamam, S.; Atta, H. Excision Repair Cross-Complementation Group 1 (ERCC1): A Prognostic and Predictive Biomarker in Patients with Colorectal Cancer Receiving Adjuvant Oxaliplatin Based Chemotherapy. J. Cancer Ther. 2016, 7, 622–634. [Google Scholar] [CrossRef]
- Choueiri, M.B.; Shen, J.P.; Gross, A.M.; Huang, J.K.; Ideker, T.; Fanta, P. ERCC1 and TS Expression as Prognostic and Predictive Biomarkers in Metastatic Colon Cancer. PLoS ONE 2015, 10, e0126898. [Google Scholar] [CrossRef] [PubMed]
- Abyarghamsari, M.; Shirazi, F.H.; Tavakoli-Ardakani, M.; Rezvani, H.; Mirzaei, H.R.; Salamzadeh, J. Study of the Relationship between Ercc1 Polymorphisms and Response to Platinum-Based Chemotherapy in Iranian Patients with Colorectal and Gastric Cancers. Iran. J. Pharm. Res. 2019, 18, 2163–2171. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.M.H.; Tzeng, C.H.; Chen, P.M.; Lin, J.K.; Lin, T.C.; Chen, W.S.; Jiang, J.K.; Wang, H.S.; Wang, W.S. ERCC1 Codon 118 C→T Polymorphism Associated with ERCC1 Expression and Outcome of FOLFOX-4 Treatment in Asian Patients with Metastatic Colorectal Carcinoma. Cancer Sci. 2009, 100, 278–283. [Google Scholar] [CrossRef] [PubMed]
- Feldheim, J.; Kessler, A.F.; Monoranu, C.M.; Ernestus, R.I.; Löhr, M.; Hagemann, C. Changes of O6-Methylguanine Dna Methyltransferase (Mgmt) Promoter Methylation in Glioblastoma Relapse—A Meta-Analysis Type Literature Review. Cancers 2019, 11, 1837. [Google Scholar] [CrossRef]
- Yuan, X.; Xu, J.; Fang, W.; Zhao, Z.; Wang, F.; Tong, Z. The Association between MGMT Promoter Methylation and Patients with Gastric Cancer: A Meta-Analysis. Genet. Test. Mol. Biomarkers 2017, 21, 213–221. [Google Scholar] [CrossRef]
- Lin, D.; Xiao, Y.; Huang, B.; Wu, X.; Chen, C.; Liang, Y.; Zeng, D. O-6-Methylguanine DNA Methyltransferase Is a Favorable Biomarker with Proliferation Suppressive Potential in Breast Cancer. J. Cancer 2020, 11, 6326–6336. [Google Scholar] [CrossRef]
- Kang, B.; Lee, H.S.; Jeon, S.W.; Park, S.Y.; Choi, G.S.; Lee, W.K.; Heo, S.; Lee, D.H.; Kim, D.S. Progressive Alteration of DNA Methylation of Alu, MGMT, MINT2, and TFPI2 Genes in Colonic Mucosa during Colorectal Cancer Development. Cancer Biomark. 2021, 32, 231–236. [Google Scholar] [CrossRef]
- Oliver, J.A.; Ortiz, R.; Melguizo, C.; Álvarez, P.J.; Gómez-Millán, J.; Prados, J. Prognostic Impact of MGMT Promoter Methylation and MGMT and CD133 Expression in Colorectal Adenocarcinoma. BMC Cancer 2014, 14, 511. [Google Scholar] [CrossRef]
- Barault, L.; Amatu, A.; Bleeker, F.E.; Moutinho, C.; Falcomatà, C.; Fiano, V.; Cassingena, A.; Siravegna, G.; Milione, M.; Cassoni, P.; et al. Digital PCR Quantification of MGMT Methylation Refines Prediction of Clinical Benefit from Alkylating Agents in Glioblastoma and Metastatic Colorectal Cancer. Ann. Oncol. 2015, 26, 1994–1999. [Google Scholar] [CrossRef]
- Kim, C.Y.; Seo, S.H.; An, M.S.; Kim, K.H.; Bae, K.B.; Hwang, J.W.; Kim, J.H.; Kim, B.M.; Kang, M.S.; Oh, M.K.; et al. ERCC1 as a Predictive Marker for FOLFOX Chemotherapy in an Adjuvant Setting. Ann. Coloproctol. 2015, 31, 92–97. [Google Scholar] [CrossRef]
