Neutral Forces and Balancing Selection Interplay to Shape the Major Histocompatibility Complex Spatial Patterns in the Striped Hamster in Inner Mongolia: Suggestive of Broad-Scale Local Adaptation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Study Area and Sampling Methods
2.3. Parasite Screening
2.4. Microsatellite Genotyping
2.5. Microsatellite Analysis
2.6. MHC Genotyping
2.7. MHC Analysis
2.7.1. Test for Positive Selection
2.7.2. MHC Diversity, Polygenetic Relationships, and Population Structure
2.8. Comparisons between Neutral and Adaptive Markers
3. Results
3.1. Parasite Communities
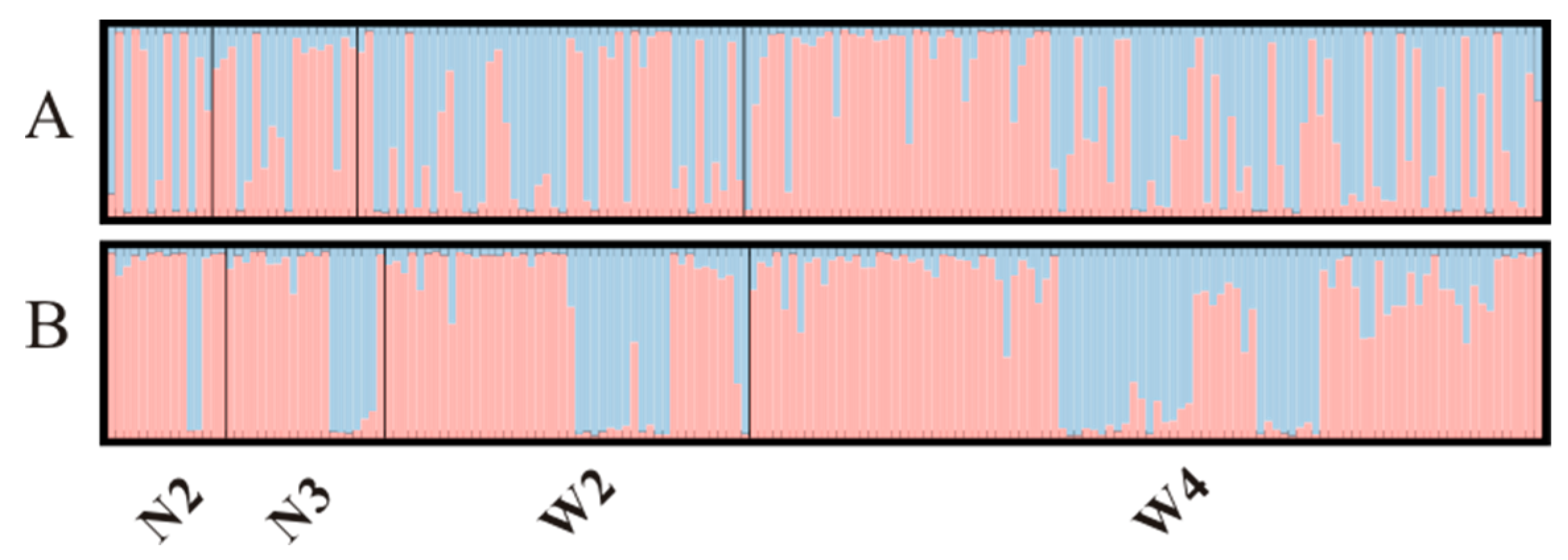
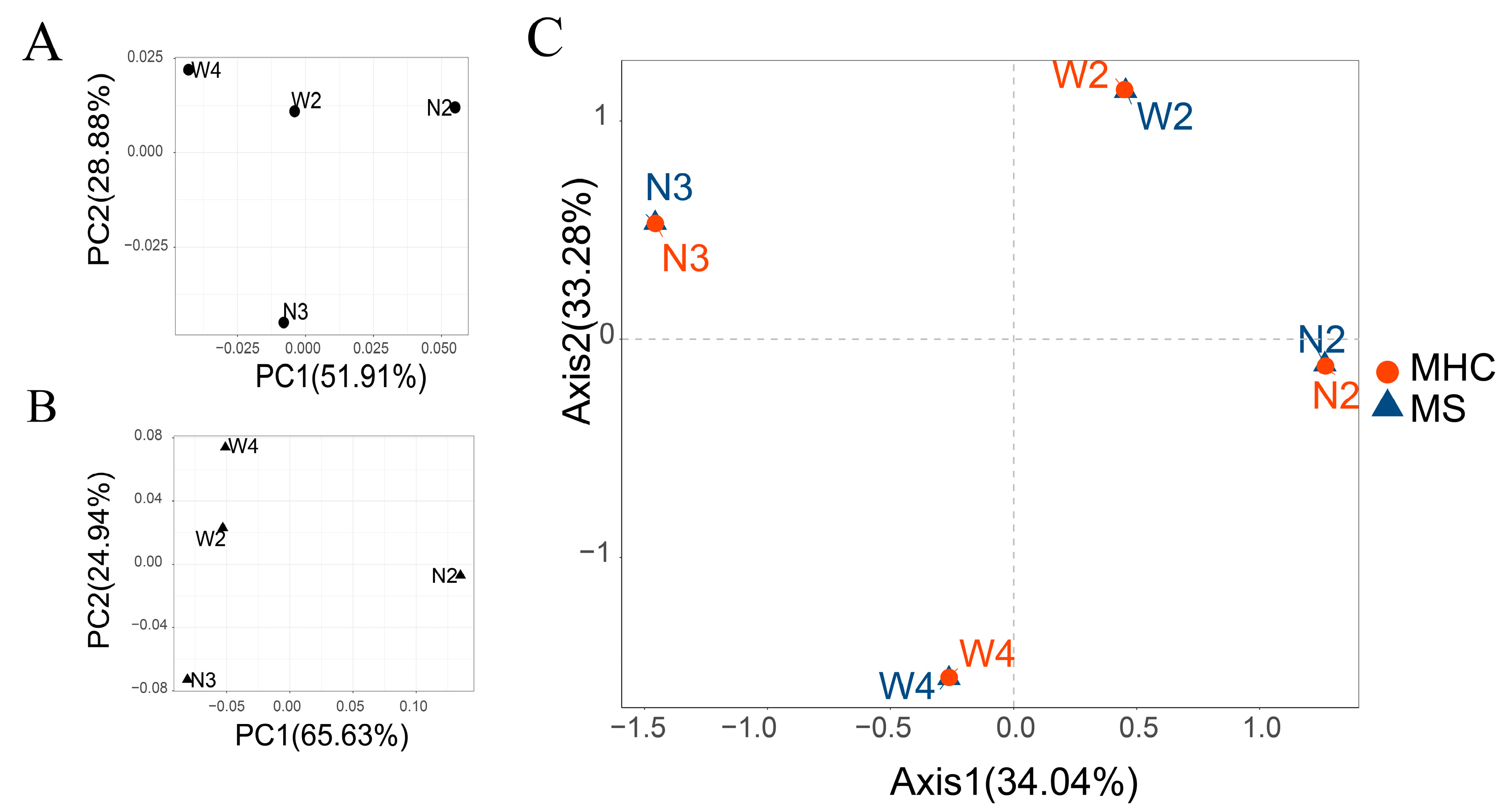
3.2. Microsatellite Genetic Diversity and Population Structure
3.3. MHC Genotyping
3.4. MHC Selection
3.5. MHC Population Diversity and Structure
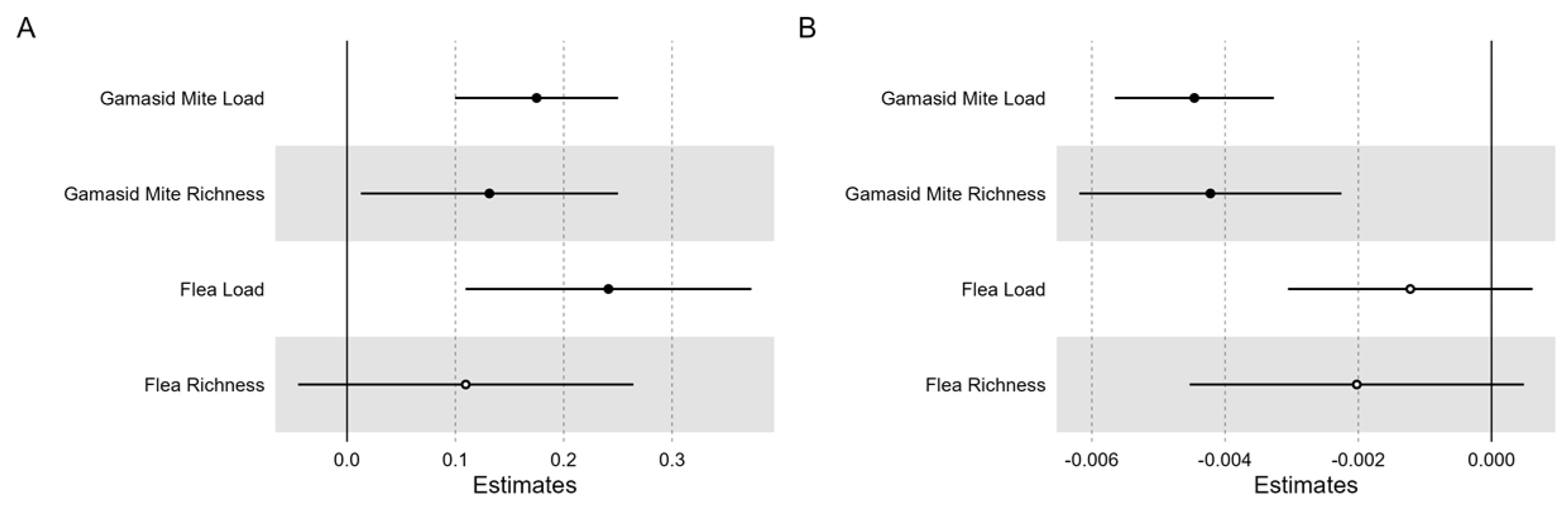
3.6. Comparing MHC and Neutral Diversity
4. Discussion
4.1. Parasite Diversity at Different Sites
4.2. Characterization of MHC Polymorphism, Historical Selection, and Parasite Resistance
4.3. Neutral Processes Mostly Shaped MHC Population Variation
4.4. The Spatial Scale of Local Adaptation
4.5. Implications for Plague Control
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Williams, G.C. Adaptation and Natural Selection: A Critique of Some Current Evolutionary Thought; Princeton University Press: Princeton, NJ, USA, 2018; ISBN 0691185506. [Google Scholar]
- Gandon, S.; Michalakis, Y. Local adaptation, evolutionary potential and host–parasite coevolution: Interactions between migration, mutation, population size and generation time. J. Evol. Biol. 2010, 15, 451–462. [Google Scholar] [CrossRef]
- Lam, D.K.; Frantz, A.C.; Burke, T.; Geffen, E.; Sin, S.Y.W. Both selection and drift drive the spatial pattern of adaptive genetic variation in a wild mammal. Evolution 2023, 77, 221–238. [Google Scholar] [CrossRef] [PubMed]
- Buzan, E.; Potušek, S.; Duniš, L.; Pokorny, B. Neutral and Selective Processes Shape MHC Diversity in Roe Deer in Slovenia. Animals 2022, 12, 723. [Google Scholar] [CrossRef] [PubMed]
- Reyna, D.L.; Fernando, L. The Ecology of Adaptive Radiation in Darwin’s Finches. Ph.D. Thesis, McGill University, Montreal, QC, Canada, 2010. [Google Scholar]
