Presence of Human DNA on Household Dogs and Its Bi-Directional Transfer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Design
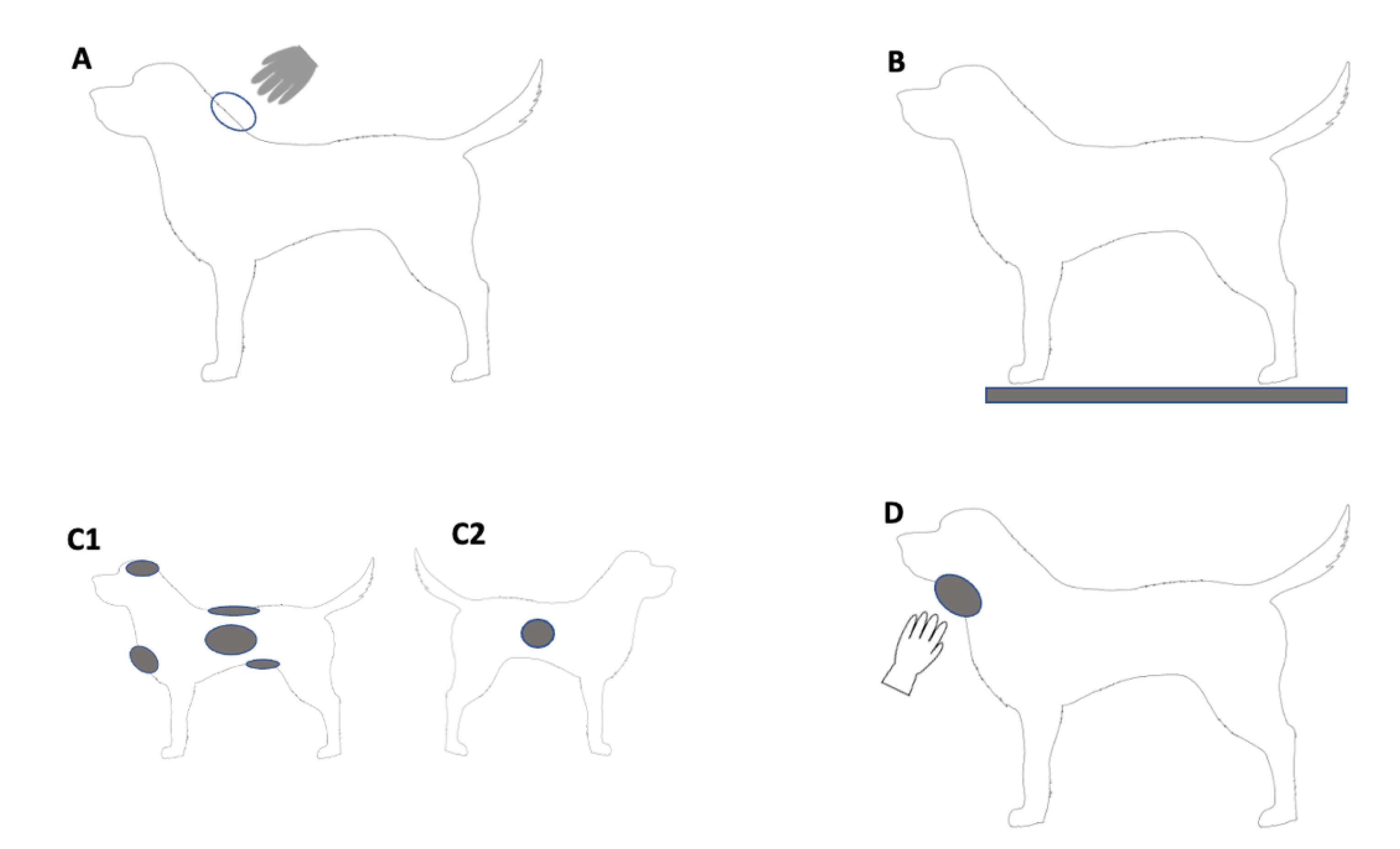
- Upon first entering the house, the researcher patted and scratched the dog on the top of its neck/scruff area with a gloved hand and a sample was collected from the glove to determine if any human DNA that was present could be transferred from the dog to another surface upon contact (Figure 1A). The gloves (InControl, Tarragindi, Australia) were taken directly from a box prior to patting and were assumed to be DNA-free. To ensure consistency across all dogs, they were scratched 4 times (hand opening and closing was considered one scratch) and then patted in the same area a further 4 times.
