Integrated Genomic Selection for Accelerating Breeding Programs of Climate-Smart Cereals
Abstract
1. Introduction
2. Methodology
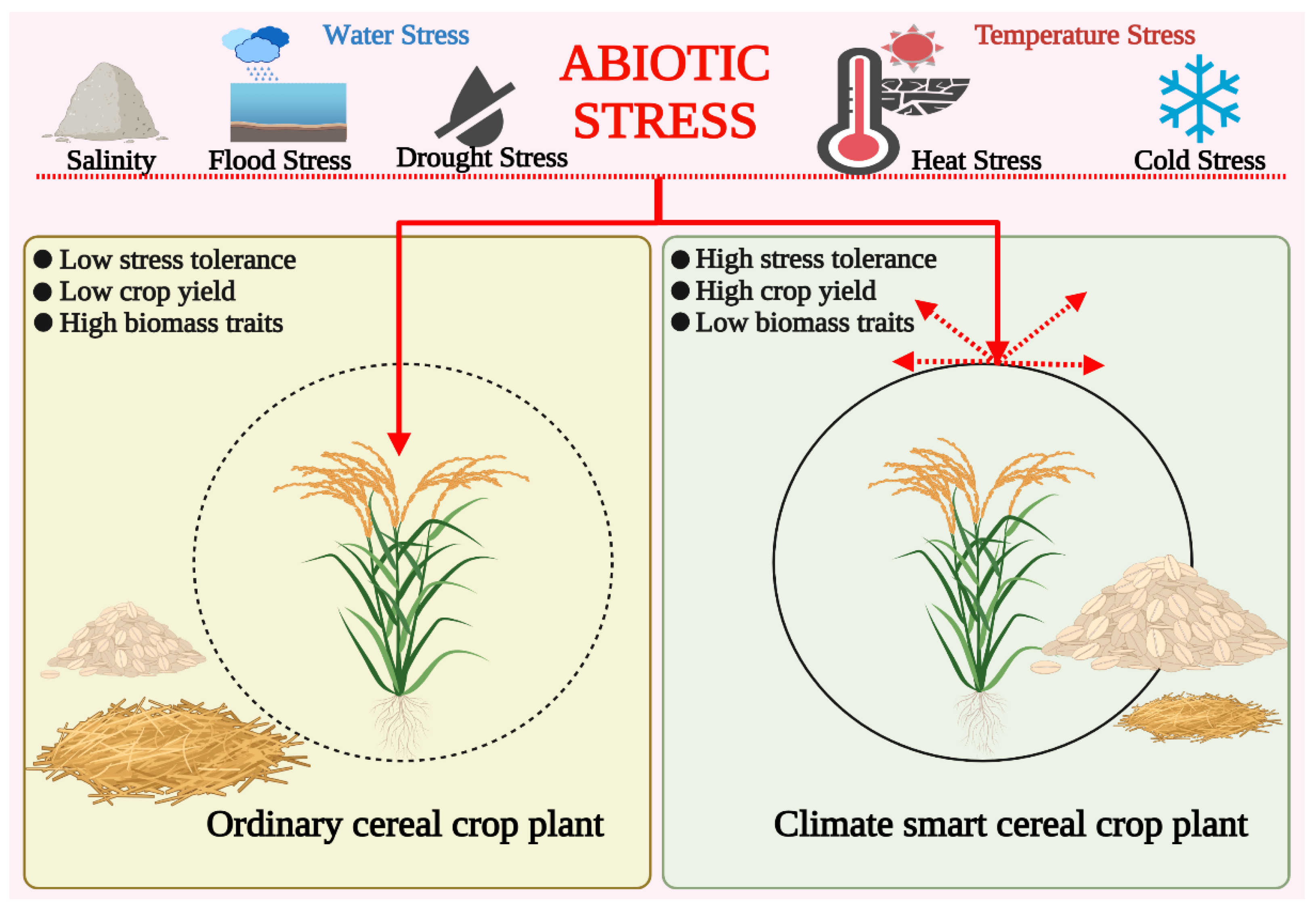
3. Climate-Smart Crops: A Promising Option for Future Food Security
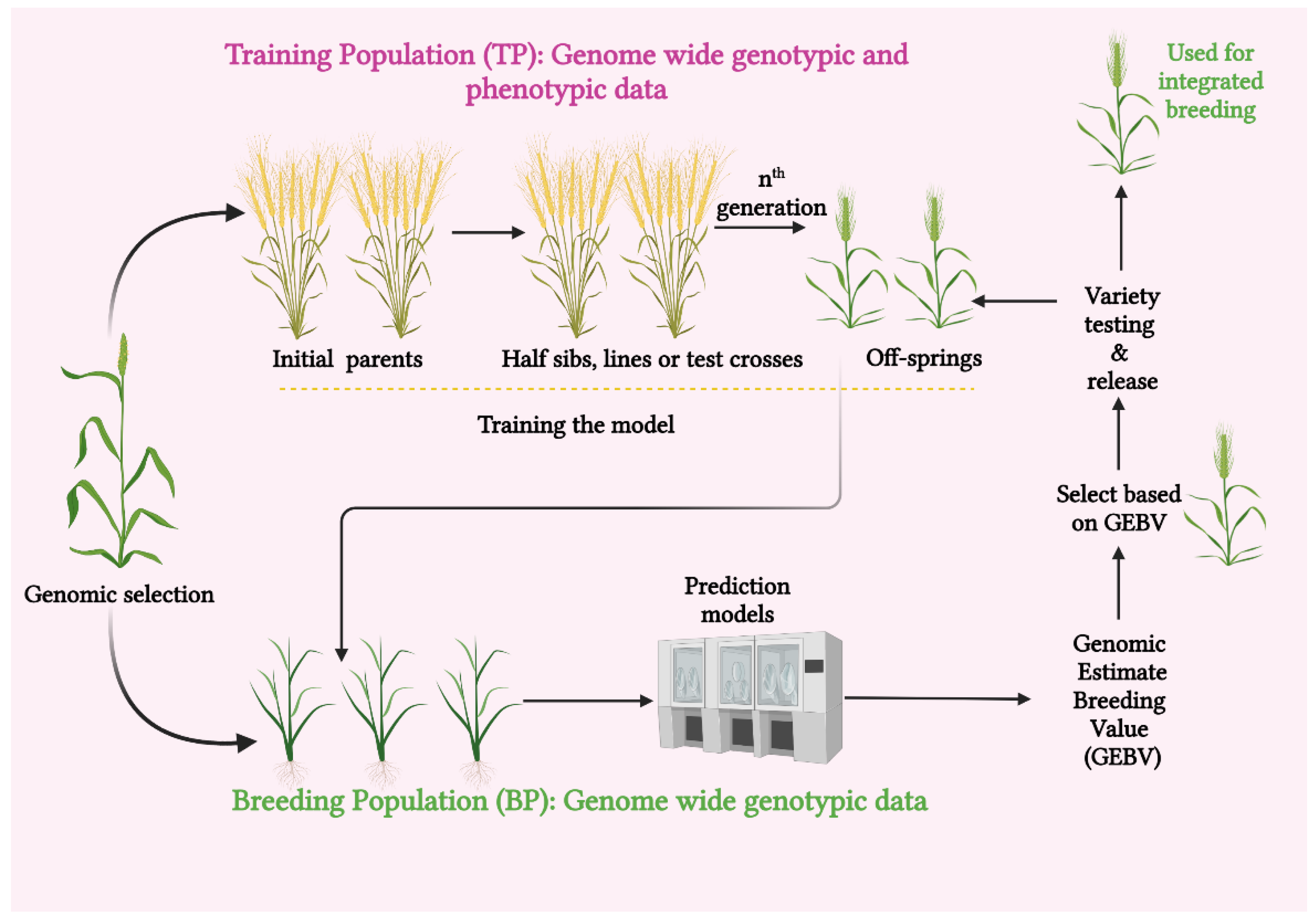
4. Integrated Genomic Selection for Making Climate-Smart Cereals
5. Genetic Gain: A Metric for Tracking Breeding Initiatives’ Forward Development
- (1)
- Genotyping and phenotyping of entities in a reference population and the building of a statistical format to study the effects of SNPs on morphological makeup, creating relational forecasting equations.
- (2)
- Newer candidates might not be phenotyped but are genotyped. Additionally, breeding values are calculated using phenotypic data and prediction models [54]. Owing to its increased genetic gain, reduced phenotyping, shorter cycle times, and improved selection accuracy, GS has been warmly accepted in breeding programs around the world over the past two decades. The feasibility of using GS in breeding crops is also being looked into, as it has given promising early evaluation results in the betterment of yield, biotic and abiotic stress resilience, and, of course, quality in cereal crops [55].
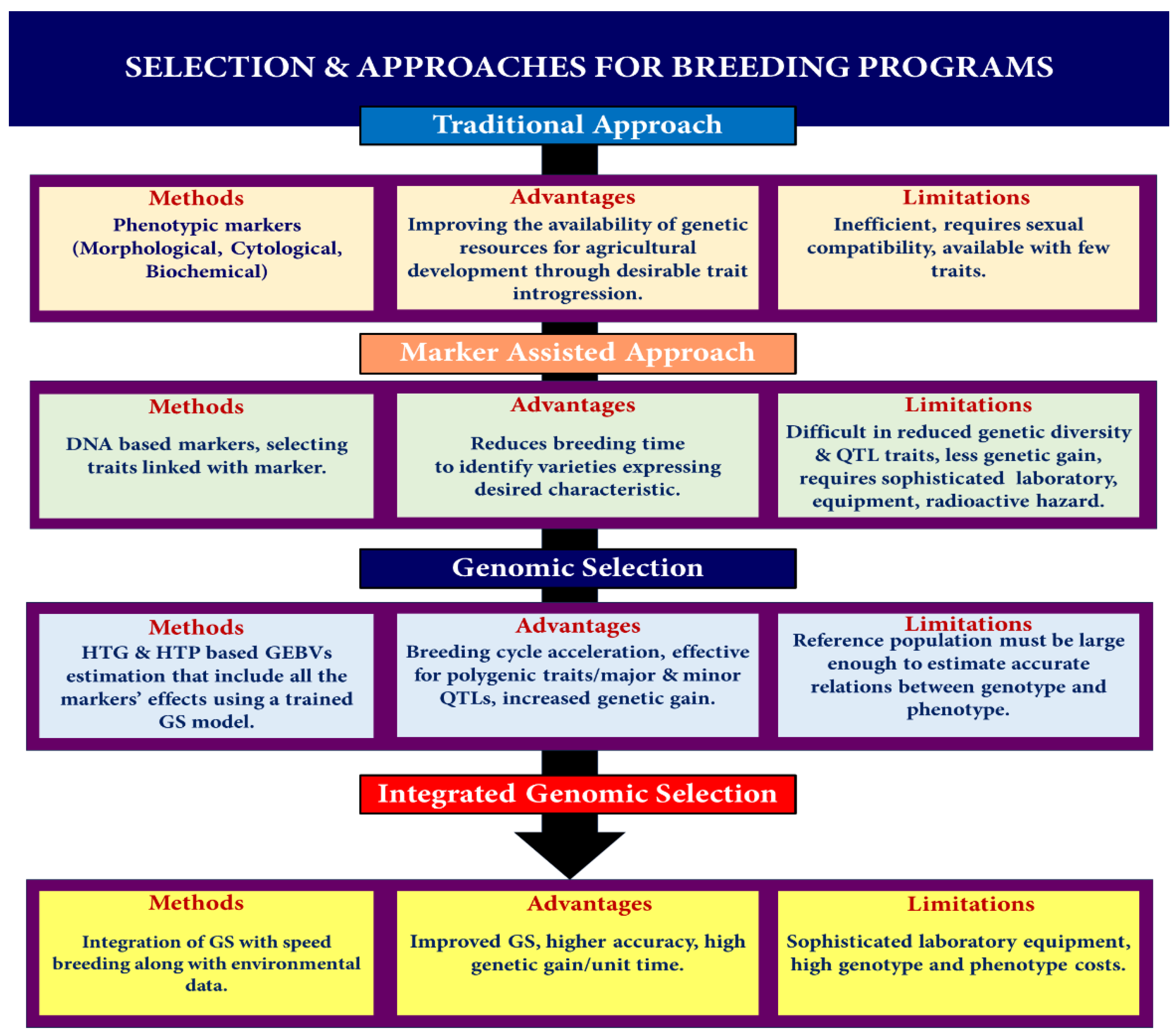
6. The Benefits of Genomic Selection over Conventional Approaches and Marker-Assisted Selection
- (a)
- To overcome the limitations of conventional phenotypic selection
- (b)
- Freedom of choice of selection at a specific stage
- (c)
- Helpful for backcross breeding
- (d)
- Pyramiding multiple monogenic traits
7. Genomic Selection Methodology
7.1. Design of Training Population
- (i)
- The size, genetic diversity, and relationship of the training population (TRN) to the test population (TST) are all critical factors determining genomic prediction precision. Specifically, the relationship between the cultivars in the TRN and those in the TST set, whether they are closely or distantly related, has been shown to impact the effectiveness of genomic prediction [108,109].
- (ii)
- The heritability of the traits under selection is another crucial factor affecting the accuracy of genomic forecasting. Characters with increased heritability, which are less complex and influenced by fewer genetic factors, can be effectively predicted using a smaller number of markers with comparatively greater effects [108,109].
- (iii)
- The accuracy or truth of genomic prediction is poorer for complicated traits that are influenced by an abundance of markers that do not exist in LD associated with QTL. Where there is a lack of correlation between markers and actual genetic factors influencing the trait, the accuracy of genomic prediction decreases [108,109].