- Weiskopf, S.R.; Rubenstein, M.A.; Crozier, L.G.; Gaichas, S.; Griffis, R.; Halofsky, J.E.; Hyde, K.J.; Morelli, T.L.; Morisette, J.T.; Muñoz, R.C. Climate change effects on biodiversity, ecosystems, ecosystem services, and natural resource management in the United States. Sci. Total Environ. 2020, 733, 137782. [Google Scholar] [CrossRef]
- Tartally, A.; Thomas, J.A.; Anton, C.; Balletto, E.; Barbero, F.; Bonelli, S.; Bräu, M.; Casacci, L.P.; Csősz, S.; Czekes, Z. Patterns of host use by brood parasitic Maculinea butterflies across Europe. Philos. Trans. R. Soc. B 2019, 374, 20180202. [Google Scholar] [CrossRef] [Green Version]
- Karvonen, A.; Seehausen, O. The Role of Parasitism in Adaptive Radiations—When Might Parasites Promote and When Might They Constrain Ecological Speciation? Int. J. Ecol. 2012, 2012, 235010. [Google Scholar] [CrossRef] [Green Version]
- Shaner, P.L.; Yu, A.Y.; Li, S.H.; Hou, C.H. The effects of food and parasitism on reproductive performance of a wild rodent. Ecol. Evol. 2018, 8, 4162–4172. [Google Scholar] [CrossRef]
- Lazzaro, B.P.; Little, T.J. Immunity in a variable world. Philos. Trans. R. Soc. B-Biol. Sci. 2009, 364, 15–26. [Google Scholar] [CrossRef] [Green Version]
- Matthews, B.; Harmon, L.J.; M’Gonigle, L.; Marchinko, K.B.; Schaschl, H. Sympatric and allopatric divergence of MHC genes in threespine stickleback. PLoS ONE 2010, 5, e10948. [Google Scholar] [CrossRef]
- Eizaguirre, C.; Lenz, T.L.; Traulsen, A.; Milinski, M. Speciation accelerated and stabilized by pleiotropic major histocompatibility complex immunogenes. Ecol. Lett. 2009, 12, 5–12. [Google Scholar] [CrossRef]
- Langmann, T.; Rehli, M. Immunobiology. Editorial. Immunobiology 2010, 215, 673. [Google Scholar] [CrossRef]
- Neefjes, J.; Jongsma, M.L.M.; Paul, P.; Bakke, O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat. Rev. Immunol. 2011, 11, 823–836. [Google Scholar] [CrossRef] [PubMed]
- Meyer, B.S.; Hablutzel, P.I.; Roose, A.K.; Hofmann, M.J.; Salzburger, W.; Raeymaekers, J. An exploration of the links between parasites, trophic ecology, morphology, and immunogenetics in the Lake Tanganyika cichlid radiation. Hydrobiologia 2019, 832, 215–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mona, S.; Crestanello, B.; Bankhead-Dronnet, S.; Pecchioli, E.; Ingrosso, S.; D’Amelio, S.; Rossi, L.; Meneguz, P.G.; Bertorelle, G. Disentangling the effects of recombination, selection, and demography on the genetic variation at a major histocompatibility complex class II gene in the alpine chamois. Mol. Ecol. 2008, 17, 4053–4067. [Google Scholar] [CrossRef] [PubMed]
