Morphology and Mitochondrial Lineage Investigations Corroborate the Systematic Status and Pliocene Colonization of Suncus niger (Mammalia: Eulipotyphla) in the Western Ghats Biodiversity Hotspot of India
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
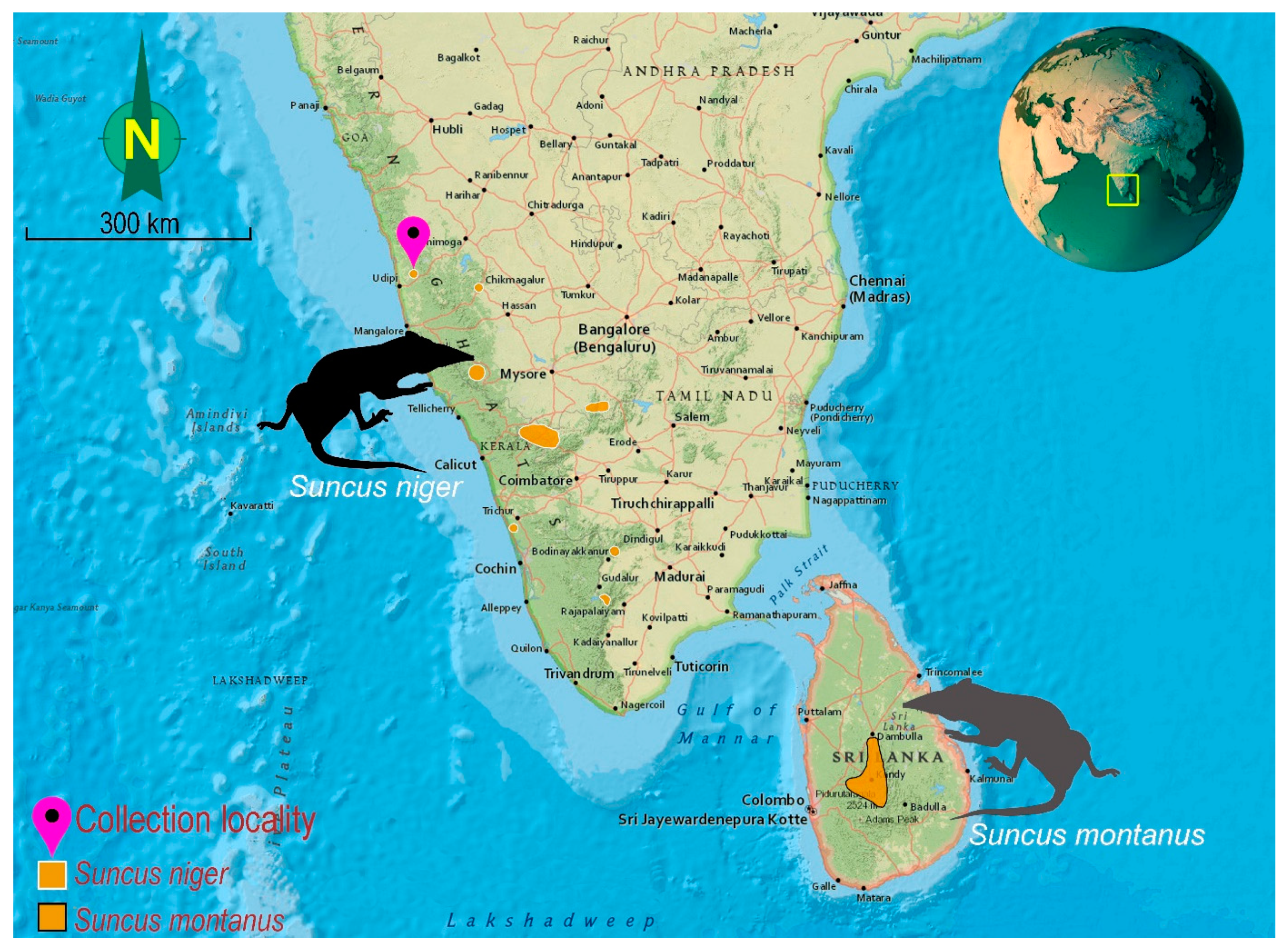
2.1. Sampling and Morphological Analyses
2.2. DNA Extraction and Amplification
2.3. Sequence Quality Check and Dataset Preparation
2.4. Phylogenetic Analyses and Time Tree
3. Results
3.1. Morphological Evidence
3.2. Molecular Identification and Genetic Divergence
3.3. Matrilineal Phylogeny and Divergence Time
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hutterer, R. Order Soricomorpha. In Mammal Species of the World: A Taxonomic and Geographic Reference; Wilson, D.E., Reeder, D.M., Eds.; The Johns Hopkins University Press: Baltimore, MD, USA, 2005; pp. 220–311. [Google Scholar]
- Nations, J.A.; Giarla, T.C.; Morni, M.A.; William Dee, J.; Swanson, M.T.; Hiller, A.E.; Khan, F.A.A.; Esselstyn, J.A. Molecular data from the holotype of the enigmatic Bornean Black Shrew, Suncus ater Medway, 1965 (Soricidae, Crocidurinae), place it in the genus Palawanosorex. Zookeys 2022, 1137, 17–31. [Google Scholar] [CrossRef] [PubMed]
- Kamalakannan, M.; Sivaperuman, C.; Kundu, S.; Gokulakrishnan, G.; Venkatraman, C.; Chandra, K. Discovery of a new mammal species (Soricidae: Eulipotyphla) from Narcondam volcanic island, India. Sci. Rep. 2021, 11, 9416. [Google Scholar] [CrossRef] [PubMed]
- Horsfield, T. A catalogue of the Mammalia in The Museum of the Hon; EastIndia Company. J. & H. Cox: London, UK, 1851; p. 135. [Google Scholar]
- Kelaart, E.F. Description of new species and varieties of mammals found in Ceylon. J. Ceylon Branch R. Asiat. Soc. 1850, 2, 321–328. [Google Scholar]
- Corbet, G.B.; Hill, J.E. Mammals of the Indomalayan Region. A Systematic Review; Oxford University Press: Oxford, UK, 1992; p. 488. [Google Scholar]
- Meegaskumbura, S.; Schneider, C.J. A taxonomic evaluation of the shrew Suncus montanus (Soricidae: Crocidurinae) of Sri Lanka and India. Ceylon J. Sci. 2008, 37, 129–136. [Google Scholar] [CrossRef] [Green Version]
- Meegaskumbura, S.; Meegaskumbura, M.; Schneider, C.J. Systematic relationships and taxonomy of Suncus montanus and S. murinus from Sri Lanka. Mol. Phylogenet. Evol. 2010, 55, 473–487. [Google Scholar] [CrossRef]
- Dubey, S.; Salamin, N.; Ohdachi, S.D.; Barrière, P.; Vogel, P. Molecular phylogenetics of shrews (Mammalia: Soricidae) reveal timing of transcontinental colonizations. Mol. Phylogenet. Evol. 2007, 44, 126–137. [Google Scholar] [CrossRef] [Green Version]