7.2. Design of Statistical Models
- (a)
- Model Structure
- (b)
- Variable Selection
- (c)
- Parameter Estimation
- (d)
- Model Evaluation
7.3. Requirement for Advanced Breeding Populations for Genomics-Assisted Breeding (GAB); NAM, MAGIC, etc.
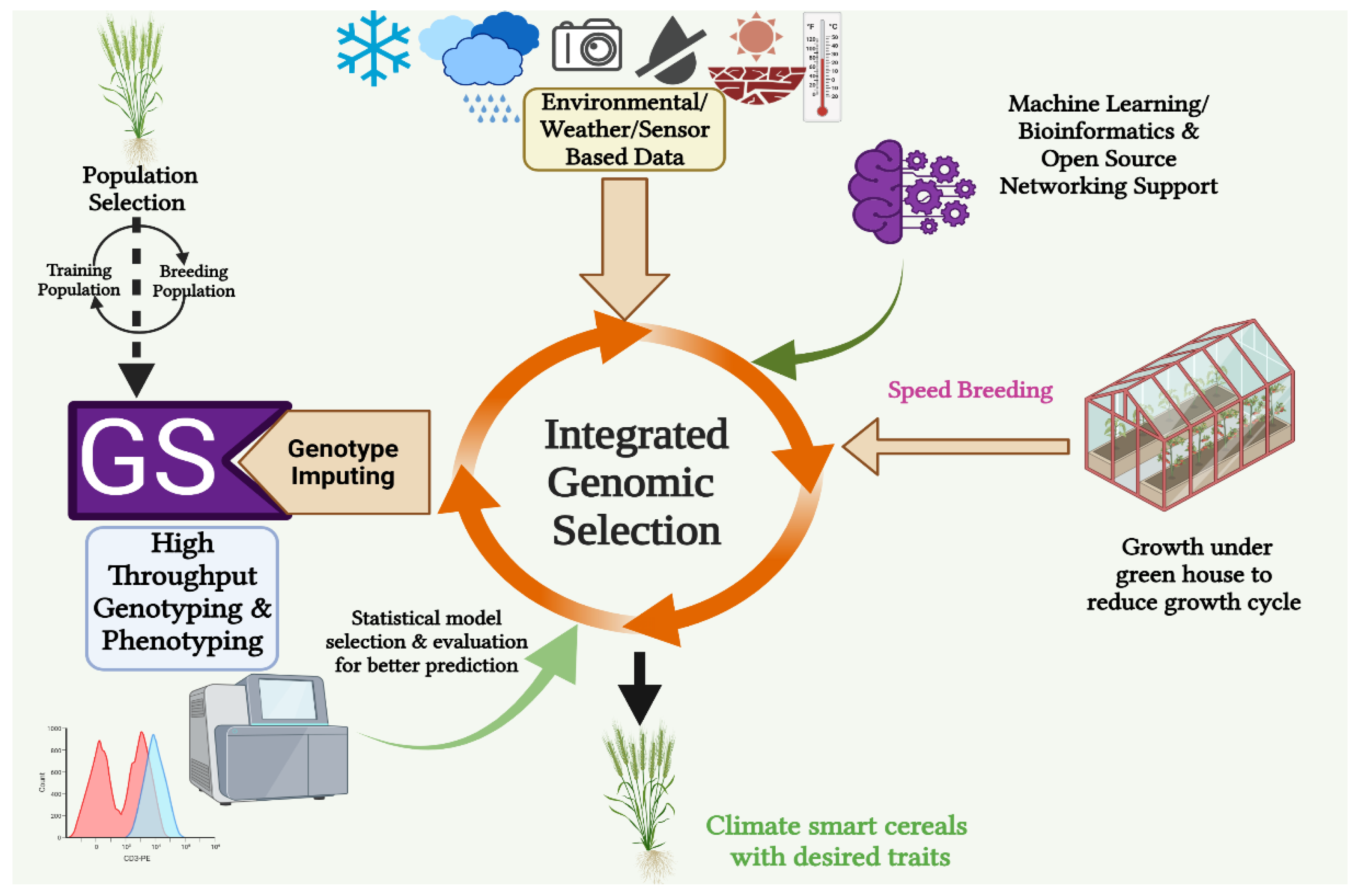
8. Integrated Genomic Selection: A Unique Approach to Boost the Capacity of Genomic Selection
8.1. Speed Breeding in Genomic Selection
8.2. Accelerating Rate of Breeding Cereals
8.3. High-Throughput Genotyping (HTG) and Genotype Imputing
8.4. High-throughput Phenotyping (HTP)
8.5. Genomic Crop Improvement by Next-Generation Sequencing (NGS)
8.6. Advances in Genotyping
8.6.1. The Illumina Golden Gate Assay
8.6.2. Genotyping by Sequencing (GBS)
8.6.3. Kompetitive Allele-Specific PCR (KASP)
8.6.4. TaqMan Assay
8.6.5. High-Resolution Melting (HRM) Analysis
8.6.6. MassARRAY
8.6.7. Restriction-Site-Associated DNA Sequencing (RAD-seq)
8.6.8. Amplicon Sequencing
8.7. Emerging Concept of Pangenomes and Super-Pangenomes
9. Statistical Tools for Integrated Genomic Selection
9.1. Breed Wheat Genomic Selection
- (1)
- Bwgs. cv, which performs replicated random model cross-validation on a training set of lines having genotypic and phenotypic data;
- (2)
- Bwgs.predict, which predicts the GEBV for those lines for which the genotype is known [272].
- (a)
- imputation of missing data;
- (b)
- dimension reduction;
- (c)
- GEBV estimation.
9.2. GMStool
- (1)
- Preparation:
- (2)
- Marker selection:
- (3)
- Final modeling.
9.3. SolGS
9.4. BGLR R-Package
9.5. GenSel
9.6. STGS
9.7. MTGS
9.8. Ime4GS
10. Issues and Challenges of Integrated Genomic Selection
11. Conclusions and Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- UN DESA|United Nations Department of Economic and Social Affairs. Growing at a Slower Pace, World Population Is Expected to Reach 9.7 Billion in 2050 and Could Peak at Nearly 11 Billion around 2100. Available online: https://www.un.org/development/desa/en/news/population/world-population-prospects-2019.html (accessed on 17 June 2023).
- This Is Why Food Security Matters Now More than Ever. Available online: https://www.weforum.org/agenda/2020/11/food-security-why-it-matters/ (accessed on 17 June 2023).
- UN Report: Global Hunger Numbers Rose to as Many as 828 Million in 2021. Available online: https://www.fao.org/newsroom/detail/un-report-global-hunger-SOFI-2022-FAO/en (accessed on 11 June 2023).
- Fact Sheets—Malnutrition. Available online: https://www.who.int/news-room/fact-sheets/detail/malnutrition (accessed on 11 June 2023).
- Global Temperatures Set to Reach New Records in Next Five Years. Available online: https://public.wmo.int/en/media/press-release/global-temperatures-set-reach-new-records-next-five-years (accessed on 17 June 2023).
- Global Temperatures Set to Break Records during Next 5 Years: WMO|UN News. Available online: https://news.un.org/en/story/2023/05/1136732 (accessed on 17 June 2023).
- Figueres, C. Take Urgent Action to Combat Climate Change and Its Impacts. UN Chron. 2015, 51, 30–31. [Google Scholar] [CrossRef]
- Waqas, M.A.; Kaya, C.; Riaz, A.; Farooq, M.; Nawaz, I.; Wilkes, A.; Li, Y. Potential Mechanisms of Abiotic Stress Tolerance in Crop Plants Induced by Thiourea. Front. Plant Sci. 2019, 10, 1336. [Google Scholar] [CrossRef]
- Kumar, S. Abiotic Stresses and Their Effects on Plant Growth, Yield and Nutritional Quality of Agricultural Produce. Int. J. Food Sci. Agric. 2020, 4, 367–378. [Google Scholar] [CrossRef]
- Suzuki, N.; Rivero, R.M.; Shulaev, V.; Blumwald, E.; Mittler, R. Abiotic and Biotic Stress Combinations. New Phytol. 2014, 203, 32–43. [Google Scholar] [CrossRef]
- Voss-Fels, K.P.; Stahl, A.; Hickey, L.T. Q&A: Modern Crop Breeding for Future Food Security. BMC Biol. 2019, 17, 18. [Google Scholar] [CrossRef]
- Hickey, L.T.; Hafeez, A.N.; Robinson, H.; Jackson, S.A.; Leal-Bertioli, S.C.M.; Tester, M.; Gao, C.; Godwin, I.D.; Hayes, B.J.; Wulff, B.B.H. Breeding Crops to Feed 10 Billion. Nat. Biotechnol. 2019, 37, 744–754. [Google Scholar] [CrossRef]
- Pradhan, P.; Fischer, G.; Van Velthuizen, H.; Reusser, D.E.; Kropp, J.P. Closing Yield Gaps: How Sustainable Can We Be? PLoS ONE 2015, 10, e0129487. [Google Scholar] [CrossRef] [PubMed]
- Hatfield, J.L.; Beres, B.L. Yield Gaps in Wheat: Path to Enhancing Productivity. Front. Plant Sci. 2019, 10, 1603. [Google Scholar] [CrossRef]
- Li, H.; Rasheed, A.; Hickey, L.T.; He, Z. Fast-Forwarding Genetic Gain. Trends Plant Sci. 2018, 23, 184–186. [Google Scholar] [CrossRef]
- Krishnappa, G.; Savadi, S.; Tyagi, B.S.; Singh, S.K.; Mamrutha, H.M.; Kumar, S.; Mishra, C.N.; Khan, H.; Gangadhara, K.; Uday, G.; et al. Integrated Genomic Selection for Rapid Improvement of Crops. Genomics 2021, 113, 1070–1086. [Google Scholar] [CrossRef]
- Climate-Smart Agriculture. Available online: https://www.worldbank.org/en/topic/climate-smart-agriculture (accessed on 12 June 2023).
- Ali, H.; Menza, M.; Hagos, F.; Haileslassie, A. Impact of Climate-Smart Agriculture Adoption on Food Security and Multidimensional Poverty of Rural Farm Households in the Central Rift Valley of Ethiopia. Agric. Food Secur. 2023, 11, 62. [Google Scholar] [CrossRef]
- Thottathil, G.P.; Jayasekaran, K.; Othman, A.S. Sequencing Crop Genomes: A Gateway to Improve Tropical Agriculture. Trop. Life Sci. Res. 2016, 27, 93–114. [Google Scholar] [PubMed]
- Desta, Z.A.; Ortiz, R. Genomic Selection: Genome-Wide Prediction in Plant Improvement. Trends Plant Sci. 2014, 19, 592–601. [Google Scholar] [CrossRef] [PubMed]
- Lande, R.; Thompson, R. Efficiency of Marker-Assisted Selection in the Improvement of Quantitative Traits. Genetics 1990, 124, 743–756. [Google Scholar] [CrossRef] [PubMed]
- Meuwissen, T.H.E.; Hayes, B.J.; Goddard, M.E. Prediction of Total Genetic Value Using Genome-Wide Dense Marker Maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [CrossRef]
- Goddard, M.E.; Hayes, B.J. Genomic Selection: Genomic Selection. J. Anim. Breed. Genet. 2007, 124, 323–330. [Google Scholar] [CrossRef]
- Li, Y.; Dungey, H.S. Expected Benefit of Genomic Selection over Forward Selection in Conifer Breeding and Deployment. PLoS ONE 2018, 13, e0208232. [Google Scholar] [CrossRef]
- Crossa, J.; Pérez-Rodríguez, P.; Cuevas, J.; Montesinos-López, O.; Jarquín, D.; De Los Campos, G.; Burgueño, J.; González-Camacho, J.M.; Pérez-Elizalde, S.; Beyene, Y.; et al. Genomic Selection in Plant Breeding: Methods, Models, and Perspectives. Trends Plant Sci. 2017, 22, 961–975. [Google Scholar] [CrossRef]
- Scheben, A.; Yuan, Y.; Edwards, D. Advances in Genomics for Adapting Crops to Climate Change. Curr. Plant Biol. 2016, 6, 2–10. [Google Scholar] [CrossRef]
- Acevedo, M.; Pixley, K.; Zinyengere, N.; Meng, S.; Tufan, H.; Cichy, K.; Bizikova, L.; Isaacs, K.; Ghezzi-Kopel, K.; Porciello, J. A Scoping Review of Adoption of Climate-Resilient Crops by Small-Scale Producers in Low- and Middle-Income Countries. Nat. Plants 2020, 6, 1231–1241. [Google Scholar] [CrossRef]
- Ray, D.K.; West, P.C.; Clark, M.; Gerber, J.S.; Prishchepov, A.V.; Chatterjee, S. Climate Change Has Likely Already Affected Global Food Production. PLoS ONE 2019, 14, e0217148. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Naeem, S.; Orlove, B.; Modi, V.; DeFries, R.S. Understanding the Causes and Consequences of Differential Decision-Making in Adaptation Research: Adapting to a Delayed Monsoon Onset in Gujarat, India. Glob. Environ. Chang. 2015, 31, 98–109. [Google Scholar] [CrossRef]
- Sandhu, K.S.; Shiv, A.; Kaur, G.; Meena, M.R.; Raja, A.K.; Vengavasi, K.; Mall, A.K.; Kumar, S.; Singh, P.K.; Singh, J.; et al. Integrated Approach in Genomic Selection to Accelerate Genetic Gain in Sugarcane. Plants 2022, 11, 2139. [Google Scholar] [CrossRef] [PubMed]
- Rashid, S.; Bin Mushtaq, M.; Farooq, I.; Khan, Z. Climate Smart Crops for Food Security. In The Nature, Causes, Effects and Mitigation of Climate Change on the Environment; Harris, S., Ed.; IntechOpen: London, UK, 2022; ISBN 978-1-83968-611-5. [Google Scholar]
- Rai, K.K. Integrating Speed Breeding with Artificial Intelligence for Developing Climate-Smart Crops. Mol. Biol. Rep. 2022, 49, 11385–11402. [Google Scholar] [CrossRef]
- Bhattacharyya, P.; Pathak, H.; Pal, S. Crop Management for Climate-Smart Agriculture. In Climate Smart Agriculture; Green Energy and Technology; Springer: Singapore, 2020; pp. 85–111. ISBN 9789811591310. [Google Scholar]
- ICARDA. Climate Smart Crops. Available online: https://www.icarda.org/research/climate-smart-crops (accessed on 12 June 2023).