- Garrigan, D.; Hedrick, P.W. Perspective: Detecting adaptive molecular polymorphism: Lessons from the MHC. Evolution 2003, 57, 1707–1722. [Google Scholar]
- Scherman, K.; Raberg, L.; Westerdahl, H. Borrelia Infection in Bank Voles Myodes glareolus Is Associated with Specific DQB Haplotypes Which Affect Allelic Divergence Within Individuals. Front. Immunol. 2021, 12, 703025. [Google Scholar] [CrossRef]
- Oliver, M.K.; Telfer, S.; Piertney, S.B. Major histocompatibility complex (MHC) heterozygote superiority to natural multi-parasite infections in the water vole (Arvicola terrestris). Proc. R. Soc. B Biol. Sci. 2009, 276, 1119–1128. [Google Scholar] [CrossRef]
- Froeschke, G.; Sommer, S.; Stoute, J.A. Insights into the complex associations between MHC class II DRB polymorphism and multiple gastrointestinal parasite infestations in the striped mouse. PLoS ONE 2012, 7, e31820. [Google Scholar] [CrossRef] [Green Version]
- Cutrera, A.P.; Mora, M.S. Selection on MHC in a Context of Historical Demographic Change in 2 Closely Distributed Species of Tuco-tucos (Ctenomys australis and C. talarum). J. Hered. 2017, 108, 628–639. [Google Scholar] [CrossRef] [Green Version]
- Peng, F.; Ballare, K.M.; Hollis, W.S.; den Haan, S.; Bolnick, D.I. What evolutionary processes maintain MHC II diversity within and among populations of stickleback? Mol. Ecol. 2021, 30, 1659–1671. [Google Scholar] [CrossRef]
- Jacek, R.; Wiesław, B.; Jim, K.; Tobias, L.L.; Jamie, W. Advances in the Evolutionary Understanding of MHC Polymorphism. Trends Genet. 2020, 36, 298–311. [Google Scholar]
- Hedrick, P.W. Pathogen resistance and genetic variation at MHC loci. Evolution 2002, 56, 1902–1908. [Google Scholar] [CrossRef] [PubMed]
- Rico, Y.; Ethier, D.M.; Davy, C.M.; Sayers, J.; Weir, R.D.; Swanson, B.J.; Nocera, J.J.; Kyle, C.J. Spatial patterns of immunogenetic and neutral variation underscore the conservation value of small, isolated American badger populations. Evol. Appl. 2016, 9, 1271–1284. [Google Scholar] [CrossRef]