- A fresh pair of gloves were donned by the researcher and a sterile plastic sheet (90 cm × 90 cm, Multigate, Villawood, Australia) with a grid-like pattern was adhered to the floor using tape in an area identified by the owner as being suitable. The owner then enticed the dog to walk over the plastic sheet four times (left to right and then from right to left twice across the sheet) and a sample was collected from areas of the sheet contacted by the paws, as identified by the researcher using the grid. As above, this was to determine if any human DNA that was present could be transferred from the dog to another surface upon contact.
- After donning new gloves, samples were collected directly from six distinct areas of the dog to assess the general presence of human DNA. The six areas, in order of collection, included the chest, top of the head, back, left and right sides, and stomach (Figure 1(C1,C2)); the swabs applied to the left side were placed under the fur to make contact with the skin; all other samples were taken from the surface of the fur. The differing samples from the left and right sides were collected to assess if there was a difference in the quantity of DNA retrieved from the base of the coat/skin surface compared to the top of the fur. Collecting the skin sample from the other side of the dog, rather than the same side as the coat sample, was done to avoid any potential impacts that taking each sample may have had on the other. While care was taken to collect samples from similarly-sized areas from each dog, due to the differences in the size of the dogs and the inability to use a template, there were some differences in the size of the areas swabbed between dogs.
- After removing gloves, the researcher patted and scratched the dog under the neck/chin area with a single bare hand (Figure 1D) to determine if their DNA could be transferred and detected on a dog during a single contact event. Here, the same scratch/pat method was applied as previously described when using a gloved hand. The area was swabbed immediately after contact.
2.2. Sample Collection and Processing
2.3. DNA Profile Interpretation
2.4. Data Analysis
3. Results
3.1. Presence of Human DNA on Dogs
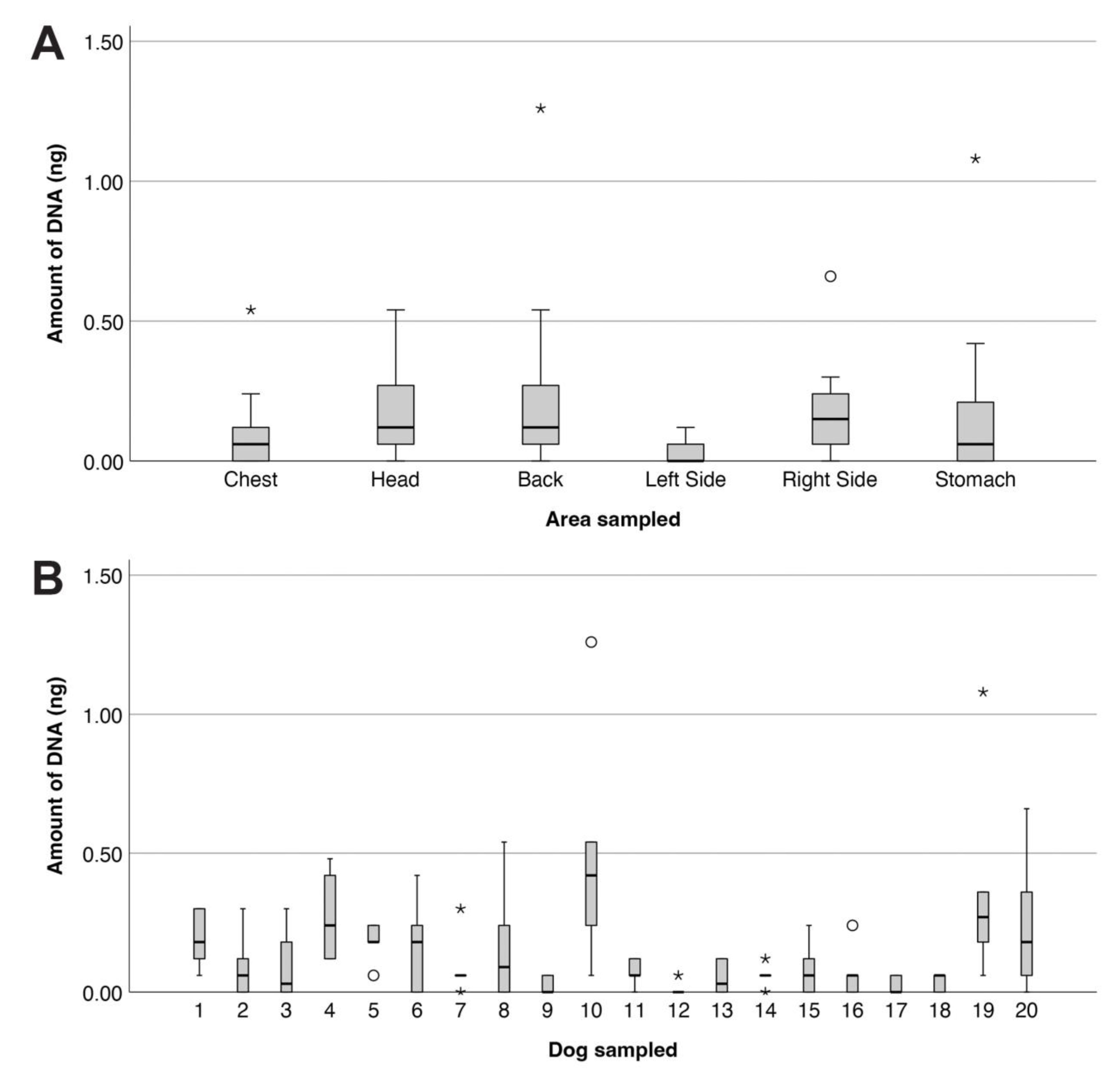
3.1.1. DNA Quantities
3.1.2. Profile Interpretation
3.1.3. Influence of Activities
3.2. Human DNA Transfer to Dogs
3.3. Human DNA Transfer from Dogs
3.3.1. To a Gloved Hand upon Contact
3.3.2. To a Sheet upon Walking
4. Discussion
4.1. Presence of Human DNA on Dogs
4.2. Human DNA Transfer to Dogs
4.3. Human DNA Transfer from Dogs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- van Oorschot, R.A.H.; Szkuta, B.; Meakin, G.E.; Kokshoorn, B.; Goray, M. DNA transfer in forensic science: A review. Forensic Sci. Int. Genet. 2019, 38, 140–166. [Google Scholar] [CrossRef]