- Dubey, S.; Salamin, N.; Ruedi, M.; Barrière, P.; Colyn, M.; Vogel, P. Biogeographic origin and radiation of the old world Crocidurine shrews (Mammalia: Soricidae) inferred from mitochondrial and nuclear genes. Mol. Phylogenet. Evol. 2008, 48, 953–963. [Google Scholar] [CrossRef] [Green Version]
- Stanhope, M.J.; Waddell, V.G.; Madsen, O.; de Jong, W.; Blair Hedges, S.; Cleven, G.C.; Kao, D.; Springer, M. Molecular evidence for multiple origins of Insectivora and for a new order of endemic African insectivore mammals. Proc. Natl. Acad. Sci. USA 1998, 95, 9967–9972. [Google Scholar] [CrossRef]
- Onuma, M.; Cao, Y.; Hasegawa, M.; Kusakabe, S. A Close Relationship of Chiroptera with Eulipotyphla (Core Insectivora) Suggested by Four Mitochondrial Genes. Zool. Sci. 2000, 17, 1327–1332. [Google Scholar] [CrossRef] [Green Version]
- Shinohara, A.; Campbell, K.L.; Suzuki, H. Molecular phylogenetic relationships of moles, shrew moles, and desmans from the new and old worlds. Mol. Phylogenet. Evol. 2003, 27, 247–258. [Google Scholar] [CrossRef]
- Willows-Munro, S. The Molecular Evolution of African Shrews (Family Soricidae). Unpublished Ph.D. Thesis, Stellenbosch University, Stellenbosch, South Africa, 2008. [Google Scholar]
- Megaaskumbura, S.; Megaaskumbura, M.; Schneider, C.J. Re-evaluation of the taxonomy of the Sri Lanka pigmy shrew Suncus fellowesgordoni (Soricidae: Crocidurinae) and its phylogenetic relationship with S. etruscus. Zootaxa 2012, 3187, 57–68. [Google Scholar] [CrossRef] [Green Version]
- Jacquet, F.; Nicolas, V.; Bonillo, C.; Cruaud, C.; Denys, C. Barcoding, molecular taxonomy, and exploration of the diversity of shrews (Soricomorpha: Soricidae) on Mount Nimba (Guinea). Zool. J. Linn. Soc. 2012, 166, 672–687. [Google Scholar] [CrossRef] [Green Version]
- Darvish, J.; Mahmoudi, A.; Pehpuri, A.; Saeidzadeh, S. New data on distribution and taxonomy of the genus Suncus (Mammalia: Soricidae) in Iran; molecular evidence. Iran. J. Anim. Biosyst. 2017, 13, 229–235. [Google Scholar]
- Quérouil, S.; Hutterer, R.; Barrière, P.; Colyn, M.; Kerbis Peterhans, J.C.; Verheyen, E. Phylogeny and evolution of African shrews (Mammalia: Soricidae) inferred from 16s rRNA sequences. Mol. Phylogenet. Evol. 2001, 20, 185–195. [Google Scholar] [CrossRef]
- Ohdachi, S.D.; Hasegawa, M.; Iwasa, M.A.; Abe, H.; Vogel, P.; Oshida, T.; Lin, L.K. Molecular phylogenetics of soricid shrews (Mammalia) based on mitochondrial cytochrome b gene sequences: With special reference to the Soricinae. J. Zool. 2006, 270, 177–191. [Google Scholar] [CrossRef]