- Joint Statement by the Heads of the Food and Agriculture Organization, International Monetary Fund, World Bank Group, World Food Programme, and World Trade Organization on the Global Food Security Crisis. Available online: https://www.imf.org/en/News/Articles/2022/07/15/pr22259-joint-statement-heads-fao-imf-wbg-wfp-wto-global-food-security-crisis (accessed on 12 June 2023).
- Food and Agriculture Organization of the United Nations. Building Climate Resilient Cropping Systems. Available online: https://www.fao.org/in-action/save-grow-climate-smart/en/ (accessed on 12 June 2023).
- Ray, D.K.; Ramankutty, N.; Mueller, N.D.; West, P.C.; Foley, J.A. Recent Patterns of Crop Yield Growth and Stagnation. Nat. Commun. 2012, 3, 1293. [Google Scholar] [CrossRef]
- Steinwand, M.A.; Ronald, P.C. Crop Biotechnology and the Future of Food. Nat. Food 2020, 1, 273–283. [Google Scholar] [CrossRef]
- Anilkumar, C.; Sunitha, N.C.; Harikrishna; Devate, N.B.; Ramesh, S. Advances in Integrated Genomic Selection for Rapid Genetic Gain in Crop Improvement: A Review. Planta 2022, 256, 87. [Google Scholar] [CrossRef]
- Onogi, A.; Watanabe, M.; Mochizuki, T.; Hayashi, T.; Nakagawa, H.; Hasegawa, T.; Iwata, H. Toward Integration of Genomic Selection with Crop Modelling: The Development of an Integrated Approach to Predicting Rice Heading Dates. Theor. Appl. Genet. 2016, 129, 805–817. [Google Scholar] [CrossRef]
- Malmberg, M.M.; Smith, C.; Thakur, P.; Drayton, M.C.; Wilson, J.; Shinozuka, M.; Clayton, W.; Inch, C.; Spangenberg, G.C.; Smith, K.F.; et al. Developing an Integrated Genomic Selection Approach beyond Biomass for Varietal Protection and Nutritive Traits in Perennial Ryegrass (Lolium perenne L.). Theor. Appl. Genet. 2023, 136, 44. [Google Scholar] [CrossRef]
- Alves, F.C.; Balmant, K.M.; Resende, M.F.R.; Kirst, M.; Los Campos, G. Accelerating Forest Tree Breeding by Integrating Genomic Selection and Greenhouse Phenotyping. Plant Genome 2020, 13, e20048. [Google Scholar] [CrossRef]
- Heslot, N.; Akdemir, D.; Sorrells, M.E.; Jannink, J.-L. Integrating Environmental Covariates and Crop Modeling into the Genomic Selection Framework to Predict Genotype by Environment Interactions. Theor. Appl. Genet. 2014, 127, 463–480. [Google Scholar] [CrossRef]
- Haile, J.K.; N’diaye, A.; Sari, E.; Walkowiak, S.; Rutkoski, J.E.; Kutcher, H.R.; Pozniak, C.J. Potential of Genomic Selection and Integrating “Omics” Data for Disease Evaluation in Wheat. Crop Breed. Genet. Genom. 2020, 2, e200016. [Google Scholar] [CrossRef]
- Kumar, N.; Rana, M.; Kumar, B.; Chand, S.; Shiv, A.; Wani, S.H.; Kumar, S. Genomic Selection for Wheat Improvement. In Physiological, Molecular, and Genetic Perspectives of Wheat Improvement; Wani, S.H., Mohan, A., Singh, G.P., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 175–207. ISBN 978-3-030-59576-0. [Google Scholar]
- Jonas, E.; De Koning, D.-J. Does Genomic Selection Have a Future in Plant Breeding? Trends Biotechnol. 2013, 31, 497–504. [Google Scholar] [CrossRef] [PubMed]
- Crossa, J. From Genotype × Environment Interaction to Gene × Environment Interaction. Curr. Genom. 2012, 13, 225–244. [Google Scholar] [CrossRef]
- Yang, D.-L.; Jing, R.-L.; Chang, X.-P.; Li, W. Identification of Quantitative Trait Loci and Environmental Interactions for Accumulation and Remobilization of Water-Soluble Carbohydrates in Wheat (Triticum aestivum L.) Stems. Genetics 2007, 176, 571–584. [Google Scholar] [CrossRef]
- Fiddy, S.; Cattermole, D.; Xie, D.; Duan, X.Y.; Mott, R. An Integrated System for Genetic Analysis. BMC Bioinform. 2006, 7, 210. [Google Scholar] [CrossRef]
- Michel, S.; Löschenberger, F.; Ametz, C.; Pachler, B.; Sparry, E.; Bürstmayr, H. Combining Grain Yield, Protein Content and Protein Quality by Multi-Trait Genomic Selection in Bread Wheat. Theor. Appl. Genet. 2019, 132, 2767–2780. [Google Scholar] [CrossRef]
- Yang, W.; Guo, T.; Luo, J.; Zhang, R.; Zhao, J.; Warburton, M.L.; Xiao, Y.; Yan, J. Target-Oriented Prioritization: Targeted Selection Strategy by Integrating Organismal and Molecular Traits through Predictive Analytics in Breeding. Genome Biol. 2022, 23, 80. [Google Scholar] [CrossRef]
- Xu, Y.; Liu, X.; Fu, J.; Wang, H.; Wang, J.; Huang, C.; Prasanna, B.M.; Olsen, M.S.; Wang, G.; Zhang, A. Enhancing Genetic Gain through Genomic Selection: From Livestock to Plants. Plant Commun. 2020, 1, 100005. [Google Scholar] [CrossRef]
- Sinha, P.; Singh, V.K.; Bohra, A.; Kumar, A.; Reif, J.C.; Varshney, R.K. Genomics and Breeding Innovations for Enhancing Genetic Gain for Climate Resilience and Nutrition Traits. Theor. Appl. Genet. 2021, 134, 1829–1843. [Google Scholar] [CrossRef]
- R2D2 Consortium; Fugeray-Scarbel, A.; Bastien, C.; Dupont-Nivet, M.; Lemarié, S. Why and How to Switch to Genomic Selection: Lessons From Plant and Animal Breeding Experience. Front. Genet. 2021, 12, 629737. [Google Scholar] [CrossRef]
- Budhlakoti, N.; Kushwaha, A.K.; Rai, A.; Chaturvedi, K.K.; Kumar, A.; Pradhan, A.K.; Kumar, U.; Kumar, R.R.; Juliana, P.; Mishra, D.C.; et al. Genomic Selection: A Tool for Accelerating the Efficiency of Molecular Breeding for Development of Climate-Resilient Crops. Front. Genet. 2022, 13, 832153. [Google Scholar] [CrossRef]
- Breseghello, F.; Coelho, A.S.G. Traditional and Modern Plant Breeding Methods with Examples in Rice (Oryza sativa L.). J. Agric. Food Chem. 2013, 61, 8277–8286. [Google Scholar] [CrossRef]
- Wang, X.; Xu, Y.; Hu, Z.; Xu, C. Genomic Selection Methods for Crop Improvement: Current Status and Prospects. Crop J. 2018, 6, 330–340. [Google Scholar] [CrossRef]
- Ansaldi, B.H.; Franks, S.J.; Weber, J.J. The Influence of Environmental Factors on Breeding System Allocation at Large Spatial Scales. AoB Plants 2018, 10, ply069. [Google Scholar] [CrossRef]
- Guo, T.; Li, X. Machine Learning for Predicting Phenotype from Genotype and Environment. Curr. Opin. Biotechnol. 2023, 79, 102853. [Google Scholar] [CrossRef]
- Wang, X.; Yang, Z.; Xu, C. A Comparison of Genomic Selection Methods for Breeding Value Prediction. Sci. Bull. 2015, 60, 925–935. [Google Scholar] [CrossRef]
- Hasan, N.; Choudhary, S.; Naaz, N.; Sharma, N.; Laskar, R.A. Recent Advancements in Molecular Marker-Assisted Selection and Applications in Plant Breeding Programmes. J. Genet. Eng. Biotechnol. 2021, 19, 128. [Google Scholar] [CrossRef]
- Jiang, G.-L. Molecular Marker-Assisted Breeding: A Plant Breeder’s Review. In Advances in Plant Breeding Strategies: Breeding, Biotechnology and Molecular Tools; Al-Khayri, J.M., Jain, S.M., Johnson, D.V., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 431–472. ISBN 978-3-319-22520-3. [Google Scholar]
- Jiang, G.-L. Molecular Markers and Marker-Assisted Breeding in Plants. In Plant Breeding from Laboratories to Fields; Andersen, S.B., Ed.; IntechOpen: London, UK, 2013; ISBN 978-953-51-1090-3. [Google Scholar]
- Sulkowska, M.K. Isoenzyme Analyses Tools Used Long Time in Forest Science. In Electrophoresis; Ghowsi, K., Ed.; IntechOpen: London, UK, 2012; pp. 157–172. ISBN 978-953-51-0846-7. [Google Scholar]
- Graham, J. Molecular Plant Breeding By Y. Xu. Wallingford, UK: CABI (2010), pp. 734, £125.00. ISBN 978-184593-392-0. Exp. Agric. 2011, 47, 173. [Google Scholar] [CrossRef]
- Nadeem, M.A.; Nawaz, M.A.; Shahid, M.Q.; Doğan, Y.; Comertpay, G.; Yıldız, M.; Hatipoğlu, R.; Ahmad, F.; Alsaleh, A.; Labhane, N.; et al. DNA Molecular Markers in Plant Breeding: Current Status and Recent Advancements in Genomic Selection and Genome Editing. Biotechnol. Biotechnol. Equip. 2018, 32, 261–285. [Google Scholar] [CrossRef]
- Manzo-Sánchez, G.; Buenrostro-Nava, M.T.; Guzmán-González, S.; Orozco-Santos, M.; Youssef, M.; Escobedo-Gracia Medrano, R.M. Genetic Diversity in Bananas and Plantains (Musa Spp.). In Molecular Approaches to Genetic Diversity; Caliskan, M., Oz, G.C., Kavakli, I.H., Ozcan, B., Eds.; IntechOpen: London, UK, 2015; ISBN 978-953-51-2042-1. [Google Scholar]
- Beckmann, J.S.; Soller, M. Restriction Fragment Length Polymorphisms and Genetic Improvement of Agricultural Species. Euphytica 1986, 35, 111–124. [Google Scholar] [CrossRef]
- Beckmann, J.S.; Soller, M. Restriction Fragment Length Polymorphisms in Genetic Improvement: Methodologies, Mapping and Costs. Theor. Appl. Genet. 1983, 67, 35–43. [Google Scholar] [CrossRef] [PubMed]
- James, K.E.; Schneider, H.; Ansell, S.W.; Evers, M.; Robba, L.; Uszynski, G.; Pedersen, N.; Newton, A.E.; Russell, S.J.; Vogel, J.C.; et al. Diversity Arrays Technology (DArT) for Pan-Genomic Evolutionary Studies of Non-Model Organisms. PLoS ONE 2008, 3, e1682. [Google Scholar] [CrossRef] [PubMed]
- Amiteye, S. Basic Concepts and Methodologies of DNA Marker Systems in Plant Molecular Breeding. Heliyon 2021, 7, e08093. [Google Scholar] [CrossRef]
- Bailey-Serres, J.; Parker, J.E.; Ainsworth, E.A.; Oldroyd, G.E.D.; Schroeder, J.I. Genetic Strategies for Improving Crop Yields. Nature 2019, 575, 109–118. [Google Scholar] [CrossRef]
- Xu, Y.; Crouch, J.H. Marker-Assisted Selection in Plant Breeding: From Publications to Practice. Crop Sci. 2008, 48, 391–407. [Google Scholar] [CrossRef]
- Na Jinda, A.; Nikornpun, M.; Jeeatid, N.; Thumdee, S.; Thippachote, K.; Pusadee, T.; Kumchai, J. Marker-Assisted Selection of Male-Sterile and Maintainer Line in Chili Improvement by Backcross Breeding. Horticulturae 2023, 9, 357. [Google Scholar] [CrossRef]
- Kulkarni, S.R.; Balachandran, S.M.; Ulaganathan, K.; Balakrishnan, D.; Praveen, M.; Prasad, A.S.H.; Fiyaz, R.A.; Senguttuvel, P.; Sinha, P.; Kale, R.R.; et al. Molecular Mapping of QTLs for Yield Related Traits in Recombinant Inbred Line (RIL) Population Derived from the Popular Rice Hybrid KRH-2 and Their Validation through SNP Genotyping. Sci. Rep. 2020, 10, 13695. [Google Scholar] [CrossRef]
- Kumawat, G.; Kanta Kumawat, C.; Chandra, K.; Pandey, S.; Chand, S.; Nandan Mishra, U.; Lenka, D.; Sharma, R. Insights into Marker Assisted Selection and Its Applications in Plant Breeding. In Plant Breeding—Current and Future Views; Abdurakhmonov, I.Y., Ed.; IntechOpen: London, UK, 2021; ISBN 978-1-83968-309-1. [Google Scholar]
- Solberg, T.R.; Sonesson, A.K.; Woolliams, J.A.; Meuwissen, T.H.E. Genomic Selection Using Different Marker Types and Densities. J. Anim. Sci. 2008, 86, 2447–2454. [Google Scholar] [CrossRef]
- Calus, M.P.L.; Meuwissen, T.H.E.; De Roos, A.P.W.; Veerkamp, R.F. Accuracy of Genomic Selection Using Different Methods to Define Haplotypes. Genetics 2008, 178, 553–561. [Google Scholar] [CrossRef]
- Pareek, C.S.; Smoczynski, R.; Tretyn, A. Sequencing Technologies and Genome Sequencing. J. Appl. Genet. 2011, 52, 413–435. [Google Scholar] [CrossRef] [PubMed]
- Kushanov, F.N.; Turaev, O.S.; Ernazarova, D.K.; Gapparov, B.M.; Oripova, B.B.; Kudratova, M.K.; Rafieva, F.U.; Khalikov, K.K.; Erjigitov, D.S.; Khidirov, M.T.; et al. Genetic Diversity, QTL Mapping, and Marker-Assisted Selection Technology in Cotton (Gossypium Spp.). Front. Plant Sci. 2021, 12, 779386. [Google Scholar] [CrossRef]
- Marks, R.A.; Hotaling, S.; Frandsen, P.B.; VanBuren, R. Representation and Participation across 20 Years of Plant Genome Sequencing. Nat. Plants 2021, 7, 1571–1578. [Google Scholar] [CrossRef] [PubMed]