- Gosch, A.; Courts, C. On DNA transfer: The lack and difficulty of systematic research and how to do it better. Forensic Sci. Int. Genet. 2019, 40, 24–36. [Google Scholar] [CrossRef] [PubMed]
- Gill, P.; Hicks, T.; Butler, J.M.; Connolly, E.; Gusmão, L.; Kokshoorn, B.; Morling, N.; van Oorschot, R.A.H.; Parson, W.; Prinz, M.; et al. DNA commission of the International society for forensic genetics: Assessing the value of forensic biological evidence—Guidelines highlighting the importance of propositions. Part II: Evaluation of biological traces considering activity level propositions. Forensic Sci. Int. Genet. 2020, 44, 102186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meakin, G.E.; Kokshoorn, B.; van Oorschot, R.A.H.; Szkuta, B. Evaluating forensic DNA evidence: Connecting the dots. WIREs Forensic Sci. 2020, 3, e1404. [Google Scholar] [CrossRef]
- van den Berge, M.; Ozcanhan, G.; Zijlstra, S.; Lindenbergh, A.; Sijen, T. Prevalence of human cell material: DNA and RNA profiling of public and private objects and after activity scenarios. Forensic Sci. Int. Genet. 2016, 21, 81–89. [Google Scholar] [CrossRef]
- Gosch, A.; Euteneuer, J.; Preuß-Wössner, J.; Courts, C. DNA transfer to firearms in alternative realistic handling scenarios. Forensic Sci. Int. Genet. 2020, 48, 102355. [Google Scholar] [CrossRef]
- Pfeifer, C.M.; Wiegand, P. Persistence of touch DNA on burglary-related tools. Int. J. Leg. Med. 2017, 131, 941–953. [Google Scholar] [CrossRef]
- Szkuta, B.; Ballantyne, K.N.; van Oorschot, R.A.H. Transfer and persistence of DNA on the hands and the influence of activities performed. Forensic Sci. Int. Genet. 2017, 28, 10–20. [Google Scholar] [CrossRef]
- Reither, J.B.; Gray, E.; Durdle, A.; Conlan, X.A.; van Oorschot, R.A.H.; Szkuta, B. Investigation into the prevalence of background DNA on flooring within houses and its transfer to a contacting surface. Forensic Sci. Int. 2021, 318, 110563. [Google Scholar] [CrossRef]
- Monkman, H.; van Oorschot, R.A.H.; Goray, M. Is there human DNA on cats. Forensic Sci. Int. Genet. Suppl. Ser. 2022, 8, 145–146. [Google Scholar] [CrossRef]
- Brower, A.; Akridge, B.; Siemens-Bradley, N. Human DNA collection from police dogs: Technique and application. Forensic Sci. Med. Pathol. 2021, 17, 230–234. [Google Scholar] [CrossRef]
- Goray, M.; Fowler, S.; Szkuta, B.; van Oorschot, R.A.H. Shedder status—An analysis of self and non-self DNA in multiple handprints deposited by the same individuals over time. Forensic Sci. Int. Genet. 2016, 23, 190–196. [Google Scholar] [CrossRef]
- Jansson, L.; Swensson, M.; Gifvars, E.; Hedell, R.; Forsberg, C.; Ansell, R.; Hedman, J. Individual shedder status and the origin of touch DNA. Forensic Sci. Int. Genet. 2022, 56, 102626. [Google Scholar] [CrossRef]