- Bracamonte, S.E.; Hofmann, M.J.; Lozano-Martin, C.; Eizaguirre, C.; Barluenga, M. Divergent and non-parallel evolution of MHC IIB in the Neotropical Midas cichlid species complex. BMC Ecol. Evol. 2022, 22, 41. [Google Scholar] [CrossRef]
- Hablutzel, P.I.; Volckaert, F.A.; Hellemans, B.; Raeymaekers, J.A. Differential modes of MHC class IIB gene evolution in cichlid fishes. Immunogenetics 2013, 65, 795–809. [Google Scholar] [CrossRef]
- Hablutzel, P.I.; Gregoir, A.F.; Vanhove, M.P.; Volckaert, F.A.; Raeymaekers, J.A. Weak link between dispersal and parasite community differentiation or immunogenetic divergence in two sympatric cichlid fishes. Mol. Ecol. 2016, 25, 5451–5466. [Google Scholar] [CrossRef] [PubMed]
- Shuai, L.; Wang, L.; Yang, Y.; Zhang, F. Effects of density dependence and climatic factors on population dynamics of Cricetulus barabensis: A 25-year field study. J. Mammal. 2020, 101, 507–514. [Google Scholar] [CrossRef]
- Poplavskaya, N.; Bannikova, A.; Neumann, K.; Pavlenko, M.; Kartavtseva, I.; Bazhenov, Y.; Bogomolov, P.; Abramov, A.; Surov, A.; Lebedev, V. Phylogeographic structure in the chromosomally polymorphic rodent Cricetulus barabensis sensu lato (Mammalia, Cricetidae). J. Zool. Syst. Evol. Res. 2019, 57, 679–694. [Google Scholar] [CrossRef]
- Li, Y.E.; Shuai, M.A.; Wang, Y.J.; Zheng, J.W.; Wang, D.P.; Gui-Jun, L.I.; Fan, J.W.; Shi, Y.S.; Zhang, X.F.; Bai, J.Y. Biological characteristics of Chinese hamster infected with Babesia. Chin. J. Comp. Med. 2016, 8, 36–41. [Google Scholar]
- Zhang, X.F.; Liu, R.; Cui, X.X.; Shuai, M.A.; Zhang, X.C.; Bai, J.Y.; Center, L.A. Changes of cytokines in the Cricetulus barabensis and their albino mutant infected with trichinella spiralis. Chin. J. Comp. Med. 2014, 23, 11–15. [Google Scholar]
- Hijmans, R.J.; Cameron, S.E.; Parra, J.L.; Jones, P.G.; Jarvis, A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. J. R. Meteorol. Soc. 2005, 25, 1965–1978. [Google Scholar] [CrossRef]
- Holland, M.M.; Parson, W. GeneMarker® HID: A reliable software tool for the analysis of forensic STR data. J. Forensic Sci. 2011, 56, 29–35. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Kalinowski, S.T. hp-rare 1.0: A computer program for performing rarefaction on measures of allelic richness. Mol. Ecol. Notes 2005, 5, 187–189. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [CrossRef]