- Uffelmann, E.; Huang, Q.Q.; Munung, N.S.; De Vries, J.; Okada, Y.; Martin, A.R.; Martin, H.C.; Lappalainen, T.; Posthuma, D. Genome-Wide Association Studies. Nat. Rev. Methods Primer 2021, 1, 59. [Google Scholar] [CrossRef]
- Singh, V.K.; Singh, A.K.; Singh, S.; Singh, B.D. Next-Generation Sequencing (NGS) Tools and Impact in Plant Breeding. In Advances in Plant Breeding Strategies: Breeding, Biotechnology and Molecular Tools; Al-Khayri, J.M., Jain, S.M., Johnson, D.V., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 563–612. ISBN 978-3-319-22520-3. [Google Scholar]
- Xiao, Q.; Bai, X.; Zhang, C.; He, Y. Advanced High-Throughput Plant Phenotyping Techniques for Genome-Wide Association Studies: A Review. J. Adv. Res. 2022, 35, 215–230. [Google Scholar] [CrossRef]
- Park, Y.-S.; Beaulieu, J.; Bousquet, J. Multi-Varietal Forestry Integrating Genomic Selection and Somatic Embryogenesis. In Vegetative Propagation of Forest Trees; National Institute of Forest Science(NiFos): Seoul, Republic of Korea, 2016; pp. 302–322. [Google Scholar]
- Heffner, E.L.; Lorenz, A.J.; Jannink, J.; Sorrells, M.E. Plant Breeding with Genomic Selection: Gain per Unit Time and Cost. Crop. Sci. 2010, 50, 1681–1690. [Google Scholar] [CrossRef]
- Ceballos, H.; Kawuki, R.S.; Gracen, V.E.; Yencho, G.C.; Hershey, C.H. Conventional Breeding, Marker-Assisted Selection, Genomic Selection and Inbreeding in Clonally Propagated Crops: A Case Study for Cassava. Theor. Appl. Genet. 2015, 128, 1647–1667. [Google Scholar] [CrossRef]
- Jannink, J.L.; Walsh, B. Association Mapping in Plant Populations. In Quantitative Genetics, Genomics and Plant Breeding; Kang, M.S., Ed.; CABI Publishing: Wallingford, UK, 2002; pp. 59–68. ISBN 978-0-85199-787-2. [Google Scholar]
- Yang, A.-Q.; Chen, B.; Ran, M.-L.; Yang, G.-M.; Zeng, C. The Application of Genomic Selection in Pig Cross Breeding. Hereditas 2020, 42, 145–152. [Google Scholar] [CrossRef]
- Parveen, R.; Kumar, M.; Swapnil; Singh, D.; Shahani, M.; Imam, Z.; Sahoo, J.P. Understanding the Genomic Selection for Crop Improvement: Current Progress and Future Prospects. Mol. Genet. Genom. 2023, 298, 813–821. [Google Scholar] [CrossRef]
- Loskutov, I.G. Advances in Cereal Crops Breeding. Plants 2021, 10, 1705. [Google Scholar] [CrossRef]
- Lau, P.Y.; Fung, W.K. Evaluation of Marker Selection Methods and Statistical Models for Chronological Age Prediction Based on DNA Methylation. Leg. Med. 2020, 47, 101744. [Google Scholar] [CrossRef] [PubMed]
- Montesinos-López, O.A.; Montesinos-López, A.; Crossa, J.; Toledo, F.H.; Pérez-Hernández, O.; Eskridge, K.M.; Rutkoski, J. A Genomic Bayesian Multi-Trait and Multi-Environment Model. G3 Genes Genomes Genet. 2016, 6, 2725–2744. [Google Scholar] [CrossRef] [PubMed]
- Cericola, F.; Jahoor, A.; Orabi, J.; Andersen, J.R.; Janss, L.L.; Jensen, J. Optimizing Training Population Size and Genotyping Strategy for Genomic Prediction Using Association Study Results and Pedigree Information. A Case of Study in Advanced Wheat Breeding Lines. PLoS ONE 2017, 12, e0169606. [Google Scholar] [CrossRef]
- Tessema, B.B.; Liu, H.; Sørensen, A.C.; Andersen, J.R.; Jensen, J. Strategies Using Genomic Selection to Increase Genetic Gain in Breeding Programs for Wheat. Front. Genet. 2020, 11, 578123. [Google Scholar] [CrossRef] [PubMed]
- Pandey, M.K.; Chaudhari, S.; Jarquin, D.; Janila, P.; Crossa, J.; Patil, S.C.; Sundravadana, S.; Khare, D.; Bhat, R.S.; Radhakrishnan, T.; et al. Genome-Based Trait Prediction in Multi- Environment Breeding Trials in Groundnut. Theor. Appl. Genet. 2020, 133, 3101–3117. [Google Scholar] [CrossRef] [PubMed]
- VanRaden, P.M. Genomic Measures of Relationship and Inbreeding. Interbull Bull. 2007, 37, 33–36. [Google Scholar]
- Paulsen, V.I.; Raghupathi, M. An Introduction to the Theory of Reproducing Kernel Hilbert Spaces, 1st ed.; Cambridge University Press: Cambridge, UK, 2016; ISBN 978-1-107-10409-9. [Google Scholar]
- Ogutu, J.O.; Schulz-Streeck, T.; Piepho, H.-P. Genomic Selection Using Regularized Linear Regression Models: Ridge Regression, Lasso, Elastic Net and Their Extensions. BMC Proc. 2012, 6, S10. [Google Scholar] [CrossRef]
- Jacquin, L.; Cao, T.-V.; Ahmadi, N. A Unified and Comprehensible View of Parametric and Kernel Methods for Genomic Prediction with Application to Rice. Front. Genet. 2016, 7, 145. [Google Scholar] [CrossRef]
- Nicolaou, N.; Constandinou, T.G. A Nonlinear Causality Estimator Based on Non-Parametric Multiplicative Regression. Front. Neuroinform. 2016, 10, 19. [Google Scholar] [CrossRef]
- Wade, P.R. Bayesian Methods in Conservation Biology. Conserv. Biol. 2000, 14, 1308–1316. [Google Scholar] [CrossRef]
- Clark, S.A.; Van Der Werf, J. Genomic Best Linear Unbiased Prediction (GBLUP) for the Estimation of Genomic Breeding Values. In Genome-Wide Association Studies and Genomic Prediction; Gondro, C., Van Der Werf, J., Hayes, B., Eds.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2013; Volume 1019, pp. 321–330. ISBN 978-1-62703-446-3. [Google Scholar]
- Stewart-Brown, B.B.; Song, Q.; Vaughn, J.N.; Li, Z. Genomic Selection for Yield and Seed Composition Traits Within an Applied Soybean Breeding Program. G3 Genes Genomes Genet. 2019, 9, 2253–2265. [Google Scholar] [CrossRef] [PubMed]
- Fernández-González, J.; Akdemir, D.; Isidro, Y.; Sánchez, J. A Comparison of Methods for Training Population Optimization in Genomic Selection. Theor. Appl. Genet. 2023, 136, 30. [Google Scholar] [CrossRef] [PubMed]
- Akdemir, D.; Isidro-Sánchez, J. Design of Training Populations for Selective Phenotyping in Genomic Prediction. Sci. Rep. 2019, 9, 1446. [Google Scholar] [CrossRef]
- Wu, X.; Lund, M.S.; Sun, D.; Zhang, Q.; Su, G. Impact of Relationships between Test and Training Animals and among Training Animals on Reliability of Genomic Prediction. J. Anim. Breed. Genet. 2015, 132, 366–375. [Google Scholar] [CrossRef] [PubMed]
- Pszczola, M.; Strabel, T.; Mulder, H.A.; Calus, M.P.L. Reliability of Direct Genomic Values for Animals with Different Relationships within and to the Reference Population. J. Dairy Sci. 2012, 95, 389–400. [Google Scholar] [CrossRef]
- Daetwyler, H.D.; Pong-Wong, R.; Villanueva, B.; Woolliams, J.A. The Impact of Genetic Architecture on Genome-Wide Evaluation Methods. Genetics 2010, 185, 1021–1031. [Google Scholar] [CrossRef]
- He, X.; Wang, J. Discovering Model Structure for Partially Linear Models. Ann. Inst. Stat. Math. 2020, 72, 45–63. [Google Scholar] [CrossRef]
- Zhang, H.H.; Cheng, G.; Liu, Y. Linear or Nonlinear? Automatic Structure Discovery for Partially Linear Models. J. Am. Stat. Assoc. 2011, 106, 1099–1112. [Google Scholar] [CrossRef]
- Söderström, T. Model Validation and Model Structure Determination. Circuits Syst. Signal Process. 2002, 21, 83–90. [Google Scholar] [CrossRef]
- Anderson, M.; Whitcomb, P. Design of Experiments: Statistical Principles of Research Design and Analysis. Technometrics 2001, 43, 236–237. [Google Scholar] [CrossRef]
- Suresh, K.; Thomas, S.; Suresh, G. Design, Data Analysis and Sampling Techniques for Clinical Research. Ann. Indian Acad. Neurol. 2011, 14, 287. [Google Scholar] [CrossRef]
- Choi, K.R.; Ryu, J.Y.; Lee, S.Y. Revisiting Statistical Design and Analysis in Scientific Research. Small 2018, 14, 1802604. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.M.; Li, Q.; Saqib, Z.; Khan, N.; Habib, T.; Khalid, N.; Majeed, M.; Tariq, A. MaxEnt Modelling and Impact of Climate Change on Habitat Suitability Variations of Economically Important Chilgoza Pine (Pinus gerardiana Wall.) in South Asia. Forests 2022, 13, 715. [Google Scholar] [CrossRef]
- Hess, K.R. Statistical Design Considerations in Animal Studies Published Recently in Cancer Research. Cancer Res. 2011, 71, 625. [Google Scholar] [CrossRef]
- Kano, Y.; Harada, A. Stepwise Variable Selection in Factor Analysis. Psychometrika 2000, 65, 7–22. [Google Scholar] [CrossRef]
- Drost, E.A. Validity and Reliability in Social Science Research. Educ. Res. Perspect. 2011, 38, 105–123. [Google Scholar]
- Barry, A.E.; Chaney, B.; Piazza-Gardner, A.K.; Chavarria, E.A. Validity and Reliability Reporting Practices in the Field of Health Education and Behavior: A Review of Seven Journals. Health Educ. Behav. 2014, 41, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Carmines, E.; Zeller, R. Reliability and Validity Assessment; SAGE Publications, Inc.: Thousand Oaks, CA, USA, 1979; ISBN 978-0-8039-1371-4. [Google Scholar]
- Cook, D.A.; Beckman, T.J. Current Concepts in Validity and Reliability for Psychometric Instruments: Theory and Application. Am. J. Med. 2006, 119, e7–e166. [Google Scholar] [CrossRef]
- Heale, R.; Twycross, A. Validity and Reliability in Quantitative Studies. Evid. Based Nurs. 2015, 18, 66–67. [Google Scholar] [CrossRef]
- Nisbet, R.; Elder, J.; Miner, G. Model Evaluation and Enhancement. In Handbook of Statistical Analysis and Data Mining Applications; Elsevier: Amsterdam, The Netherlands, 2009; pp. 285–312. ISBN 978-0-12-374765-5. [Google Scholar]
- Wallach, D.; Makowski, D.; Jones, J.W.; Brun, F. Model Evaluation. In Working with Dynamic Crop Models; Elsevier: Amsterdam, The Netherlands, 2014; pp. 345–406. ISBN 978-0-12-397008-4. [Google Scholar]
- Williams, G. Model Performance Evaluation. In Data Mining with Rattle and R; Springer: New York, NY, USA, 2011; pp. 307–321. ISBN 978-1-4419-9889-7. [Google Scholar]
- Xin, Y.; Vasquez, V.R.; Whiting, W.B. Effect of Regression Approach in the Estimation of Nonlinear Model Parameters on Process Design and Simulation: Applications to Kinetic and Thermodynamic Models. Comput. Chem. Eng. 2000, 24, 1269–1274. [Google Scholar] [CrossRef]
- David, I.J.; Asiribo, O.E.; Dikko, H.G. Nonlinear Split-Plot Design Model in Parameters Estimation Using Estimated Generalized Least Square—Maximum Likelihood Estimation. ComTech Comput. Math. Eng. Appl. 2018, 9, 65. [Google Scholar] [CrossRef]
- Basavarajaiah, D.M.; Narasimha Murthy, B. Statistical Implications and Its Practical Approach to Research Methodology. In Design of Experiments and Advanced Statistical Techniques in Clinical Research; Springer: Singapore, 2020; pp. 223–244. ISBN 9789811582097. [Google Scholar]
- Pires Camargo, C.; Gemperli, R. Statistical Basic Steps to Be Considered on Planning a Research. Plast. Reconstr. Surg. Glob. Open 2018, 6, e1718. [Google Scholar] [CrossRef]
- Zhao, Y.; Mette, M.F.; Reif, J.C. Genomic Selection in Hybrid Breeding. Plant Breed. 2015, 134, 1–10. [Google Scholar] [CrossRef]
- Liu, X.; Wang, H.; Wang, H.; Guo, Z.; Xu, X.; Liu, J.; Wang, S.; Li, W.-X.; Zou, C.; Prasanna, B.M.; et al. Factors Affecting Genomic Selection Revealed by Empirical Evidence in Maize. Crop J. 2018, 6, 341–352. [Google Scholar] [CrossRef]
- Rio, S.; Mary-Huard, T.; Moreau, L.; Charcosset, A. Genomic Selection Efficiency and a Priori Estimation of Accuracy in a Structured Dent Maize Panel. Theor. Appl. Genet. 2019, 132, 81–96. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, J.P.R.; Pires, L.P.M.; De Castro Vasconcellos, R.C.; Pereira, G.S.; Von Pinho, R.G.; Balestre, M. Genomic Selection to Resistance to Stenocarpella Maydis in Maize Lines Using DArTseq Markers. BMC Genet. 2016, 17, 86. [Google Scholar] [CrossRef]