- Kanokwongnuwut, P.; Martin, B.; Kirkbride, K.P.; Linacre, A. Shedding light on shedders. Forensic Sci. Int. Genet. 2018, 36, 20–25. [Google Scholar] [CrossRef] [Green Version]
- Utz, R.L. Walking the Dog: The Effect of Pet Ownership on Human Health and Health Behaviors. Soc. Indic. Res. 2014, 116, 327–339. [Google Scholar] [CrossRef]
- Joosten, P.; Van Cleven, A.; Sarrazin, S.; Paepe, D.; De Sutter, A.; Dewulf, J. Dogs and Their Owners Have Frequent and Intensive Contact. Int. J. Environ. Res. Public. Health 2020, 17, 4300. [Google Scholar] [CrossRef]
- Szkuta, B.; Ansell, R.; Boiso, L.; Connolly, E.; Kloosterman, A.D.; Kokshoorn, B.; McKenna, L.G.; Steensma, K.; van Oorschot, R.A.H. Assessment of the transfer, persistence, prevalence and recovery of DNA traces from clothing: An inter-laboratory study on worn upper garments. Forensic Sci. Int. Genet. 2019, 42, 56–68. [Google Scholar] [CrossRef] [PubMed]
- Goray, M.; Kokshoorn, B.; Steensma, K.; Szkuta, B.; van Oorschot, R.A.H. DNA detection of a temporary and original user of an office space. Forensic Sci. Int. Genet. 2020, 44, 102203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fonneløp, A.E.; Ramse, M.; Egeland, T.; Gill, P. The implications of shedder status and background DNA on direct and secondary transfer in an attack scenario. Forensic Sci. Int. Genet. 2017, 29, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Manoli, P.; Antoniou, A.; Bashiardes, E.; Xenophontos, S.; Photiades, M.; Stribley, V.; Mylona, M.; Demetriou, C.; Cariolou, M.A. Sex-specific age association with primary DNA transfer. Int. J. Leg. Med. 2016, 130, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Daly, D.J.; Murphy, C.; McDermott, S.D. The transfer of touch DNA from hands to glass, fabric and wood. Forensic Sci. Int. Genet. 2012, 6, 41–46. [Google Scholar] [CrossRef]
- Sessa, F.; Salerno, M.; Bertozzi, G.; Messina, G.; Ricci, P.; Ledda, C.; Rapisarda, V.; Cantatore, S.; Turillazzi, E.; Pomara, C. Touch DNA: Impact of handling time on touch deposit and evaluation of different recovery techniques: An experimental study. Sci. Rep. 2019, 9, 9542. [Google Scholar] [CrossRef] [Green Version]
- Buckingham, A.K.; Harvey, M.L.; van Oorschot, R.A.H. The origin of unknown source DNA from touched objects. Forensic Sci. Int. Genet. 2016, 25, 26–33. [Google Scholar] [CrossRef]
- Cale, C.M.; Earll, M.E.; Latham, K.E.; Bush, G.L. Could secondary DNA transfer falsely place someone at the scene of a crime? J. Forensic Sci. 2016, 61, 196–203. [Google Scholar] [CrossRef]
- Helmus, J.; Bajanowski, T.; Poetsch, M. DNA transfer—A never ending story. A study on scenarios involving a second person as carrier. Int. J. Leg. Med. 2016, 130, 121–125. [Google Scholar] [CrossRef]