- Earl, D.A.; Vonholdt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Jakobsson, M.; Rosenberg, N.A. CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 2007, 23, 1801–1806. [Google Scholar] [CrossRef] [Green Version]
- Rosenberg, N.A. DISTRUCT: A program for the graphical display of population structure. Mol. Ecol. Notes 2004, 4, 137–138. [Google Scholar] [CrossRef]
- Hughes, A.L.; Yeager, M. Natural selection at major histocompatibility complex loci of vertebrates. Annu. Rev. Genet. 1998, 32, 415. [Google Scholar] [CrossRef] [Green Version]
- Schad, J.; Sommer, S.; Ganzhorn, J.U. MHC variability of a small lemur in the littoral forest fragments of southeastern Madagascar. Conserv. Genet. 2004, 5, 299–309. [Google Scholar] [CrossRef]
- Sebastian, A.; Herdegen, M.; Migalska, M.; Radwan, J. amplisas: A web server for multilocus genotyping using next-generation amplicon sequencing data. Mol. Ecol. Resour. 2016, 16, 498–510. [Google Scholar] [CrossRef] [Green Version]
- Nei, M.; Gojobori, T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 1986, 3, 418–426. [Google Scholar]
- Gao, F.; Chen, C.; Arab, D.A.; Du, Z.; He, Y.; Ho, S.Y.W. EasyCodeML: A visual tool for analysis of selection using CodeML. Ecol. Evol. 2019, 9, 3891–3898. [Google Scholar] [CrossRef] [Green Version]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [Green Version]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weaver, S.; Shank, S.D.; Spielman, S.J.; Li, M.; Muse, S.V.; Kosakovsky Pond, S.L. Datamonkey 2.0: A modern web application for characterizing selective and other evolutionary processes. Mol. Biol. Evol. 2018, 35, 773–777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [Green Version]
- Huson, D.H. SplitsTree: Analyzing and visualizing evolutionary data. Bioinformatics 1998, 14, 68–73. [Google Scholar] [CrossRef] [Green Version]
- Hardy, O.J.; Vekemans, X. SPAGeDI: A versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol. Ecol. Notes 2002, 2, 618–620. [Google Scholar] [CrossRef] [Green Version]
- Dixon, P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Hijmans, R.J.; Van Etten, J.; Mattiuzzi, M.; Sumner, M.; Greenberg, J.A.; Lamigueiro, O.P.; Bevan, A.; Racine, E.B.; Shortridge, A. Raster package in R. Version. 2013. Available online: https://mirrors.sjtug.sjtu.edu.cn/cran/web/packages/raster/raster.pdf (accessed on 25 June 2023).