- Badu-Apraku, B.; Talabi, A.O.; Fakorede, M.A.B.; Fasanmade, Y.; Gedil, M.; Magorokosho, C.; Asiedu, R. Yield Gains and Associated Changes in an Early Yellow Bi-Parental Maize Population Following Genomic Selection for Striga Resistance and Drought Tolerance. BMC Plant Biol. 2019, 19, 129. [Google Scholar] [CrossRef]
- Beyene, Y.; Semagn, K.; Mugo, S.; Tarekegne, A.; Babu, R.; Meisel, B.; Sehabiague, P.; Makumbi, D.; Magorokosho, C.; Oikeh, S.; et al. Genetic Gains in Grain Yield Through Genomic Selection in Eight Bi-parental Maize Populations under Drought Stress. Crop. Sci. 2015, 55, 154–163. [Google Scholar] [CrossRef]
- Das, R.R.; Vinayan, M.T.; Patel, M.B.; Phagna, R.K.; Singh, S.B.; Shahi, J.P.; Sarma, A.; Barua, N.S.; Babu, R.; Seetharam, K.; et al. Genetic Gains with Rapid-Cycle Genomic Selection for Combined Drought and Waterlogging Tolerance in Tropical Maize (Zea Mays L.). Plant Genome 2020, 13, e20035. [Google Scholar] [CrossRef]
- Sallam, A.H.; Smith, K.P. Genomic Selection Performs Similarly to Phenotypic Selection in Barley. Crop Sci. 2016, 56, 2871–2881. [Google Scholar] [CrossRef]
- Abed, A.; Pérez-Rodríguez, P.; Crossa, J.; Belzile, F. When Less Can Be Better: How Can We Make Genomic Selection More Cost-Effective and Accurate in Barley? Theor. Appl. Genet. 2018, 131, 1873–1890. [Google Scholar] [CrossRef] [PubMed]
- Tiede, T.; Smith, K.P. Evaluation and Retrospective Optimization of Genomic Selection for Yield and Disease Resistance in Spring Barley. Mol. Breed. 2018, 38, 55. [Google Scholar] [CrossRef]
- Grenier, C.; Cao, T.-V.; Ospina, Y.; Quintero, C.; Châtel, M.H.; Tohme, J.; Courtois, B.; Ahmadi, N. Accuracy of Genomic Selection in a Rice Synthetic Population Developed for Recurrent Selection Breeding. PLoS ONE 2015, 10, e0136594. [Google Scholar] [CrossRef]
- Spindel, J.; Begum, H.; Akdemir, D.; Virk, P.; Collard, B.; Redoña, E.; Atlin, G.; Jannink, J.-L.; McCouch, S.R. Genomic Selection and Association Mapping in Rice (Oryza sativa): Effect of Trait Genetic Architecture, Training Population Composition, Marker Number and Statistical Model on Accuracy of Rice Genomic Selection in Elite, Tropical Rice Breeding Lines. PLoS Genet. 2015, 11, e1004982. [Google Scholar] [CrossRef]
- Yabe, S.; Yoshida, H.; Kajiya-Kanegae, H.; Yamasaki, M.; Iwata, H.; Ebana, K.; Hayashi, T.; Nakagawa, H. Description of Grain Weight Distribution Leading to Genomic Selection for Grain-Filling Characteristics in Rice. PLoS ONE 2018, 13, e0207627. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Wang, X.; Ding, X.; Zheng, X.; Yang, Z.; Xu, C.; Hu, Z. Genomic Selection of Agronomic Traits in Hybrid Rice Using an NCII Population. Rice 2018, 11, 32. [Google Scholar] [CrossRef]
- Cui, Y.; Li, R.; Li, G.; Zhang, F.; Zhu, T.; Zhang, Q.; Ali, J.; Li, Z.; Xu, S. Hybrid Breeding of Rice via Genomic Selection. Plant Biotechnol. J. 2020, 18, 57–67. [Google Scholar] [CrossRef]
- Ben Hassen, M.; Cao, T.V.; Bartholomé, J.; Orasen, G.; Colombi, C.; Rakotomalala, J.; Razafinimpiasa, L.; Bertone, C.; Biselli, C.; Volante, A.; et al. Rice Diversity Panel Provides Accurate Genomic Predictions for Complex Traits in the Progenies of Biparental Crosses Involving Members of the Panel. Theor. Appl. Genet. 2018, 131, 417–435. [Google Scholar] [CrossRef]
- Huang, M.; Balimponya, E.G.; Mgonja, E.M.; McHale, L.K.; Luzi-Kihupi, A.; Wang, G.-L.; Sneller, C.H. Use of Genomic Selection in Breeding Rice (Oryza sativa L.) for Resistance to Rice Blast (Magnaporthe oryzae). Mol. Breed. 2019, 39, 114. [Google Scholar] [CrossRef]
- Bhandari, A.; Bartholomé, J.; Cao-Hamadoun, T.-V.; Kumari, N.; Frouin, J.; Kumar, A.; Ahmadi, N. Selection of Trait-Specific Markers and Multi-Environment Models Improve Genomic Predictive Ability in Rice. PLoS ONE 2019, 14, e0208871. [Google Scholar] [CrossRef]
- Leng, P.; Lübberstedt, T.; Xu, M. Genomics-Assisted Breeding—A Revolutionary Strategy for Crop Improvement. J. Integr. Agric. 2017, 16, 2674–2685. [Google Scholar] [CrossRef]
- Varshney, R.K.; Graner, A.; Sorrells, M.E. Genomics-Assisted Breeding for Crop Improvement. Trends Plant Sci. 2005, 10, 621–630. [Google Scholar] [CrossRef]
- Varshney, R.K.; Bohra, A.; Yu, J.; Graner, A.; Zhang, Q.; Sorrells, M.E. Designing Future Crops: Genomics-Assisted Breeding Comes of Age. Trends Plant Sci. 2021, 26, 631–649. [Google Scholar] [CrossRef] [PubMed]
- Bohra, A.; Pandey, M.K.; Jha, U.C.; Singh, B.; Singh, I.P.; Datta, D.; Chaturvedi, S.K.; Nadarajan, N.; Varshney, R.K. Genomics-Assisted Breeding in Four Major Pulse Crops of Developing Countries: Present Status and Prospects. Theor. Appl. Genet. 2014, 127, 1263–1291. [Google Scholar] [CrossRef]
- Ferrão, M.A.G.; da Fonseca, A.F.A.; Volpi, P.S.; de Souza, L.C.; Comério, M.; Filho, A.C.V.; Riva-Souza, E.M.; Munoz, P.R.; Ferrão, R.G.; Ferrão, L.F.V. Genomic-Assisted Breeding for Climate-Smart Coffee. Plant Genome 2023, e20321. [Google Scholar] [CrossRef] [PubMed]
- Cruet-Burgos, C.; Morris, G.P.; Rhodes, D.H. Characterization of Grain Carotenoids in Global Sorghum Germplasm to Guide Genomics-Assisted Breeding Strategies. BMC Plant Biol. 2023, 23, 165. [Google Scholar] [CrossRef]
- Newman, C.S.; Andres, R.J.; Youngblood, R.C.; Campbell, J.D.; Simpson, S.A.; Cannon, S.B.; Scheffler, B.E.; Oakley, A.T.; Hulse-Kemp, A.M.; Dunne, J.C. Initiation of Genomics-Assisted Breeding in Virginia-Type Peanuts through the Generation of a de Novo Reference Genome and Informative Markers. Front. Plant Sci. 2022, 13, 1073542. [Google Scholar] [CrossRef]
- Garcia-Abadillo, J.; Morales, L.; Buerstmayr, H.; Michel, S.; Lillemo, M.; Holzapfel, J.; Hartl, L.; Akdemir, D.; Carvalho, H.F.; Isidro-Sánchez, J. Alternative Scoring Methods of Fusarium Head Blight Resistance for Genomic Assisted Breeding. Front. Plant Sci. 2022, 13, 1057914. [Google Scholar] [CrossRef]
- Michel, S.; Löschenberger, F.; Ametz, C.; Pachler, B.; Sparry, E.; Bürstmayr, H. Simultaneous Selection for Grain Yield and Protein Content in Genomics-Assisted Wheat Breeding. Theor. Appl. Genet. 2019, 132, 1745–1760. [Google Scholar] [CrossRef]
- Subedi, M.; Ghimire, B.; Bagwell, J.W.; Buck, J.W.; Mergoum, M. Wheat End-Use Quality: State of Art, Genetics, Genomics-Assisted Improvement, Future Challenges, and Opportunities. Front. Genet. 2022, 13, 1032601. [Google Scholar] [CrossRef]
- Bohra, A.; Saxena, K.B.; Varshney, R.K.; Saxena, R.K. Genomics-Assisted Breeding for Pigeonpea Improvement. Theor. Appl. Genet. 2020, 133, 1721–1737. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Mohan, S.M.; Gaur, P.M.; Gangarao, N.V.P.R.; Pandey, M.K.; Bohra, A.; Sawargaonkar, S.L.; Chitikineni, A.; Kimurto, P.K.; Janila, P.; et al. Achievements and Prospects of Genomics-Assisted Breeding in Three Legume Crops of the Semi-Arid Tropics. Biotechnol. Adv. 2013, 31, 1120–1134. [Google Scholar] [CrossRef] [PubMed]
- Bekele, W.A.; Itaya, A.; Boyle, B.; Yan, W.; Mitchell Fetch, J.; Tinker, N.A. A Targeted Genotyping-by-Sequencing Tool (Rapture) for Genomics-Assisted Breeding in Oat. Theor. Appl. Genet. 2020, 133, 653–664. [Google Scholar] [CrossRef]
- Swamy, B.P.M.; Kumar, A. Genomics-Based Precision Breeding Approaches to Improve Drought Tolerance in Rice. Biotechnol. Adv. 2013, 31, 1308–1318. [Google Scholar] [CrossRef] [PubMed]
- Jiao, C.; Hao, C.; Li, T.; Bohra, A.; Wang, L.; Hou, J.; Liu, H.; Liu, H.; Zhao, J.; Wang, Y.; et al. Fast Integration and Accumulation of Beneficial Breeding Alleles through an AB–NAMIC Strategy in Wheat. Plant Commun. 2023, 4, 100549. [Google Scholar] [CrossRef]
- Stadlmeier, M.; Hartl, L.; Mohler, V. Usefulness of a Multiparent Advanced Generation Intercross Population With a Greatly Reduced Mating Design for Genetic Studies in Winter Wheat. Front. Plant Sci. 2018, 9, 1825. [Google Scholar] [CrossRef]
- Pandey, M.K.; Pandey, A.K.; Kumar, R.; Nwosu, C.V.; Guo, B.; Wright, G.C.; Bhat, R.S.; Chen, X.; Bera, S.K.; Yuan, M.; et al. Translational Genomics for Achieving Higher Genetic Gains in Groundnut. Theor. Appl. Genet. 2020, 133, 1679–1702. [Google Scholar] [CrossRef]
- Scott, M.F.; Ladejobi, O.; Amer, S.; Bentley, A.R.; Biernaskie, J.; Boden, S.A.; Clark, M.; Dell’Acqua, M.; Dixon, L.E.; Filippi, C.V.; et al. Multi-Parent Populations in Crops: A Toolbox Integrating Genomics and Genetic Mapping with Breeding. Heredity 2020, 125, 396–416. [Google Scholar] [CrossRef]
- Oren, E.; Dafna, A.; Tzuri, G.; Halperin, I.; Isaacson, T.; Elkabetz, M.; Meir, A.; Saar, U.; Ohali, S.; La, T.; et al. Pan-Genome and Multi-Parental Framework for High-Resolution Trait Dissection in Melon (Cucumis melo). Plant J. 2022, 112, 1525–1542. [Google Scholar] [CrossRef]
- Arrones, A.; Vilanova, S.; Plazas, M.; Mangino, G.; Pascual, L.; Díez, M.J.; Prohens, J.; Gramazio, P. The Dawn of the Age of Multi-Parent MAGIC Populations in Plant Breeding: Novel Powerful Next-Generation Resources for Genetic Analysis and Selection of Recombinant Elite Material. Biology 2020, 9, 229. [Google Scholar] [CrossRef]
- Huynh, B.-L.; Ehlers, J.D.; Huang, B.E.; Muñoz-Amatriaín, M.; Lonardi, S.; Santos, J.R.P.; Ndeve, A.; Batieno, B.J.; Boukar, O.; Cisse, N.; et al. A Multi-Parent Advanced Generation Inter-Cross (MAGIC) Population for Genetic Analysis and Improvement of Cowpea (Vigna unguiculata L. Walp.). Plant J. 2018, 93, 1129–1142. [Google Scholar] [CrossRef] [PubMed]
- Bandillo, N.; Raghavan, C.; Muyco, P.A.; Sevilla, M.A.L.; Lobina, I.T.; Dilla-Ermita, C.J.; Tung, C.-W.; McCouch, S.; Thomson, M.; Mauleon, R.; et al. Multi-Parent Advanced Generation Inter-Cross (MAGIC) Populations in Rice: Progress and Potential for Genetics Research and Breeding. Rice 2013, 6, 11. [Google Scholar] [CrossRef] [PubMed]
- Samantara, K.; Reyes, V.P.; Agrawal, N.; Mohapatra, S.R.; Jena, K.K. Advances and Trends on the Utilization of Multi-Parent Advanced Generation Intercross (MAGIC) for Crop Improvement. Euphytica 2021, 217, 189. [Google Scholar] [CrossRef]
- Juliana, P.; Montesinos-López, O.A.; Crossa, J.; Mondal, S.; González Pérez, L.; Poland, J.; Huerta-Espino, J.; Crespo-Herrera, L.; Govindan, V.; Dreisigacker, S.; et al. Integrating Genomic-Enabled Prediction and High-Throughput Phenotyping in Breeding for Climate-Resilient Bread Wheat. Theor. Appl. Genet. 2019, 132, 177–194. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Cao, Y. Genetic Dissection of Grain Yield of Maize and Yield-Related Traits Through Association Mapping and Genomic Prediction. Front. Plant Sci. 2021, 12, 690059. [Google Scholar] [CrossRef]
- Bernardo, R.; Yu, J. Prospects for Genomewide Selection for Quantitative Traits in Maize. Crop. Sci. 2007, 47, 1082–1090. [Google Scholar] [CrossRef]
- Ghimire, B.; Sapkota, S.; Bahri, B.A.; Martinez-Espinoza, A.D.; Buck, J.W.; Mergoum, M. Fusarium Head Blight and Rust Diseases in Soft Red Winter Wheat in the Southeast United States: State of the Art, Challenges and Future Perspective for Breeding. Front. Plant Sci. 2020, 11, 1080. [Google Scholar] [CrossRef]