- Omar, H.; Adamson, E.A.S.; Bhassu, S.; Goodman, S.M.; Soarimalala, V.; Hashim, R.; Ruedi, M. Phylogenetic relationships of Malayan and Malagasy pygmy shrews of the genus Suncus (Soricomorpha: Soricidae) inferred from mitochondrial cytochrome b gene sequences. Raffles Bull. Zool. 2011, 59, 237–243. [Google Scholar]
- Willows-Munro, S.; Matthee, C.A. Exploring the diversity and molecular evolution of shrews (family Soricidae) using mtDNA cytochrome b data, Afr. Zool. 2011, 46, 246–262. [Google Scholar]
- Meegaskumbura, S.; Meegaskumbura, M.; Schneider, C. Phylogenetic position of Suncus fellowesgordoni with pigmy shrews from Madagascar and Southeast Asia inferred from cytochrome-b. Ceylon J. Sci. 2012, 41, 83–87. [Google Scholar] [CrossRef] [Green Version]
- Ohdachi, S.D.; Kinoshita, G.; Oda, S.; Motokawa, M.; Jogahara, T.; Arai, S.; Nguyen, S.T.; Suzuki, H.; Katakura, K.; Bawm, S.; et al. Intraspecific phylogeny of the house shrews, Suncus murinus-S. montanus species complex, based on the mitochondrial cyt b gene. Mammal. Study 2016, 41, 229–238. [Google Scholar] [CrossRef] [Green Version]
- Ohdachi, S.D.; Kinoshita, G.; Nasher, A.K.; Yonezawa, T.; Arai, S.; Kikuchi, F.; Lin, K.S.; Bawm, S. Re-evaluation of the phylogeny based on mitochondrial cytochrome b gene in the house shrew, Suncus murinus-S. montanus species complex, with special reference to Yemen and Myanmar populations. J. Wildl. Biodivers. 2017, 1, 79–87. [Google Scholar]
- Shpirer, E.; Haddas-Sasson, M.; Spivak-Glater, M.; Feldstein, T.; Meiri, S.; Huchon, D. Molecular relationships of the Israeli shrews (Eulipotyphla: Soricidae) based on cytochrome b sequences. Mammalia 2021, 85, 79–89. [Google Scholar] [CrossRef]
- Meegaskumbura, S.; Meegaskumbura, M.; Schneider, C.J. Phylogenetic Relationships of the Endemic Sri Lankan Shrew Genera: Solisorex and Feroculus. Ceylon J Sci (Biol.. Sci.) 2014, 43, 65–71. [Google Scholar] [CrossRef] [Green Version]
- Yamagata, T.; Ohishi, K.; Faruque, M.O.; Masangkay, J.S.; Ba-Loc, C.; Vu-Binh, D.; Mansjoer, S.S.; Ikeda, H.; Namikawa, T. Genetic variation and geographic distribution on the mitochondrial DNA in local population of the musk shrew, Suncus murinus. Jpn. J. Genet. 1995, 70, 321–337. [Google Scholar] [CrossRef] [Green Version]
- Ruedi, M.; Courvoisier, C.; Vogel, P.; Catzeflis, F.M. Genetic differentiation and zoogeography of Asian Suncus murinus (Mammalia: Soricidae). Biol. J. Linn. Soc. 1996, 57, 307–316. [Google Scholar] [CrossRef] [Green Version]
- Kurachi, M.; Chau, B.L.; Dang, V.B.; Dorji, T.; Yamamoto, Y.; Nyunt, M.M.; Maeda, Y.; Chhum-Phith, L.; Namikawa, T.; Yamagata, T. Population structure of wild musk shrew (Suncus murinus) in Asia based on mitochondrial DNA variation, with research in Cambodia and Bhutan. Biochem. Genet. 2007, 45, 165–183. [Google Scholar] [CrossRef]