- Albers, C.N.; Jensen, A.; Bælum, J.; Jacobsen, C.S. Inhibition of DNA Polymerases Used in Q-PCR by Structurally Different Soil-Derived Humic Substances. Geomicrobiol. J. 2013, 30, 675–681. [Google Scholar] [CrossRef]
- Diegoli, T.M.; Moore, T.; Weitz, S.; Bille, T.W. Prevalence and impact of PCR artifacts from soil samples using the Applied Biosystems™ GlobalFiler™ PCR amplification kit: Interference of non-human artifacts in DNA profiles from a homicide investigation. Forensic Sci. Int. Genet. Suppl. Ser. 2019, 7, 41–43. [Google Scholar] [CrossRef]
- Thornbury, D.; Goray, M.; van Oorschot, R.A.H. Transfer of DNA without contact from used clothing, pillowcases and towels by shaking agitation. Sci. Justice 2021, 61, 797–805. [Google Scholar] [CrossRef]
- Thornbury, D.; Goray, M.; van Oorschot, R.A.H. Indirect DNA transfer without contact from dried biological materials on various surfaces. Forensic Sci. Int. Genet. 2021, 51, 102457. [Google Scholar] [CrossRef]
| Profile Composition | DNA Presence | Transfer from Dog | Transfer to Dog | |
|---|---|---|---|---|
| Gloved Hand after Contact | Sheet after Walking | Dog after Patting | ||
| No profile | 16 | 1 | 4 | 1 |
| Limited information | 23 | 5 | 9 | 4 |
| Complex family mixture | 5 | 1 | 0 | 0 |
| Maj MH + Min Complex | 2 | 0 | 0 | 0 |
| Maj MH + Min P + Min U | 0 | 0 | 0 | 1 |
| Maj MH + Min 2U | 1 | 1 | 0 | 0 |
| Maj MH + Min H2 | 1 | 0 | 0 | 0 |
| Maj MH + Min U | 7 | 2 | 0 | 2 |
| Maj H2 + Min H3 | 2 | 0 | 0 | 0 |
| Maj H2 + Min MH | 1 | 0 | 0 | 0 |
| Maj H2 + Min U | 4 | 0 | 1 | 0 |
| Maj H3 + Min H2 + Min U | 1 | 0 | 0 | 0 |
| Maj H3 + Min H2 | 1 | 0 | 1 | 0 |
| Maj H3 + Min U | 0 | 0 | 0 | 1 |
| Maj P + Min U | 0 | 0 | 0 | 2 |
| Maj U + Min U | 3 | 2 | 1 | 0 |
| NE MH + H2 | 7 | 1 | 0 | 1 |
| NE MH + H2 + U | 3 | 0 | 0 | 0 |
| NE MH + U | 12 | 1 | 0 | 0 |
| NE H2 + 2U | 1 | 0 | 0 | 0 |
| NE H2 + H3 | 3 | 1 | 0 | 0 |
| NE H2 + U | 3 | 0 | 0 | 0 |
| NE H3 + 2U | 1 | 0 | 0 | 0 |
| NE H3 + U | 0 | 0 | 1 | 0 |
| NE P + H2 + H3 | 0 | 0 | 0 | 1 |
| NE P + H2 + U | 0 | 0 | 0 | 1 |
| NE P + H3 | 0 | 0 | 0 | 1 |
| NE P + H3 + U | 0 | 0 | 0 | 1 |
| NE P + U | 0 | 0 | 0 | 3 |
| NE 2U | 5 | 1 | 0 | 0 |
| SS H2 | 0 | 0 | 1 | 0 |
| SS H3 | 3 | 0 | 0 | 0 |
| SS MH | 9 | 1 | 0 | 1 |
| SS U | 6 | 3 | 2 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Monkman, H.; Szkuta, B.; van Oorschot, R.A.H. Presence of Human DNA on Household Dogs and Its Bi-Directional Transfer. Genes 2023, 14, 1486. https://doi.org/10.3390/genes14071486
Monkman H, Szkuta B, van Oorschot RAH. Presence of Human DNA on Household Dogs and Its Bi-Directional Transfer. Genes. 2023; 14(7):1486. https://doi.org/10.3390/genes14071486
Chicago/Turabian StyleMonkman, Heidi, Bianca Szkuta, and Roland A. H. van Oorschot. 2023. "Presence of Human DNA on Household Dogs and Its Bi-Directional Transfer" Genes 14, no. 7: 1486. https://doi.org/10.3390/genes14071486