- Odilbekov, F.; Armoniené, R.; Koc, A.; Svensson, J.; Chawade, A. GWAS-Assisted Genomic Prediction to Predict Resistance to Septoria Tritici Blotch in Nordic Winter Wheat at Seedling Stage. Front. Genet. 2019, 10, 1224. [Google Scholar] [CrossRef]
- Kokhmetova, A.; Sehgal, D.; Ali, S.; Atishova, M.; Kumarbayeva, M.; Leonova, I.; Dreisigacker, S. Genome-Wide Association Study of Tan Spot Resistance in a Hexaploid Wheat Collection From Kazakhstan. Front. Genet. 2020, 11, 581214. [Google Scholar] [CrossRef]
- AlTameemi, R.; Gill, H.S.; Ali, S.; Ayana, G.; Halder, J.; Sidhu, J.S.; Gill, U.S.; Turnipseed, B.; Hernandez, J.L.G.; Sehgal, S.K. Genome-Wide Association Analysis Permits Characterization of Stagonospora Nodorum Blotch (SNB) Resistance in Hard Winter Wheat. Sci. Rep. 2021, 11, 12570. [Google Scholar] [CrossRef]
- Speck, A.; Trouvé, J.-P.; Enjalbert, J.; Geffroy, V.; Joets, J.; Moreau, L. Genetic Architecture of Powdery Mildew Resistance Revealed by a Genome-Wide Association Study of a Worldwide Collection of Flax (Linum usitatissimum L.). Front. Plant Sci. 2022, 13, 871633. [Google Scholar] [CrossRef] [PubMed]
- Abdul Fiyaz, R.; Ajay, B.C.; Ramya, K.T.; Aravind Kumar, J.; Sundaram, R.M.; Subba Rao, L.V. Speed Breeding: Methods and Applications. In Accelerated Plant Breeding, Volume 1; Gosal, S.S., Wani, S.H., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 31–49. ISBN 978-3-030-41865-6. [Google Scholar]
- Balaji, B.; Dharani, E.; Shricharan, S.; Shakespear, S.; Singh, A.K.; Pillai, M.A.; Saini, J.K.Y. Genome Editing for Speed Breeding of Horticultural Crops. J. AgriSearch 2021, 9, 196–200. [Google Scholar] [CrossRef]
- Watson, A.; Ghosh, S.; Williams, M.J.; Cuddy, W.S.; Simmonds, J.; Rey, M.-D.; Asyraf Md Hatta, M.; Hinchliffe, A.; Steed, A.; Reynolds, D.; et al. Speed Breeding Is a Powerful Tool to Accelerate Crop Research and Breeding. Nat. Plants 2018, 4, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Majeed, M.; Bhatti, K.H.; Pieroni, A.; Sõukand, R.; Bussmann, R.W.; Khan, A.M.; Chaudhari, S.K.; Aziz, M.A.; Amjad, M.S. Gathered Wild Food Plants among Diverse Religious Groups in Jhelum District, Punjab, Pakistan. Foods 2021, 10, 594. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Singh, A.V.; Singh, A.; Singh, S.P. Speed Breeding: An Innovative Method for Crop Improvement. EC Agric. 2021, 7, 42–48. [Google Scholar] [CrossRef]
- Majeed, M.; Bhatti, K.H.; Amjad, M.S. Impact of climatic variations on the flowering phenology of plant species in jhelum district, Punjab, Pakistan. Appl. Ecol. Environ. Res. 2021, 19, 3343–3376. [Google Scholar] [CrossRef]
- Kole, C.; Muthamilarasan, M.; Henry, R.; Edwards, D.; Sharma, R.; Abberton, M.; Batley, J.; Bentley, A.; Blakeney, M.; Bryant, J.; et al. Application of Genomics-Assisted Breeding for Generation of Climate Resilient Crops: Progress and Prospects. Front. Plant Sci. 2015, 6, 563. [Google Scholar] [CrossRef]
- Samantara, K.; Bohra, A.; Mohapatra, S.R.; Prihatini, R.; Asibe, F.; Singh, L.; Reyes, V.P.; Tiwari, A.; Maurya, A.K.; Croser, J.S.; et al. Breeding More Crops in Less Time: A Perspective on Speed Breeding. Biology 2022, 11, 275. [Google Scholar] [CrossRef]
- Delmer, D.P. Agriculture in the Developing World: Connecting Innovations in Plant Research to Downstream Applications. Proc. Natl. Acad. Sci. USA 2005, 102, 15739–15746. [Google Scholar] [CrossRef]
- Liu, W.; Yuan, J.S.; Stewart, C.N., Jr. Advanced Genetic Tools for Plant Biotechnology. Nat. Rev. Genet. 2013, 14, 781–793. [Google Scholar] [CrossRef]
- Jighly, A.; Lin, Z.; Pembleton, L.W.; Cogan, N.O.I.; Spangenberg, G.C.; Hayes, B.J.; Daetwyler, H.D. Boosting Genetic Gain in Allogamous Crops via Speed Breeding and Genomic Selection. Front. Plant Sci. 2019, 10, 1364. [Google Scholar] [CrossRef] [PubMed]
- Ahmar, S.; Gill, R.A.; Jung, K.-H.; Faheem, A.; Qasim, M.U.; Mubeen, M.; Zhou, W. Conventional and Molecular Techniques from Simple Breeding to Speed Breeding in Crop Plants: Recent Advances and Future Outlook. Int. J. Mol. Sci. 2020, 21, 2590. [Google Scholar] [CrossRef] [PubMed]
- Ribaut, J.M.; Hoisington, D. Marker-assisted selection: New tools and strategies. Trends Plant Sci. 1998, 3, 236–239. [Google Scholar] [CrossRef]
- Majeed, M.; Khan, A.M.; Habib, T.; Anwar, M.M.; Sahito, H.A.; Khan, N.; Ali, K. Vegetation Analysis and Environmental Indicators of an Arid Tropical Forest Ecosystem of Pakistan. Ecol. Indic. 2022, 142, 109291. [Google Scholar] [CrossRef]
- Bohar, R.; Chitkineni, A.; Varshney, R.K. Genetic Molecular Markers to Accelerate Genetic Gains in Crops. BioTechniques 2020, 69, 158–160. [Google Scholar] [CrossRef]
- Lee, M. DNA Markers and Plant Breeding Programs. In Advances in Agronomy; Elsevier: Amsterdam, The Netherlands, 1995; Volume 55, pp. 265–344. ISBN 978-0-12-000755-4. [Google Scholar]
- Cappetta, E.; Andolfo, G.; Di Matteo, A.; Barone, A.; Frusciante, L.; Ercolano, M.R. Accelerating Tomato Breeding by Exploiting Genomic Selection Approaches. Plants 2020, 9, 1236. [Google Scholar] [CrossRef]
- Paterson, A.H.; Tanksley, S.D.; Sorrells, M.E. DNA Markers in Plant Improvement. In Advances in Agronomy; Elsevier: Amsterdam, The Netherlands, 1991; Volume 46, pp. 39–90. ISBN 978-0-12-000746-2. [Google Scholar]
- Arshad, F.; Waheed, M.; Harun, N.; Fatima, K.; Ali Khan, B.; Fatima, K.; Abbas, Z.; Jabeen, S.; Majeed, M. Indigenous Farmer’s Perception about Fodder and Foraging Species of Semi-Arid Lowlands of Pakistan: A Case Study of District Kasur, Pakistan. Taiwania 2022, 67, 510–523. [Google Scholar]
- Forster, B.P.; Till, B.J.; Ghanim, A.M.A.; Huynh, H.O.A.; Burstmayr, H.; Caligari, P.D.S. Accelerated Plant Breeding. CABI Rev. 2014, 2014, 1–16. [Google Scholar] [CrossRef]
- Shamshad, M.; Sharma, A. The Usage of Genomic Selection Strategy in Plant Breeding. In Next Generation Plant Breeding; Çiftçi, Y.Ö., Ed.; InTech: London, UK, 2018; ISBN 978-1-78923-925-6. [Google Scholar]
- Haider, S.; Kueffer, C.; Bruelheide, H.; Seipel, T.; Alexander, J.M.; Rew, L.J.; Arévalo, J.R.; Cavieres, L.A.; McDougall, K.L.; Milbau, A.; et al. Mountain Roads and Non-Native Species Modify Elevational Patterns of Plant Diversity. Glob. Ecol. Biogeogr. 2018, 27, 667–678. [Google Scholar] [CrossRef]
- Tang, L.; Li, T.; Li, D.; Meng, X. Elevational Patterns of Plant Richness in the Taibai Mountain, China. Sci. World J. 2014, 2014, 309053. [Google Scholar] [CrossRef]
- Naud, L.; Måsviken, J.; Freire, S.; Angerbjörn, A.; Dalén, L.; Dalerum, F. Altitude Effects on Spatial Components of Vascular Plant Diversity in a Subarctic Mountain Tundra. Ecol. Evol. 2019, 9, 4783–4795. [Google Scholar] [CrossRef] [PubMed]
- Oishi, Y. Factors That Shape the Elevational Patterns of Plant Diversity in the Yatsugatake Mountains, Japan. Ecol. Evol. 2021, 11, 4887–4897. [Google Scholar] [CrossRef] [PubMed]
- Contaldi, F.; Cappetta, E.; Esposito, S. Practical Workflow from High-Throughput Genotyping to Genomic Estimated Breeding Values (GEBVs). In Crop Breeding; Tripodi, P., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2021; Volume 2264, pp. 119–135. ISBN 978-1-07-161200-2. [Google Scholar]
- Tost, J.; Gut, I.G. Genotyping Single Nucleotide Polymorphisms by MALDI Mass Spectrometry in Clinical Applications. Clin. Biochem. 2005, 38, 335–350. [Google Scholar] [CrossRef]
- Bhat, J.A.; Yu, D. High-throughput NGS-based Genotyping and Phenotyping: Role in Genomics-assisted Breeding for Soybean Improvement. Legume Sci. 2021, 3, e81. [Google Scholar] [CrossRef]
- Goretti, E.; Wagner, D.R.; Devaux, Y. MiRNAs as Biomarkers of Myocardial Infarction: A Step Forward towards Personalized Medicine? Trends Mol. Med. 2014, 20, 716–725. [Google Scholar] [CrossRef]
- Phocas, F. Genotyping, the Usefulness of Imputation to Increase SNP Density, and Imputation Methods and Tools. In Genomic Prediction of Complex Traits; Ahmadi, N., Bartholomé, J., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2022; Volume 2467, pp. 113–138. ISBN 978-1-07-162204-9. [Google Scholar]
- Marchini, J.; Howie, B. Genotype Imputation for Genome-Wide Association Studies. Nat. Rev. Genet. 2010, 11, 499–511. [Google Scholar] [CrossRef]
- Evangelou, E.; Ioannidis, J.P.A. Meta-Analysis Methods for Genome-Wide Association Studies and Beyond. Nat. Rev. Genet. 2013, 14, 379–389. [Google Scholar] [CrossRef]
- Lee, S.; Abecasis, G.R.; Boehnke, M.; Lin, X. Rare-Variant Association Analysis: Study Designs and Statistical Tests. Am. J. Hum. Genet. 2014, 95, 5–23. [Google Scholar] [CrossRef]
- Rabbani, B.; Nakaoka, H.; Akhondzadeh, S.; Tekin, M.; Mahdieh, N. Next Generation Sequencing: Implications in Personalized Medicine and Pharmacogenomics. Mol. Biosyst. 2016, 12, 1818–1830. [Google Scholar] [CrossRef]
- Chandra, A.; Mitry, D.; Wright, A.; Campbell, H.; Charteris, D.G. Genome-Wide Association Studies: Applications and Insights Gained in Ophthalmology. Eye 2014, 28, 1066–1079. [Google Scholar] [CrossRef]
- Singh, A.; Ganapathysubramanian, B.; Singh, A.K.; Sarkar, S. Machine Learning for High-Throughput Stress Phenotyping in Plants. Trends Plant Sci. 2016, 21, 110–124. [Google Scholar] [CrossRef] [PubMed]
- Mahlein, A.-K.; Kuska, M.T.; Thomas, S.; Wahabzada, M.; Behmann, J.; Rascher, U.; Kersting, K. Quantitative and Qualitative Phenotyping of Disease Resistance of Crops by Hyperspectral Sensors: Seamless Interlocking of Phytopathology, Sensors, and Machine Learning Is Needed! Curr. Opin. Plant Biol. 2019, 50, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Batley, J.; Edwards, D. SNP Applications in Plants. In Association Mapping in Plants; Oraguzie, N.C., Rikkerink, E.H.A., Gardiner, S.E., De Silva, H.N., Eds.; Springer: New York, NY, USA, 2007; pp. 95–102. ISBN 978-0-387-35844-4. [Google Scholar]
- Goggin, F.L.; Lorence, A.; Topp, C.N. Applying High-Throughput Phenotyping to Plant–Insect Interactions: Picturing More Resistant Crops. Curr. Opin. Insect Sci. 2015, 9, 69–76. [Google Scholar] [CrossRef]
- Li, D.; Quan, C.; Song, Z.; Li, X.; Yu, G.; Li, C.; Muhammad, A. High-Throughput Plant Phenotyping Platform (HT3P) as a Novel Tool for Estimating Agronomic Traits From the Lab to the Field. Front. Bioeng. Biotechnol. 2021, 8, 623705. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Feng, H.; Zhang, X.; Zhang, J.; Doonan, J.H.; Batchelor, W.D.; Xiong, L.; Yan, J. Crop Phenomics and High-Throughput Phenotyping: Past Decades, Current Challenges, and Future Perspectives. Mol. Plant 2020, 13, 187–214. [Google Scholar] [CrossRef] [PubMed]
- Casto, A.L.; Schuhl, H.; Tovar, J.C.; Wang, Q.; Bart, R.S.; Fahlgren, N.; Gehan, M.A. Picturing the Future of Food. Plant Phenome J. 2021, 4, e20014. [Google Scholar] [CrossRef]
- Brown, T.B.; Cheng, R.; Sirault, X.R.; Rungrat, T.; Murray, K.D.; Trtilek, M.; Furbank, R.T.; Badger, M.; Pogson, B.J.; Borevitz, J.O. TraitCapture: Genomic and Environment Modelling of Plant Phenomic Data. Curr. Opin. Plant Biol. 2014, 18, 73–79. [Google Scholar] [CrossRef]
- Mammadov, J.; Aggarwal, R.; Buyyarapu, R.; Kumpatla, S. SNP Markers and Their Impact on Plant Breeding. Int. J. Plant Genom. 2012, 2012, 728398. [Google Scholar] [CrossRef]