- Dolédec, S.; Chessel, D. Co-inertia analysis: An alternative method for studying species–environment relationships. Freshw. Biol. 1994, 31, 277–294. [Google Scholar] [CrossRef]
- Altizer, S.; Dobson, A.; Hosseini, P.; Hudson, P.; Pascual, M.; Rohani, P. Seasonality and the dynamics of infectious diseases. Ecol. Lett. 2006, 9, 467–484. [Google Scholar] [CrossRef] [Green Version]
- Froeschke, G.; Harf, R.; Sommer, S.; Matthee, S. Effects of precipitation on parasite burden along a natural climatic gradient in southern Africa—Implications for possible shifts in infestation patterns due to global changes. Oikos 2010, 119, 1029–1039. [Google Scholar] [CrossRef]
- Guernier, V.; Hochberg, M.E.; Guegan, J.F. Ecology drives the worldwide distribution of human diseases. PLoS Biol. 2004, 2, e141. [Google Scholar] [CrossRef] [PubMed]
- Johnson, P.; Haas, S.E. Why do parasites exhibit reverse latitudinal diversity gradients? Testing the roles of host diversity, habitat and climate. Glob. Ecol. Biogeogr. 2021, 30, 1810–1821. [Google Scholar] [CrossRef] [PubMed]
- Nunn, C.L.; Altizer, S.M.; Sechrest, W.; Cunningham, A.A. Latitudinal gradients of parasite species richness in primates. Divers. Distrib. 2005, 11, 249–256. [Google Scholar] [CrossRef]
- Samuel, M.D.; Poje, J.E.; Rocke, T.E.; Metzger, M.E. Potential Effects of Environmental Conditions on Prairie Dog Flea Development and Implications for Sylvatic Plague Epizootics. Ecohealth 2022, 19, 365–377. [Google Scholar] [CrossRef]
- Hammond, T.T.; Hendrickson, C.I.; Maxwell, T.L.; Petrosky, A.L.; Palme, R.; Pigage, J.C.; Pigage, H.K. Host biology and environmental variables differentially predict flea abundances for two rodent hosts in a plague-relevant system. Int. J. Parasitol. Parasites Wildl. 2019, 9, 174–183. [Google Scholar] [CrossRef]
- Pietrock, M.; Marcogliese, D.J. Free-living endohelminth stages: At the mercy of environmental conditions. Trends Parasitol. 2003, 19, 293–299. [Google Scholar] [CrossRef]
- Brooks, D.R.; Mclennan, D.A.; León-Règagnon, V.; Hoberg, E. Phylogeny, ecological fitting and lung flukes: Helping solve the problem of emerging infectious diseases. Rev. Mex. Biodivers. 2006, 77, 225–233. [Google Scholar]
- Yin, P.; Guo, X.; Jin, D.; Song, W.; Zhang, L.; Zhao, C.; Fan, R.; Zhang, Z.; Mao, K. Infestation and Seasonal Fluctuation of Gamasid Mites (Parasitiformes: Gamasida) on Indochinese Forest Rat, Rattus andamanensis (Rodentia: Muridae) in Southern Yunnan of China. Biology 2021, 10, 1297. [Google Scholar] [CrossRef] [PubMed]
- Makarova, O.L. A review of gamasid mites (Parasitiformes, Mesostigmata) dwelling in the taiga of the Pechoro-Ilychskii Nature Reserve (northern Cis-Ural Region) with analysis of their assemblages in spruce forests. Entomol. Rev. 2011, 91, 915–931. [Google Scholar] [CrossRef]
- Pham, H.V.; Dang, D.T.; Tran, M.N.; Nguyen, N.D.; Nguyen, T.V. Correlates of environmental factors and human plague: An ecological study in Vietnam. Int. J. Epidemiol. 2009, 38, 1634–1641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krasnov, B.R.; Khokhlova, I.S.; Fielden, L.J.; Burdelova, N.I. Time of survival under starvation in two flea species (Siphonaptera: Pulicidae) at different air temperatures and relative humidities. J. Vector Ecol. 2002, 27, 70–81. [Google Scholar]
- Loiseau, C.; Harrigan, R.J.; Robert, A.; Bowie, R.C.; Thomassen, H.A.; Smith, T.B.; Sehgal, R.N. Host and habitat specialization of avian malaria in Africa. Mol. Ecol. 2012, 21, 431–441. [Google Scholar] [CrossRef] [Green Version]
- Guo, T.; Xu, R. Study on flea communities of the rodent in Dongling Mountain in Beijing. Chin. J. Vector Biol. Control 2002, 13, 355–357. [Google Scholar]
- Yang, M.; Wei, J.; Li, P.; Wei, S.; Huang, Y.; Qin, Q. MHC polymorphism and disease resistance to Singapore grouper iridovirus (SGIV) in the orange-spotted grouper, Epinephelus coioides. Sci. Bull. 2016, 61, 693–699. [Google Scholar] [CrossRef] [Green Version]