- Campana, D.; Walter, T.; Pusceddu, S.; Gelsomino, F.; Graillot, E.; Prinzi, N.; Spallanzani, A.; Fiorentino, M.; Barritault, M.; Dall’Olio, F.; et al. Correlation between MGMT Promoter Methylation and Response to Temozolomide-Based Therapy in Neuroendocrine Neoplasms: An Observational Retrospective Multicenter Study. Endocrine 2018, 60, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Pietrantonio, F.; Lobefaro, R.; Antista, M.; Lonardi, S.; Raimondi, A.; Morano, F.; Mosconi, S.; Rimassa, L.; Murgioni, S.; Sartore-Bianchi, A.; et al. Capecitabine and Temozolomide versus FOLFIRI in RAS-Mutated, MGMT-Methylated Metastatic Colorectal Cancer. Clin. Cancer Res. 2020, 26, 1017–1024. [Google Scholar] [CrossRef] [PubMed]
- Ishibashl, K.; Okada, N.; Ishiguro, T.; Kuwabara, K.; Ohsawa, T.; Yokoyama, M.; Kumamoto, K.; Haga, N.; Mori, T.; Yamada, H.; et al. The Expression of Thymidylate Synthase (TS) and Excision Repair Complementing-1 (ERCC-1) Protein in Patients with Unresectable Colorectal Cancer Treated with MFOLFOX6 Therapy. Jpn. J. Cancer Chemother. 2010, 37, 2532–2535. [Google Scholar]
| Primer | Sequence | Annealing Temperature | |
|---|---|---|---|
| MSP ERCC1 | UF | 5′-TGGAATTGTTGGTGAGGGTTTTG-3′ | 55 |
| UR | 5′-ACCTTCCCCTCCTCTCAACTT-3′ | ||
| MF | 5′-CGGAATTGTCGGTGAGGGTTTCG-3′ | 58.5 | |
| MR | 5′-ACCTTCCCCTCCTCTCAACTT-3′ | ||
| MSP MGMT | UF | 5′-TTGTGTTTTGATGTTTGTAGGTTTTTGT-3′ | 55 |
| UR | 5′-AACTCCAGACTCTTCCAAAAACAAAACA-3′ | ||
| MF | 5′-TTTCGACGTTCGTAGGTTTTCGC-3′ | 57 | |
| MR | 5′-GCACTCTTCCGAAAACGAAACG-3′ | ||
| ERCC1 SNP | F | 5′-GCAGAGCTCACCTGAGGAAC-3′ | 56.5 |
| R | 5′-GAGGTGCAAGAAGAGGTGGA-3′ | ||
| Clinicopathological parameters | Factors | N | % | |
| All cases | 111 | |||
| Gender | Men | 65 | 41.4 | |
| Women | 46 | 58.6 | ||
| Age | ≤50 | 35 | 31.5 | |
| >50 | 76 | 68.5 | ||
| Anatomic site | Colon | 96 | 86.5 | |
| Rectum | 15 | 13.5 | ||
| Differentiation | Well | 75 | 67.6 | |
| Moderate–poor | 36 | 32.4 | ||
| Lymph node metastasis | No | 45 | 40.5 | |
| Yes | 66 | 59.5 | ||
| Lymph invasion | No | 39 | 35.1 | |
| Yes | 72 | 64.9 | ||
| Perineural invasion | No | 55 | 49.5 | |
| Yes | 56 | 50.5 | ||
| Venous invasion | No | 75 | 67.6 | |
| Yes | 36 | 32.4 | ||
| T Stage | T1–T2 | 58 | 53.7 | |
| T3–T4 | 50 | 46.3 | ||
| Lymph node metastasis | No | 45 | 40.5 | |
| Yes | 66 | 59.5 | ||
| Distant metastasis | No | 45 | 45.5 | |
| Yes | 55 | 55.5 | ||
| Cancer relapse | No | 19 | 31.7 | |
| Yes | 41 | 68.3 | ||
| FOLFOX | Non-responder | 10 | 74.1 | |
| Responder | 17 | 25.9 | ||
| IHC | ERCC1 expression | Negative | 50 | 47.2 |
| Positive | 56 | 52.8 | ||
| ERCC1 methylation | Negative | 56 | 55.4 | |
| Positive | 45 | 44.6 | ||
| MGMT methylation | Negative | 31 | 31 | |
| Positive | 69 | 69 | ||
| ERCC1 rs11615 | CC | 12 | 32.4 | |
| CT | 19 | 51.4 | ||
| TT | 6 | 16.2 | ||
| Factors | ERCC1 Expression | ERCC1 Methylation | MGMT Methylation | |||
|---|---|---|---|---|---|---|
| r | p | r | p | r | p | |
| Gender | −0.080 | 0.417 | 0.059 | 0.558 | 0.057 | 0.575 |
| Differentiation | −0.148 | 0.129 | 0.199 | 0.047 * | 0.070 | 0.478 |
| Lymph invasion | 0.021 | 0.827 | −0.112 | 0.276 | 0.282 | 0.005 * |
| Venous invasion | 0.030 | 0.764 | 0.006 | 0.950 | 0.286 | 0.004 * |