- Chen, S.; Wei, H.; Peng, H.; Yong, B. The complete mitogenome of Asian House Shrews, Suncus murinus (Soricidae). Mitochondrial DNA Part A DNA Mapp. Seq. Anal. 2016, 27, 1127–1128. [Google Scholar] [CrossRef]
- Murphy, W.J.; Eizirik, E.; O’Brien, S.J.; Madsen, O.; Scally, M.; Douady, C.J.; Teeling, E.; Ryder, O.A.; Stanhope, M.J.; de Jong, W.W.; et al. Resolution of the early placental mammal radiation using bayesian phylogenetics. Science 2001, 294, 2348–2351. [Google Scholar] [CrossRef]
- Kennerley, R.J.; Lacher, T.E.; Hudson, M.A.; Long, B.; McCay, S.D.; Roach, N.S.; Turvey, S.T.; Young, R.P. Global patterns of extinction risk and conservation needs for Rodentia and Eulipotyphla. Divers. Distrib. 2021, 27, 1792–1806. [Google Scholar] [CrossRef]
- Hutterer, R.; Balete, D.S.; Giarla, T.C.; Heaney, L.R.; Esselstyn, J.A. A new genus and species of shrew (Mammalia: Soricidae) from Palawan Island, Philippines. J. Mammal. 2018, 99, 518–536. [Google Scholar] [CrossRef] [Green Version]
- Menon, V. Indian Mammals—A Field Guide; Hachette Book Publishing India Pvt. Ltd.: Gurgaon, India, 2014; pp. 336–341. [Google Scholar]
- Jenkins, P.; Ruedi, M.; Catzeflis, M. A biochemical and morphological investigation of Suncus dayi (Dobson, 1888) and discussion of relationship in Suncus Hemprich & Ehrenberg, 1833, Crocidura Wagler, 1832, and Sylvisorex Thomas, 1904 (Insectivora: Soricidae). Bonn. Zool. Bull. 1998, 47, 257–276. [Google Scholar]
- Percie du Sert, N.; Hurst, V.; Ahluwalia, A.; Alam, S.; Avey, M.T.; Baker, M.; Browne, W.J.; Clark, A.; Cuthill, I.C.; Dirnagl, U.; et al. The ARRIVE guidelines 2.0: Updated guidelines for reporting animal research. PLoS Biol. 2020, 18, e3000410. [Google Scholar]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001; Volume 1. [Google Scholar]
- Verma, S.K.; Singh, L. Novel universal primers establish identity of an enormous number of animal species for forensic application. Mol. Ecol. Notes 2002, 3, 28–31. [Google Scholar] [CrossRef]
- Palumbi, S.R. Nucleic Acids II: The Polymerase Chain Reaction. In Molecular Systematics; Hillis, D.M., Moritz, C., Mable, B.K., Eds.; Sinauer Associates: Sunderland, MA, USA, 1996; pp. 205–207. [Google Scholar]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. JModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [Green Version]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [Green Version]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Battistuzzi, F.U.; Billing-Ross, P.; Murillo, O.; Filipski, A.; Kumar, S. Estimating Divergence Times in Large Molecular Phylogenies. Proc. Natl. Acad. Sci. USA 2012, 109, 19333–19338. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Tao, Q.; Kumar, S. Theoretical Foundation of the RelTime Method for Estimating Divergence Times from Variable Evolutionary Rates. Mol. Biol. Evol. 2018, 35, 1770–1782. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar]
- Teixeira, J.C.; Huber, C.D. The inflated significance of neutral genetic diversity in conservation genetics. Proc. Natl. Acad. Sci. USA 2021, 118, e2015096118. [Google Scholar] [CrossRef] [PubMed]
- Cowie, R.H.; Bouchet, P.; Fontaine, B. The Sixth Mass Extinction: Fact, fiction or speculation? Biol. Rev. Camb. Philos. Soc. 2022, 97, 640–663. [Google Scholar] [CrossRef] [PubMed]
- Pimm, S.L.; Jenkins, C.N.; Abell, R.; Brooks, T.M.; Gittleman, J.L.; Joppa, L.N.; Raven, P.H.; Roberts, C.M.; Sexton, J.O. The biodiversity of species and their rates of extinction, distribution, and protection. Science 2014, 344, 1246752. [Google Scholar] [CrossRef] [PubMed]
- Costello, M.J.; May, R.M.; Stork, N.E. Can we name Earth’s species before they go extinct? Science 2013, 339, 413–416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lees, A.C.; Pimm, S.L. Species, extinct before we know them? Curr. Biol. 2015, 25, R177–R180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abu, A.; Leow, L.K.; Ramli, R.; Omar, H. Classification of Suncus murinus species complex (Soricidae: Crocidurinae) in Peninsular Malaysia using image analysis and machine learning approaches. BMC Bioinform. 2016, 17, 505. [Google Scholar] [CrossRef] [Green Version]
- Sagar, H.S.S.C.; Girish, D.V.; Bharath, C.V.; Madhusudhan, M.C.; Sharath, I.M.; Kumar, K.; Amarnath, B.; Surya, M.M. Indian Shrew: First record of Hill Shrew Suncus niger from the Bababudan Hills, Chikkamagaluru District, Western Ghats, Karnataka, India. Small Mammal Mail#414. Zoo’s Print 2017, 32, 34–36. [Google Scholar]
- Burgin, C.J.; He, K. Family Soricidae (shrews). In Handbook of the Mammals of the World, Insectivores, Sloths, and Colugos; Wilson, D.E., Mittermeier, R.A., Eds.; Lynx Edicions: Barcelona, Spain, 2018; Volume 8, pp. 332–551. [Google Scholar]
- Bradley, R.D.; Baker, R.J. A Test of the Genetic Species Concept: Cytochrome-b Sequences and Mammals. J. Mammal. 2001, 82, 960–973. [Google Scholar] [CrossRef]
- Baker, R.J.; Bradley, R.D. Speciation in Mammals and the Genetic Species Concept. J. Mammal. 2006, 87, 643–662. [Google Scholar] [CrossRef] [Green Version]