- Schenekar, T.; Weiss, S. Selection and genetic drift in captive versus wild populations: An assessment of neutral and adaptive (MHC-linked) genetic variation in wild and hatchery brown trout (Salmo trutta) populations. Conserv. Genet. 2017, 18, 1011–1022. [Google Scholar] [CrossRef] [Green Version]
- Keller, L.F.; Waller, D.M. Inbreeding effects in wild populations. Trends Ecol. Evol. 2002, 17, 230–241. [Google Scholar] [CrossRef]
- Xie, X.H.; Dong, X.B.; Qin, Z.; Kong, F.H.; Lai-Xiang, X.U. Cloning and Sequence Analysis of MHC II Exon 2 of DQA Gene in Cricetulus Barabensis. J. Qufu Norm. Univ. Nat. Sci. 2009, 2, 98–103. [Google Scholar]
- Monzón-Argüello, C.; Garcia, D.; Gajardo, G.; Consuegra, S. Eco-immunology of fish invasions: The role of MHC variation. Immunogenetics 2014, 66, 393–402. [Google Scholar] [CrossRef] [Green Version]
- Winternitz, J.C.; Wares, J.P. Duplication and population dynamics shape historic patterns of selection and genetic variation at the major histocompatibility complex in rodents. Ecol. Evol. 2013, 3, 1552–1568. [Google Scholar] [CrossRef] [PubMed]
- Winternitz, J.C.; Wares, J.P.; Yabsley, M.J.; Altizer, S. Wild cyclic voles maintain high neutral and MHC diversity without strong evidence for parasite-mediated selection. Evol. Ecol. 2014, 28, 957–975. [Google Scholar] [CrossRef]
- Scherman, K.; Råberg, L.; Westerdahl, H. Positive Selection on MHC Class II DRB and DQB Genes in the Bank Vole (Myodes glareolus). J. Mol. Evol. 2014, 78, 293–305. [Google Scholar] [CrossRef]
- Oliver, M.K.; Piertney, S.B. Selection Maintains MHC Diversity through a Natural Population Bottleneck. Mol. Biol. Evol. 2012, 29, 1713–1720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milinski, M. The Major Histocompatibility Complex, Sexual Selection, and Mate Choice. Annu. Rev. Ecol. Evol. Syst. 2006, 37, 159–186. [Google Scholar] [CrossRef]
- Eizaguirre, C.; Lenz, T.L.; Kalbe, M.; Milinski, M. Rapid and adaptive evolution of MHC genes under parasite selection in experimental vertebrate populations. Nat. Commun. 2012, 3, 621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klitz, W.; Hedrick, P.; Louis, E.J. New reservoirs of HLA alleles: Pools of rare variants enhance immune defense. Trends Genet. 2012, 28, 480–486. [Google Scholar] [CrossRef]
- Hansen, M.M.; Skaala, O.; Jensen, L.F.; Bekkevold, D.; Mensberg, K. Gene flow, effective population size and selection at major histocompatibility complex genes: Brown trout in the Hardanger Fjord, Norway. Mol. Ecol. 2010, 16, 1413–1425. [Google Scholar] [CrossRef] [Green Version]
- Oliver, M.K.; Lambin, X.; Cornulier, T.; Piertney, S.B. Spatio-temporal variation in the strength and mode of selection acting on major histocompatibility complex diversity in water vole (Arvicola terrestris) metapopulations. Mol. Ecol. 2010, 18, 80–92. [Google Scholar] [CrossRef]
- Del Real-Monroy, M.; Ortega, J. Spatial distribution of microsatellite and MHC-DRB exon 2 gene variability in the Jamaican fruit bat (Artibeus jamaicensis) in Mexico. Mamm. Biol. 2017, 84, 1–11. [Google Scholar] [CrossRef]
- Spurgin, L.G.; Richardson, D.S. How pathogens drive genetic diversity: MHC, mechanisms and misunderstandings. Proc. R. Soc. B Biol. Sci. 2010, 277, 979–988. [Google Scholar] [CrossRef] [PubMed]
- Nadachowska-Brzyska, K.; Zieliński, P.; Radwan, J.; Babik, W. Interspecific hybridization increases MHC class II diversity in two sister species of newts. Mol. Ecol. 2011, 21, 887–906. [Google Scholar] [CrossRef] [PubMed]
- Rico, Y.; Morris-Pocock, J.; Zigouris, J.; Nocera, J.J.; Kyle, C.J. Lack of Spatial Immunogenetic Structure among Wolverine (Gulo gulo) Populations Suggestive of Broad Scale Balancing Selection. PLoS ONE 2015, 10, e140170. [Google Scholar] [CrossRef] [Green Version]
- Kamath, P.L.; Getz, W.M. Unraveling the effects of selection and demography on immune gene variation in free-ranging plains zebra (Equus quagga) populations. PLoS ONE 2012, 7, e50971. [Google Scholar] [CrossRef] [Green Version]
- Fraser, D.J.; Weir, L.K.; Bernatchez, L.; Hansen, M.M.; Taylor, E.B. Extent and scale of local adaptation in salmonid fishes: Review and meta-analysis. Heredity 2011, 106, 404–420. [Google Scholar] [CrossRef] [Green Version]