- Reynolds, M.; Chapman, S.; Crespo-Herrera, L.; Molero, G.; Mondal, S.; Pequeno, D.N.L.; Pinto, F.; Pinera-Chavez, F.J.; Poland, J.; Rivera-Amado, C.; et al. Breeder Friendly Phenotyping. Plant Sci. 2020, 295, 110396. [Google Scholar] [CrossRef]
- Zenda, T.; Liu, S.; Dong, A.; Li, J.; Wang, Y.; Liu, X.; Wang, N.; Duan, H. Omics-Facilitated Crop Improvement for Climate Resilience and Superior Nutritive Value. Front. Plant Sci. 2021, 12, 774994. [Google Scholar] [CrossRef]
- Davey, J.W.; Hohenlohe, P.A.; Etter, P.D.; Boone, J.Q.; Catchen, J.M.; Blaxter, M.L. Genome-Wide Genetic Marker Discovery and Genotyping Using next-Generation Sequencing. Nat. Rev. Genet. 2011, 12, 499–510. [Google Scholar] [CrossRef] [PubMed]
- Deschamps, S.; Llaca, V.; May, G.D. Genotyping-by-Sequencing in Plants. Biology 2012, 1, 460–483. [Google Scholar] [CrossRef] [PubMed]
- Bohra, A. Emerging Paradigms in Genomics-Based Crop Improvement. Sci. World J. 2013, 2013, 585467. [Google Scholar] [CrossRef] [PubMed]
- Dahui, Q. Next-Generation Sequencing and Its Clinical Application. Cancer Biol. Med. 2019, 16, 4–10. [Google Scholar] [CrossRef]
- Cavanagh, C.; Morell, M.; Mackay, I.; Powell, W. From Mutations to MAGIC: Resources for Gene Discovery, Validation and Delivery in Crop Plants. Curr. Opin. Plant Biol. 2008, 11, 215–221. [Google Scholar] [CrossRef]
- Tindall, E.A.; Petersen, D.C.; Nikolaysen, S.; Miller, W.; Schuster, S.C.; Hayes, V.M. Interpretation of Custom Designed Illumina Genotype Cluster Plots for Targeted Association Studies and Next-Generation Sequence Validation. BMC Res. Notes 2010, 3, 39. [Google Scholar] [CrossRef]
- Durstewitz, G.; Polley, A.; Plieske, J.; Luerssen, H.; Graner, E.M.; Wieseke, R.; Ganal, M.W. SNP Discovery by Amplicon Sequencing and Multiplex SNP Genotyping in the Allopolyploid Species Brassica NapusThis Article Is One of a Selection of Papers from the Conference “Exploiting Genome-Wide Association in Oilseed Brassicas: A Model for Genetic Improvement of Major OECD Crops for Sustainable Farming”. Genome 2010, 53, 948–956. [Google Scholar] [CrossRef]
- Appleby, N.; Edwards, D.; Batley, J. New Technologies for Ultra-High Throughput Genotyping in Plants. In Plant Genomics; Gustafson, J.P., Langridge, P., Somers, D.J., Eds.; Methods in Molecular BiologyTM; Humana Press: Totowa, NJ, USA, 2009; Volume 513, pp. 19–39. ISBN 978-1-58829-997-0. [Google Scholar]
- Dagnall, C.L.; Morton, L.M.; Hicks, B.D.; Li, S.; Zhou, W.; Karlins, E.; Teshome, K.; Chowdhury, S.; Lashley, K.S.; Sampson, J.N.; et al. Successful Use of Whole Genome Amplified DNA from Multiple Source Types for High-Density Illumina SNP Microarrays. BMC Genom. 2018, 19, 182. [Google Scholar] [CrossRef]
- Patel, D.A.; Zander, M.; Dalton-Morgan, J.; Batley, J. Advances in Plant Genotyping: Where the Future Will Take Us. In Plant Genotyping; Batley, J., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2015; Volume 1245, pp. 1–11. ISBN 978-1-4939-1965-9. [Google Scholar]
- Mir, R.R.; Varshney, R.K. Future Prospects of Molecular Markers in Plants. In Molecular markers in plants; Blackwell Publishing Ltd.: Oxford, UK, 2012; pp. 169–190. ISBN 978-1-118-47302-3. [Google Scholar]
- Semagn, K.; Babu, R.; Hearne, S.; Olsen, M. Single Nucleotide Polymorphism Genotyping Using Kompetitive Allele Specific PCR (KASP): Overview of the Technology and Its Application in Crop Improvement. Mol. Breed. 2014, 33, 1–14. [Google Scholar] [CrossRef]
- Chagné, D.; Batley, J.; Edwards, D.; Forster, J.W. Single Nucleotide Polymorphism Genotyping in Plants. In Association Mapping in Plants; Oraguzie, N.C., Rikkerink, E.H.A., Gardiner, S.E., De Silva, H.N., Eds.; Springer: New York, NY, USA, 2007; pp. 77–94. ISBN 978-0-387-35844-4. [Google Scholar]
- Druml, B.; Cichna-Markl, M. High Resolution Melting (HRM) Analysis of DNA—Its Role and Potential in Food Analysis. Food Chem. 2014, 158, 245–254. [Google Scholar] [CrossRef]
- Jurinke, C.; Van Den Boom, D.; Cantor, C.R.; Köster, H. The Use of MassARRAY Technology for High Throughput Genotyping. In Chip Technology; Hoheisel, J., Brazma, A., Büssow, K., Cantor, C.R., Christians, F.C., Chui, G., Diaz, R., Drmanac, R., Drmanac, S., Eickhoff, H., et al., Eds.; Advances in Biochemical Engineering/Biotechnology; Springer: Berlin/Heidelberg, Germany, 2002; Volume 77, pp. 57–74. ISBN 978-3-540-43215-9. [Google Scholar]
- Kumar, S.; Banks, T.W.; Cloutier, S. SNP Discovery through Next-Generation Sequencing and Its Applications. Int. J. Plant Genom. 2012, 2012, 831460. [Google Scholar] [CrossRef] [PubMed]
- Ganal, M.W.; Altmann, T.; Röder, M.S. SNP Identification in Crop Plants. Curr. Opin. Plant Biol. 2009, 12, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; He, Q.; Wang, J.; Wang, B.; Zhao, J.; Huang, S.; Yang, T.; Tang, Y.; Yang, S.; Aisimutuola, P.; et al. Super-Pangenome Analyses Highlight Genomic Diversity and Structural Variation across Wild and Cultivated Tomato Species. Nat. Genet. 2023, 55, 852–860. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yang, W.; Zhang, S.; Hu, H.; Yuan, Y.; Dong, J.; Chen, L.; Ma, Y.; Yang, T.; Zhou, L.; et al. A Pangenome Analysis Pipeline Provides Insights into Functional Gene Identification in Rice. Genome Biol. 2023, 24, 19. [Google Scholar] [CrossRef] [PubMed]
- Petereit, J.; Bayer, P.E.; Thomas, W.J.W.; Tay Fernandez, C.G.; Amas, J.; Zhang, Y.; Batley, J.; Edwards, D. Pangenomics and Crop Genome Adaptation in a Changing Climate. Plants 2022, 11, 1949. [Google Scholar] [CrossRef] [PubMed]
- Bayer, P.E.; Golicz, A.A.; Scheben, A.; Batley, J.; Edwards, D. Plant pan-genomes are the new reference. Nat. Plants 2020, 6, 914–920. [Google Scholar] [CrossRef] [PubMed]
- Bohra, A.; Bansal, K.C.; Graner, A. The 3366 Chickpea Genomes for Research and Breeding. Trends Plant Sci. 2022, 27, 217–219. [Google Scholar] [CrossRef]
- Brockhurst, M.A.; Harrison, E.; Hall, J.P.J.; Richards, T.; McNally, A.; MacLean, C. The Ecology and Evolution of Pangenomes. Curr. Biol. 2019, 29, R1094–R1103. [Google Scholar] [CrossRef]
- Chaudhari, N.M.; Gupta, V.K.; Dutta, C. BPGA- an Ultra-Fast Pan-Genome Analysis Pipeline. Sci. Rep. 2016, 6, 24373. [Google Scholar] [CrossRef]
- Della Coletta, R.; Qiu, Y.; Ou, S.; Hufford, M.B.; Hirsch, C.N. How the Pan-Genome Is Changing Crop Genomics and Improvement. Genome Biol. 2021, 22, 3. [Google Scholar] [CrossRef]
- Pronozin, A.Y.; Bragina, M.K.; Salina, E.A. Crop Pangenomes. Vavilov J. Genet. Breed. 2021, 25, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Mohd Saad, N.S.; Neik, T.X.; Thomas, W.J.W.; Amas, J.C.; Cantila, A.Y.; Craig, R.J.; Edwards, D.; Batley, J. Advancing Designer Crops for Climate Resilience through an Integrated Genomics Approach. Curr. Opin. Plant Biol. 2022, 67, 102220. [Google Scholar] [CrossRef] [PubMed]
- Naqvi, R.Z.; Siddiqui, H.A.; Mahmood, M.A.; Najeebullah, S.; Ehsan, A.; Azhar, M.; Farooq, M.; Amin, I.; Asad, S.; Mukhtar, Z.; et al. Smart Breeding Approaches in Post-Genomics Era for Developing Climate-Resilient Food Crops. Front. Plant Sci. 2022, 13, 972164. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Zhao, X.; Mace, E.; Henry, R.; Jordan, D. Exploring and Exploiting Pan-Genomics for Crop Improvement. Mol. Plant 2019, 12, 156–169. [Google Scholar] [CrossRef]
- Wang, W.; Mauleon, R.; Hu, Z.; Chebotarov, D.; Tai, S.; Wu, Z.; Li, M.; Zheng, T.; Fuentes, R.R.; Zhang, F.; et al. Genomic Variation in 3,010 Diverse Accessions of Asian Cultivated Rice. Nature 2018, 557, 43–49. [Google Scholar] [CrossRef]
- Zhao, Q.; Feng, Q.; Lu, H.; Li, Y.; Wang, A.; Tian, Q.; Zhan, Q.; Lu, Y.; Zhang, L.; Huang, T.; et al. Pan-Genome Analysis Highlights the Extent of Genomic Variation in Cultivated and Wild Rice. Nat. Genet. 2018, 50, 278–284. [Google Scholar] [CrossRef]
- Jayakodi, M.; Padmarasu, S.; Haberer, G.; Bonthala, V.S.; Gundlach, H.; Monat, C.; Lux, T.; Kamal, N.; Lang, D.; Himmelbach, A.; et al. The Barley Pan-Genome Reveals the Hidden Legacy of Mutation Breeding. Nature 2020, 588, 284–289. [Google Scholar] [CrossRef]
- Montenegro, J.D.; Golicz, A.A.; Bayer, P.E.; Hurgobin, B.; Lee, H.; Chan, C.K.; Visendi, P.; Lai, K.; Doležel, J.; Batley, J.; et al. The Pangenome of Hexaploid Bread Wheat. Plant J. 2017, 90, 1007–1013. [Google Scholar] [CrossRef]
- Raza, A.; Bohra, A.; Varshney, R.K. Pan-Genome for Pearl Millet That Beats the Heat. Trends Plant Sci. 2023, 28, 857–860. [Google Scholar] [CrossRef]
- Liu, Y.; Du, H.; Li, P.; Shen, Y.; Peng, H.; Liu, S.; Zhou, G.-A.; Zhang, H.; Liu, Z.; Shi, M.; et al. Pan-Genome of Wild and Cultivated Soybeans. Cell 2020, 182, 162–176.e13. [Google Scholar] [CrossRef]
- Rijzaani, H.; Bayer, P.E.; Rouard, M.; Doležel, J.; Batley, J.; Edwards, D. The Pangenome of Banana Highlights Differences between Genera and Genomes. Plant Genome 2022, 15, e20100. [Google Scholar] [CrossRef] [PubMed]
- Jha, U.C.; Nayyar, H.; von Wettberg, E.J.B.; Naik, Y.D.; Thudi, M.; Siddique, K.H.M. Legume Pangenome: Status and Scope for Crop Improvement. Plants 2022, 11, 3041. [Google Scholar] [CrossRef] [PubMed]
- Shang, L.; Li, X.; He, H.; Yuan, Q.; Song, Y.; Wei, Z.; Lin, H.; Hu, M.; Zhao, F.; Zhang, C.; et al. A Super Pan-Genomic Landscape of Rice. Cell Res. 2022, 32, 878–896. [Google Scholar] [CrossRef] [PubMed]
- Leonard, A.S.; Crysnanto, D.; Mapel, X.M.; Bhati, M.; Pausch, H. Graph Construction Method Impacts Variation Representation and Analyses in a Bovine Super-Pangenome. Genome Biol. 2023, 24, 124. [Google Scholar] [CrossRef]
- Wang, S.; Qian, Y.-Q.; Zhao, R.-P.; Chen, L.-L.; Song, J.-M. Graph-Based Pan-Genomes: Increased Opportunities in Plant Genomics. J. Exp. Bot. 2023, 74, 24–39. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, Z.; Bao, Z.; Li, H.; Lyu, Y.; Zan, Y.; Wu, Y.; Cheng, L.; Fang, Y.; Wu, K.; et al. Graph Pangenome Captures Missing Heritability and Empowers Tomato Breeding. Nature 2022, 606, 527–534. [Google Scholar] [CrossRef]
- Khan, A.W.; Garg, V.; Roorkiwal, M.; Golicz, A.A.; Edwards, D.; Varshney, R.K. Super-Pangenome by Integrating the Wild Side of a Species for Accelerated Crop Improvement. Trends Plant Sci. 2020, 25, 148–158. [Google Scholar] [CrossRef]
- Roy, A. Relevance of Genomic Selection for Development of Crops with Climate Change Resilience. Pharma Innov. J. 2023, 12, 1837–1847. [Google Scholar]
- Whittaker, J.C.; Thompson, R.; Denham, M.C. Marker-Assisted Selection Using Ridge Regression. Genet. Res. 2000, 75, 249–252. [Google Scholar] [CrossRef]
- Handerson, C.R. Applications of Linear Models in Animal Breeding; University of Guelph: Guelph, ON, Canada, 1984. [Google Scholar]
- Endelman, J.B. Ridge Regression and Other Kernels for Genomic Selection with R Package RrBLUP. Plant Genome 2011, 4, 250–255. [Google Scholar] [CrossRef]
- Charmet, G.; Tran, L.-G.; Auzanneau, J.; Rincent, R.; Bouchet, S. BWGS: A R Package for Genomic Selection and Its Application to a Wheat Breeding Programme. PLoS ONE 2020, 15, e0222733. [Google Scholar] [CrossRef] [PubMed]
- GitHub—JaeYoonKim72/GMStool. Available online: https://github.com/JaeYoonKim72/GMStool (accessed on 13 June 2023).