- Ali, J.R.; Aitchison, J.C. Gondwana to Asia: Plate tectonics, paleogeography and the biological connectivity of the Indian sub-continent from the Middle Jurassic through latest Eocene (166–35 Ma). Earth Sci. Rev. 2008, 88, 145–166. [Google Scholar] [CrossRef]
- Dubey, S.; Koyasu, K.; Parapanov, R.; Ribi, M.; Hutterer, R.; Vogel, P. Molecular phylogenetics reveals Messinian, Pliocene, and Pleistocene colonizations of islands by North African shrews. Mol. Phylogenet. Evol. 2008, 47, 877–882. [Google Scholar] [CrossRef] [Green Version]
- Warren, B.H.; Strasberg, D.; Bruggemann, J.H.; Prys-Jones, R.P.; Thébaud, C. Why does the biota of the Madagascar region have such a strong Asiatic flavour? Cladistics 2010, 26, 526–538. [Google Scholar] [CrossRef] [PubMed]
- Dittus, W.P.J. The biogeography and ecology of Sri Lankan mammals point to conservation priorities. Ceylon J. Sci. 2017, 46, 33–64. [Google Scholar] [CrossRef]
- Bossuyt, F.; Milinkovitch, M.C. Amphibians as indicators of early Tertiary “out of India” dispersal of vertebrates. Science 2001, 292, 93–95. [Google Scholar] [CrossRef] [Green Version]
- Bossuyt, F.; Meegaskumbura, M.; Beenaerts, N.; Gower, D.J.; Pethiyagoda, R.; Roelants, K.; Mannaert, A.; Wilkinson, M.; Bahir, M.M.; Manamendra-Arachchi, K.; et al. Local endemism within the western Ghats-Sri Lanka Biodiversity hotspot. Science 2004, 306, 479–481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinya, S.; Bover, P.; Jurado-Rivera, J.A.; Trenado, S.; Parpal, L.; Férriz, I.; Talavera, A.; Hinckley, A.; Pons, J.; López-Fuster, M.J. Recent island colonization by an introduced shrew in the western Mediterranean. Hystrix Ital. J. Mammal. 2018, 29, 232–235. [Google Scholar]
- Esselstyn, J.A.; Achmadi, A.S.; Handika, H.; Swanson, M.T.; Giarla, T.C.; Rowe, K.C. Fourteen new, endemic species of shrew (genus Crocidura) from Sulawesi reveal a spectacular island radiation. Bull. Am. Mus. Nat. Hist. 2021, 454, 1–108. [Google Scholar] [CrossRef]
- Molur, S.; Srinivasulu, C.; Srinivasulu, B.; Walker, S.; Nameer, P.O.; Ravikumar, L. Status of Non-volant Small Mammals: Conservation Assessment and Management Plan (CAMP) Workshop Report; Zoo Outreach Organizaton/CBSG South Asia: Coimbatore, India, 2005; p. 618. [Google Scholar]
- Molur, S.; Singh, M. Non-volant small mammals of the Western Ghats of Coorg District, southern India. J. Threat. Taxa 2009, 1, 589–608. [Google Scholar] [CrossRef] [Green Version]
- Molur, S.; Nameer, P.O.; de A. Goonatilake, W.I.L.D.P.T.S. Suncus montanus. The IUCN Red List of Threatened Species 2008: E.T21147A9251556. 2008. Available online: https://www.iucnredlist.org/species/21147/9251556 (accessed on 6 June 2023).