| Perineural invasion | 0.015 | 0.876 | 0.006 | 0.952 | 0.409 | ≤0.001 ** |
| Stage | −0.056 | 0.574 | 0.005 | 0.965 | 0.232 | 0.022 * |
| Lymph node metastasis | 0.052 | 0.594 | 0.079 | 0.436 | 0.322 | 0.001 * |
| Distant metastasis | −0.213 | 0.039 * | 0.105 | 0.323 | 0.606 | ≤0.001 ** |
| Cancer relapse | −0.234 | 0.074 | 0.196 | 0.156 | 0.356 | 0.008 * |
| Cancer resistance | 0.054 | 0.688 | −0.575 | 0.002 * | −0.420 | 0.021 * |
| ERCC1 expression | −0.700 | ≤0.001 ** | 0.255 | 0.013 * | ||
| MGMT methylation | −0.215 | 0.037 * | 0.376 | ≤0.001 ** | ||
| ERCC1 methylation | −0.693 | ≤0.001 ** | ||||
| Factors | Combined Methylation of MGMT and ERCC1 | |
|---|---|---|
| R | p | |
| Differentiation | 0.232 | 0.08 * |
| Lymph invasion | 0.187 | 0.170 |
| Venous invasion | 0.283 | 0.036 * |
| Perineural invasion | 0.417 | 0.002 * |
| Stage | 0.283 | 0.036 * |
| Lymph node metastasis | 0.244 | 0.07 * |
| Distant metastasis | 0.605 | ≤0.001 ** |
| Cancer relapse | 0.446 | 0.014 * |
| Cancer resistance | 0.655 | 0.006 * |
| Factor | Multivariate Analysis (p) |
|---|---|
| Stage | 0.351 |
| Differentiation | 0.507 |
| Lymph node metastasis | 0.684 |
| Lymph invasion | 0.912 |
| Venous invasion | 0.124 |
| Perineural invasion | 0.048 * |
| Distant metastasis | ≤0.001 ** |
| Cancer relapse | 0.018 * |
| Survival | ≤0.001 ** |
| Factor | Univariate Analysis | Multivariate Analysis | ||
|---|---|---|---|---|
| Overall Survival | 3-Year OS | 5-Year OS | ||
| p | p | p | p | |
| Lymph node metastasis | 0.210 | 0.333 | 0.210 | 0.362 |
| Lymph invasion | 0.207 | 0.235 | 0.207 | 0.9 |
| Vascular invasion | 0.691 | 0.8 | 0.691 | - |
| Perineural invasion | 0.003 * | 0.008 * | 0.003 * | 0.444 |
| Venous invasion | 350 | 0.404 | 0.350 | - |
| Distant metastasis | ≤0.001 ** | ≤0.001 ** | ≤0.001 ** | 0.001 ** |
| ERCC1 expression | ≤0.001 ** | 0.002 * | ≤0.001 ** | 0.05 * |
| ERCC1 methylation | 0.039 * | 0.062 | 0.039 * | 0.276 |
| MGMT methylation | ≤0.001 ** | ≤0.001 ** | ≤0.001 ** | ≤0.001 ** |
| Combined methylation | ≤0.001 ** | ≤0.001 ** | ≤0.001 ** | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jamai, D.; Gargouri, R.; Selmi, B.; Khabir, A. ERCC1 and MGMT Methylation as a Predictive Marker of Relapse and FOLFOX Response in Colorectal Cancer Patients from South Tunisia. Genes 2023, 14, 1467. https://doi.org/10.3390/genes14071467
Jamai D, Gargouri R, Selmi B, Khabir A. ERCC1 and MGMT Methylation as a Predictive Marker of Relapse and FOLFOX Response in Colorectal Cancer Patients from South Tunisia. Genes. 2023; 14(7):1467. https://doi.org/10.3390/genes14071467
Chicago/Turabian StyleJamai, Dhouha, Raja Gargouri, Boulbaba Selmi, and Abdelmajid Khabir. 2023. "ERCC1 and MGMT Methylation as a Predictive Marker of Relapse and FOLFOX Response in Colorectal Cancer Patients from South Tunisia" Genes 14, no. 7: 1467. https://doi.org/10.3390/genes14071467
APA StyleJamai, D., Gargouri, R., Selmi, B., & Khabir, A. (2023). ERCC1 and MGMT Methylation as a Predictive Marker of Relapse and FOLFOX Response in Colorectal Cancer Patients from South Tunisia. Genes, 14(7), 1467. https://doi.org/10.3390/genes14071467