- Jeong, S.; Kim, J.-Y.; Kim, N. GMStool: GWAS-Based Marker Selection Tool for Genomic Prediction from Genomic Data. Sci. Rep. 2020, 10, 19653. [Google Scholar] [CrossRef] [PubMed]
- Tecle, I.Y.; Edwards, J.D.; Menda, N.; Egesi, C.; Rabbi, I.Y.; Kulakow, P.; Kawuki, R.; Jannink, J.-L.; Mueller, L.A. SolGS: A Web-Based Tool for Genomic Selection. BMC Bioinform. 2014, 15, 398. [Google Scholar] [CrossRef] [PubMed]
- Pérez, P.; de los Campos, G. Genome-Wide Regression and Prediction with the BGLR Statistical Package. Genetics 2014, 198, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, C.S.; Choudhary, R.K.; Kumar, D. Genomic Selection Hands On: Using GenSel and RrBLUP Package; NBAGR: Karnal, India, 2016.
- Guha Majumdar, S.; Rai, A.; Mishra, D.C. Integrated Framework for Selection of Additive and Nonadditive Genetic Markers for Genomic Selection. J. Comput. Biol. 2020, 27, 845–855. [Google Scholar] [CrossRef]
- Caamal-Pat, D.; Pérez-Rodríguez, P.; Crossa, J.; Velasco-Cruz, C.; Pérez-Elizalde, S.; Vázquez-Peña, M. Lme4GS: An R-Package for Genomic Selection. Front. Genet. 2021, 12, 680569. [Google Scholar] [CrossRef]
- Milani, P.; Torres-Aguilar, P.; Hamaker, B.; Manary, M.; Abushamma, S.; Laar, A.; Steiner, R.; Ehsani, M.; De La Parra, J.; Skaven-Ruben, D.; et al. The Whole Grain Manifesto: From Green Revolution to Grain Evolution. Glob. Food Secur. 2022, 34, 100649. [Google Scholar] [CrossRef]
- Shah, T.I.; Shah, A.M.; Bangroo, S.A.; Sharma, M.P.; Aezum, A.M.; Kirmani, N.A.; Lone, A.H.; Jeelani, M.I.; Rai, A.P.; Wani, F.J.; et al. Soil Quality Index as Affected by Integrated Nutrient Management in the Himalayan Foothills. Agronomy 2022, 12, 1870. [Google Scholar] [CrossRef]
- Pazhamala, L.; Saxena, R.K.; Singh, V.K.; Sameerkumar, C.V.; Kumar, V.; Sinha, P.; Patel, K.; Obala, J.; Kaoneka, S.R.; Tongoona, P.; et al. Genomics-Assisted Breeding for Boosting Crop Improvement in Pigeonpea (Cajanus cajan). Front. Plant Sci. 2015, 6, 50. [Google Scholar] [CrossRef]
- Salgotra, R.K.; Stewart, C.N. Functional Markers for Precision Plant Breeding. Int. J. Mol. Sci. 2020, 21, 4792. [Google Scholar] [CrossRef]
- Dulakakharia, B.; Longkho, K.; Sharma, V.; Verma, R.K. Rice Drought Tolerance: Emerging Molecular Breeding Strategies in the Post-Genomic Era. In Smart Plant Breeding for Field Crops in Post-Genomics Era; Sharma, D., Singh, S., Sharma, S.K., Singh, R., Eds.; Springer Nature: Singapore, 2023; pp. 99–135. ISBN 978-981-19821-7-0. [Google Scholar]
- Zenda, T.; Wang, N.; Dong, A.; Zhou, Y.; Duan, H. Reproductive-Stage Heat Stress in Cereals: Impact, Plant Responses and Strategies for Tolerance Improvement. Int. J. Mol. Sci. 2022, 23, 6929. [Google Scholar] [CrossRef]
- Pathirana, R.; Carimi, F. Management and Utilization of Plant Genetic Resources for a Sustainable Agriculture. Plants 2022, 11, 2038. [Google Scholar] [CrossRef] [PubMed]
- Nandini, B.; Venkatesh; Reddy, U.G.; Mallikarjuna, B.P.; Manu, B.; Vaijayanthi, P.V.; Ashwini, M.; Surendra, P.; Vijayakumar, A.G.; Kumar, C.J.; et al. Genomic Design for Abiotic Stress Resistance in Pigeonpea. In Genomic Designing for Abiotic Stress Resistant Pulse Crops; Kole, C., Ed.; Springer International Publishing: Cham, Switzerland, 2022; pp. 169–248. ISBN 978-3-030-91038-9. [Google Scholar]
- Arya, M.; Mishra, S.B. Kamaluddin Transgenic and Molecular Approaches for Pigeonpea and Chick Pea Improvement. In Technologies in Plant Biotechnology and Breeding of Field Crops; Kamaluddin, Kiran, U., Abdin, M.Z., Eds.; Springer Nature: Singapore, 2022; pp. 239–272. ISBN 9789811657665. [Google Scholar]
- Sinha, S.; Kushwaha, B.K.; Deshmukh, R.K. QTL Mapping Using Advanced Mapping Populations and High-throughput Genotyping. In Genotyping by Sequencing for Crop Improvement; Sonah, H., Goyal, V., Shivaraj, S.M., Deshmukh, R.K., Eds.; Wiley: Hoboken, NJ, USA, 2022; pp. 52–79. ISBN 978-1-119-74565-5. [Google Scholar]
- Merrick, L.F.; Herr, A.W.; Sandhu, K.S.; Lozada, D.N.; Carter, A.H. Utilizing Genomic Selection for Wheat Population Development and Improvement. Agronomy 2022, 12, 522. [Google Scholar] [CrossRef]
- Egea, I.; Estrada, Y.; Faura, C.; Egea-Fernández, J.M.; Bolarin, M.C.; Flores, F.B. Salt-Tolerant Alternative Crops as Sources of Quality Food to Mitigate the Negative Impact of Salinity on Agricultural Production. Front. Plant Sci. 2023, 14, 1092885. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, A. Effect of Low Temperature Stress on Photosynthesis and Allied Traits: A Review. In Physiological Processes in Plants Under Low Temperature Stress; Springer: Singapore, 2022; pp. 199–297. ISBN 9789811690365. [Google Scholar]
- Soualiou, S.; Wang, Z.; Sun, W.; De Reffye, P.; Collins, B.; Louarn, G.; Song, Y. Functional–Structural Plant Models Mission in Advancing Crop Science: Opportunities and Prospects. Front. Plant Sci. 2021, 12, 747142. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Zhang, S.; Poleksic, A.; Xie, L. Heterogeneous Multi-Layered Network Model for Omics Data Integration and Analysis. Front. Genet. 2020, 10, 1381. [Google Scholar] [CrossRef] [PubMed]
| S. No. | Character | MAS | GS |
|---|---|---|---|
| 1. | Marker number | Phenotype trait selected indirectly using genetic marker linked to the genomic region controlling trait of interest. | MAS variant based on GEBVs estimated using all the markers’ effects using a trained GS model. |
| 2. | Trait nature | Effective for oligogenic traits/major QTL traits with major effects. | Effective for traits with small effects along with major effects, i.e., polygenic traits/major and minor QTLs. |
| 3. | Prerequisite | Mapping and confirmation of markers connected with trait-associated QTL. | Training a good GS model in a TP utilizing genotype and phenotype data. |
| 4. | Approach | It is a targeted approach where only markers linked to a few validated major QTLs are used to implement MAS. | It is a holistic approach where all the markers used in training a GS model are used to implement GS in the BP. |
| 5. | Population nature | Applied on any population of a given crop if the QTL is validated, which is very rare. Relatively less effective for improving quantitative traits. | Workable on a BP that is related to or a derivative of the TP. Highly effective in improving quantitative traits. |
| 6. | Implementation | To complement any of the conventional breeding strategies like MA-Backcross, MA-Pedigree, MA-Recurrent selection. | More appropriately implemented in line with development breeding. |
| 7. | Genetic gain | Genetic gain per unit of time is less and much time is spent on QTL detection and validation. | Genetic gain per unit of time is relatively high as all the QTLs with major and minor effects are considered. |
| 8. | Limitations | Linkage drag, background noise, and environmental instability, especially for quantitative traits. | Factors influencing prediction accuracy. |
| 9. | Suitability | Complex genome, high polyploidy, heterozygosity, varied chromosome number, low/medium-density markers. | Improves the breeding efficiency and prediction, covers the entire genome, is preferred in purebred breeding across many animal species, forecasts the breeding potential of individual lines, and increases heritability estimation. |
| S.No. | Crops | Model | Trait | References |
|---|---|---|---|---|
| 1 | Maize | GBLUP | Grain yield | [131] |
| RRBLUP | Grain yield | [132] | ||
| 100 kernel weight | [132] | |||
| Bayes A, Bayes B, Bayes C, LASSO, and RKHS GBLUP and multigroup GBLUP | Grain yield | [133] | ||
| RRBLUP and BSSV (Bayesian stochastic search variable) | Ear rot | [134] | ||
| BLUP | Striga resistance Drought tolerance | [135] | ||
| GBLUP | Drought tolerance | [136] | ||
| RRBLUP and GBLUP | Water-logging tolerance | [137] | ||
| 2 | Barley | RRBLUP | Grain yield | [138] |
| GBLUP and RKHS | Thousand kernel weight (TKW) | [139] | ||
| GBLUP | DON resistance | [140] | ||
| 3 | Rice | Bayesian LASSO | Grain yield | [141] |
| RRBLUP | Panicle weight | [142] | ||
| GBLUP | Grain yield, Field grain, Field grain weight, The variance of field grain | [143] | ||
| GBLUP, SVM, LASSO, and PLS | Field grain | [144] | ||
| GBLUP | Field grain weight | [145] | ||
| GBLUP, RKHS, and Bayes B | Panicle weight Nitrogen balance index | [146] | ||
| GBLUP | Thousand-grain weight (TGW), Grain yield | [57] | ||
| RRBLUP and LUP | Blast resistance | [147] | ||
| GBLUP and RKHS | Drought tolerance | [148] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sinha, D.; Maurya, A.K.; Abdi, G.; Majeed, M.; Agarwal, R.; Mukherjee, R.; Ganguly, S.; Aziz, R.; Bhatia, M.; Majgaonkar, A.; et al. Integrated Genomic Selection for Accelerating Breeding Programs of Climate-Smart Cereals. Genes 2023, 14, 1484. https://doi.org/10.3390/genes14071484
Sinha D, Maurya AK, Abdi G, Majeed M, Agarwal R, Mukherjee R, Ganguly S, Aziz R, Bhatia M, Majgaonkar A, et al. Integrated Genomic Selection for Accelerating Breeding Programs of Climate-Smart Cereals. Genes. 2023; 14(7):1484. https://doi.org/10.3390/genes14071484
Chicago/Turabian StyleSinha, Dwaipayan, Arun Kumar Maurya, Gholamreza Abdi, Muhammad Majeed, Rachna Agarwal, Rashmi Mukherjee, Sharmistha Ganguly, Robina Aziz, Manika Bhatia, Aqsa Majgaonkar, and et al. 2023. "Integrated Genomic Selection for Accelerating Breeding Programs of Climate-Smart Cereals" Genes 14, no. 7: 1484. https://doi.org/10.3390/genes14071484
APA StyleSinha, D., Maurya, A. K., Abdi, G., Majeed, M., Agarwal, R., Mukherjee, R., Ganguly, S., Aziz, R., Bhatia, M., Majgaonkar, A., Seal, S., Das, M., Banerjee, S., Chowdhury, S., Adeyemi, S. B., & Chen, J.-T. (2023). Integrated Genomic Selection for Accelerating Breeding Programs of Climate-Smart Cereals. Genes, 14(7), 1484. https://doi.org/10.3390/genes14071484