- Shanker, K. The role of competition and habitat in structuring small mammal communities in a tropical montane ecosystem in southern India. J. Zool., Lond. 2001, 253, 15–24. [Google Scholar] [CrossRef]
- Wijesinghe, M.R.; del Brooke, M. Impact of habitat disturbance on the distribution of endemic species of small mammals and birds in a tropical rain forest in Sri Lanka. J. Trop. Ecol. 2005, 21, 661–668. [Google Scholar] [CrossRef]
- CEPF. Ecosystem Profile: Western Ghats and Sri Lanka Biodiversity Hotspot—Western Ghats Region; Critical Ecosystem Partnership Fund: Arlington, TX, USA, 2007; 100p. [Google Scholar]
- Nicolas, V.; Jacquet, F.; Hutterer, R.; Konečný, A.; Kouassi, S.K.; Durnez, L.; Lalis, A.; Colyn, M.; Denys, C. Multilocus Phylogeny of the Crocidura Poensis Species Complex (Mammalia, Eulipotyphla): Influences of the Palaeoclimate on Its Diversification and Evolution. J. Biogeogr. 2019, 46, 871–883. [Google Scholar] [CrossRef] [Green Version]
- Demos, T.C.; Kerbis Peterhans, J.C.; Agwanda, B.; Hickerson, M.J. Uncovering cryptic diversity and refugial persistence among small mammal lineages across the Eastern Afromontane biodiversity hotspot. Mol. Phylogenetics Evol. 2014, 71, 41–54. [Google Scholar] [CrossRef] [PubMed]




| Measurements | Sn [This Study] | Sn [34] | Smo [8] | Smu [8] | Smu [ZSI] | Se [22] | Sf [22] | Sd [35] | Ss [35] | Ss [ZSI] | Sz (GBIF) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| India | India | Sri Lanka | Sri Lanka | India | Sri Lanka | Sri Lanka | India | Sri Lanka | India | Sri Lanka | |
| (n = 3) | (n ≥ 1) | (n = 18) | (n = 8) | (n = 5) | (n = 3) | (n = 5) | (n = 7) | (n = 11) | (n = 4) | (n = 2) | |
| HBL | 80–110 | 80–105 | 87.3–121.5 | 100–160 | 107–120 | 42.1–42.4 | 44.0–49.4 | 70–78 | 68–85 | 59–75 | 108–114 |
| TL | 45–63.51 | 45–65 | 56.1–75.5 | 72–85 | 62–69 | 27.4–31.1 | 33.5–38.0 | 83–88 | 44–55 | 37–54 | 92–97 |
| EH | 11.2–12.69 | - | 8–12.6 | 11.4–13.6 | 0.9–13 | 5.9–6.2 | 5.6–7.0 | - | - | 0.9–10 | 10–13 |
| HFL | 14–18 | 14–17 | 15.6–19.1 | 20.1–23.4 | 16–18.5 | 7.4–7.6 | 9.6–11.0 | 15.5–16.5 | 10.5–15 | 11–14 | 20 |
| GL | 26.3–27.3 | - | 23.1–27.8 | 27–34.3 | 24–27.3 | 11.9 | 12.9–13.7 | - | - | 14.4–15.2 | - |
| BL | 22.9–24.2 | - | 20.5–25.3 | 24.3–31.7 | 21.2–25.6 | 11.0–11.1 | 11.7–12.3 | - | - | 13.5–14.2 | - |
| CL | 26–27.5 | - | 22.9–27.8 | 27–34.7 | 24.1–28.7 | 12.0–12.1 | 12.7–13.6 | 18.9–20.2 | 18.6–22.2 | 18.6–22.2 | - |
| MTR | 6.9–7.9 | - | 6.8–8.1 | 7.5–9.4 | 6.9–7.8 | 2.8–3.1 | 3.6–3.9 | 8.5–8.8 | 8.1–10.3 | 6.1–6.6 | - |
| PL | 11.6–12.2 | - | 10.2–12.5 | 11.8–15.5 | 10.8–12.5 | 3.8–4.5 | 4.8–5.4, | - | - | 8.2–9.1 | - |
| LR | 9.5–10.1 | - | 8.4–10.3 | 10.2–12.4 | 9.8–11.9 | 3.8–4.0 | 4.2–4.7 | - | - | 6.7–6.5 | - |
| BB | 11.3–12.1 | - | 9.7–12.3 | 11.3–15.3 | 10.7–13.3 | 5.3 | 5.5–5.7 | 8.3–9.8 | 8.9–9.6 | 8.9–9.6 | - |
| PW | 6.9–7.4 | - | 6.4–7.7 | 8.2–9.5 | 8.1–8.9 | 3.0–3.1 | 3.3–3.9 | 5.6–6.0 | 5.2–6.9 | 5.2–6.9 | - |
| HB | 5.8–6.1 | - | 5.4–6.5 | 6.3–7.9 | 6.2–7.5 | 2.5–2.6 | 3.0–3.2 | 5.0–5.3 | 3.9–5.0 | 3.9–5.0 | - |
| ML | 13.6–14.7 | - | 13.2–15.3 | 14.7–19.1 | 13.7–15.2 | 6.0–6.3 | 6.9–7.3 | - | - | 10.5–11.2 | - |
| LDT | 7.0–8.0 | - | 7.0–8.4 | 8.1–9.8 | 7.8–8.3 | 3.0–3.2 | 3.8–3.9 | - | - | 6.1–6.7 | - |
| DD | 6.4–7.7 | - | 5.9–8.5 | 8–11 | 7.9–9.1 | 2.7–3.0 | 3.2–3.5 | - | - | 5.6–5.8 | - |
| Species | Inter-Species | Intra-Species | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S. niger | 1.14 | ||||||||||||
| S. montanus | 8.66 | 1.65 | |||||||||||
| S. murinus | 9.85 | 7.87 | 3.68 | ||||||||||
| S. stoliczkanus | 8.49 | 9.50 | 9.67 | 0.24 | |||||||||
| S. malayanus | 15.78 | 14.59 | 15.32 | 17.25 | 1.47 | ||||||||
| S. madagascariensis | 16.17 | 16.42 | 16.14 | 18.08 | 7.31 | 0.97 | |||||||
| S. etruscus | 16.30 | 15.48 | 16.50 | 17.66 | 8.66 | 3.15 | 0.16 | ||||||
| S. fellowesgordoni | 16.45 | 18.12 | 17.34 | 18.27 | 9.85 | 10.83 | 12.24 | 0.32 | |||||
| S. dayi | 18.19 | 19.20 | 19.42 | 17.50 | 21.52 | 18.02 | 15.86 | 22.30 | 0.73 | ||||
| S. varilla | 21.18 | 21.22 | 22.23 | 22.66 | 22.15 | 20.17 | 17.88 | 20.07 | 22.06 | 0.00 | |||
| S. megalura | 21.85 | 19.83 | 19.61 | 21.07 | 27.03 | 23.88 | 23.18 | 26.19 | 19.23 | 22.60 | n/c | ||
| S. remyi | 24.63 | 23.66 | 24.59 | 22.03 | 21.71 | 22.33 | 19.69 | 21.37 | 22.17 | 20.69 | 25.72 | n/c | |
| S. hututsi | 26.29 | 24.40 | 25.45 | 24.93 | 22.88 | 24.36 | 24.49 | 24.56 | 21.58 | 23.44 | 21.86 | 14.11 | 0.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kundu, S.; Kamalakannan, M.; Kim, A.R.; Hegde, V.D.; Banerjee, D.; Jung, W.-K.; Kim, Y.-M.; Kim, H.-W. Morphology and Mitochondrial Lineage Investigations Corroborate the Systematic Status and Pliocene Colonization of Suncus niger (Mammalia: Eulipotyphla) in the Western Ghats Biodiversity Hotspot of India. Genes 2023, 14, 1493. https://doi.org/10.3390/genes14071493
Kundu S, Kamalakannan M, Kim AR, Hegde VD, Banerjee D, Jung W-K, Kim Y-M, Kim H-W. Morphology and Mitochondrial Lineage Investigations Corroborate the Systematic Status and Pliocene Colonization of Suncus niger (Mammalia: Eulipotyphla) in the Western Ghats Biodiversity Hotspot of India. Genes. 2023; 14(7):1493. https://doi.org/10.3390/genes14071493
Chicago/Turabian StyleKundu, Shantanu, Manokaran Kamalakannan, Ah Ran Kim, Vishwanath D. Hegde, Dhriti Banerjee, Won-Kyo Jung, Young-Mog Kim, and Hyun-Woo Kim. 2023. "Morphology and Mitochondrial Lineage Investigations Corroborate the Systematic Status and Pliocene Colonization of Suncus niger (Mammalia: Eulipotyphla) in the Western Ghats Biodiversity Hotspot of India" Genes 14, no. 7: 1493. https://doi.org/10.3390/genes14071493
APA StyleKundu, S., Kamalakannan, M., Kim, A. R., Hegde, V. D., Banerjee, D., Jung, W.-K., Kim, Y.-M., & Kim, H.-W. (2023). Morphology and Mitochondrial Lineage Investigations Corroborate the Systematic Status and Pliocene Colonization of Suncus niger (Mammalia: Eulipotyphla) in the Western Ghats Biodiversity Hotspot of India. Genes, 14(7), 1493. https://doi.org/10.3390/genes14071493