- Tack, A.J.M.; Horns, F.; Laine, A. The impact of spatial scale and habitat configuration on patterns of trait variation and local adaptation in a wild plant parasite. Evolution 2014, 68, 176–189. [Google Scholar] [CrossRef] [Green Version]
- O’Malley, K.G.; Banks, M.A. A latitudinal cline in the Chinook salmon (Oncorhynchus tshawytscha) Clock gene: Evidence for selection on PolyQ length variants. Proc. R. Soc. B-Biol. Sci. 2008, 275, 2813–2821. [Google Scholar] [CrossRef]
- Eckert, A.J.; Maloney, P.E.; Vogler, D.R.; Jensen, C.E.; Mix, A.D.; Neale, D.B. Local adaptation at fine spatial scales: An example from sugar pine (Pinus lambertiana, Pinaceae). Tree Genet. Genomes 2015, 11, 42. [Google Scholar] [CrossRef]
- Schradin, C.; Lindholm, A.K.; Johannesen, J.; Schoepf, I.; Yuen, C.H.; Konig, B.; Pillay, N. Social flexibility and social evolution in mammals: A case study of the African striped mouse (Rhabdomys pumilio). Mol. Ecol. 2012, 21, 541–553. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, Y.; Liu, F.; Shen, X.; Wang, Y.; Fan, M.; Peng, Y.; Wang, S.; Feng, Y.; Zhang, W.; et al. Genetic source tracking of human plague cases in Inner Mongolia-Beijing. PLoS Negl. Trop. Dis. 2021, 15, e9558. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, J.J. Fleas and the epidemiology of plague in Inner Mongolia, China. Chin. J. Vector Biol. Control 2011, 6, 576–578. [Google Scholar]



| N2 | N3 | W2 | W4 | |
|---|---|---|---|---|
| Gamasid Mite Load | 0.59 | 1.29 | 1.32 | 2.89 |
| Flea Load | 0.47 | 0.42 | 1.19 | 0.92 |
| Gamasid Mite Richness | 0.31 | 0.45 | 0.47 | 1.09 |
| Flea Richness | 0.35 | 0.29 | 0.40 | 0.50 |
| Gamasid Mite Prevalence | 24% | 38% | 37% | 55% |
| Flea Prevalence | 47% | 21% | 34% | 35% |
| Annual Mean Temperature | 1.78 | 2.14 | 7.18 | 4.98 |
| Annual Precipitation | 242 | 249 | 440 | 201 |
| Pop | N | Na | Ne | I | Ho | He | uHe | F | Ar | HWE |
|---|---|---|---|---|---|---|---|---|---|---|
| N2 | 17 | 9.286 | 6.094 | 1.975 | 0.496 | 0.828 | 0.862 | 0.390 | 4.84 | 0.167 ns |
| N3 | 24 | 10.571 | 7.708 | 2.116 | 0.477 | 0.850 | 0.874 | 0.428 | 5.02 | 0.092 ns |
| W2 | 62 | 15.143 | 9.176 | 2.362 | 0.552 | 0.873 | 0.883 | 0.358 | 5.15 | 0.104 ns |
| W4 | 111 | 21.429 | 12.146 | 2.594 | 0.600 | 0.890 | 0.894 | 0.317 | 5.34 | 0.000 * |
| Total | 214 | 14.107 | 8.781 | 2.262 | 0.531 | 0.860 | 0.878 | 0.373 | 5.08 | / |
| N2 | N3 | W2 | W4 | |
|---|---|---|---|---|
| 0.006 | 0.005 | 0.043 | N2 | |
| 0.014 | 0.004 | 0.020 | N3 | |
| 0.020 | 0.003 | 0.022 | W2 | |
| 0.029 | 0.018 | 0.016 | W4 |
| Models | 5 | 9 | 16 | 17 | 23 | 30 | 36 | 40 | 46 | 49 | 53 | 56 | 57 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FEL | √ | √ | |||||||||||
| MEME | √ | √ | √ | √ | √ | √ | √ | √ | |||||
| FUBAR | √ | √ | √ | √ | √ | √ | |||||||
| SLAC | √ | √ | √ | √ | |||||||||
| M8 vs. M7 | √ | √ | √ |
| Pop | h | S | Hd | K | π | An |
|---|---|---|---|---|---|---|
| N2 | 47 | 66 | 0.972 | 18.49 | 0.108 | 10.80 |
| N3 | 51 | 67 | 0.971 | 18.12 | 0.106 | 10.45 |
| W2 | 66 | 76 | 0.971 | 18.01 | 0.105 | 11.91 |
| W4 | 70 | 77 | 0.970 | 17.86 | 0.104 | 12.52 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, P.; Li, G.; Zhao, N.; Song, X.; Wang, J.; Shi, X.; Wang, B.; Zhang, L.; Dong, L.; Li, Q.; et al. Neutral Forces and Balancing Selection Interplay to Shape the Major Histocompatibility Complex Spatial Patterns in the Striped Hamster in Inner Mongolia: Suggestive of Broad-Scale Local Adaptation. Genes 2023, 14, 1500. https://doi.org/10.3390/genes14071500
Liu P, Li G, Zhao N, Song X, Wang J, Shi X, Wang B, Zhang L, Dong L, Li Q, et al. Neutral Forces and Balancing Selection Interplay to Shape the Major Histocompatibility Complex Spatial Patterns in the Striped Hamster in Inner Mongolia: Suggestive of Broad-Scale Local Adaptation. Genes. 2023; 14(7):1500. https://doi.org/10.3390/genes14071500
Chicago/Turabian StyleLiu, Pengbo, Guichang Li, Ning Zhao, Xiuping Song, Jun Wang, Xinfei Shi, Bin Wang, Lu Zhang, Li Dong, Qingduo Li, and et al. 2023. "Neutral Forces and Balancing Selection Interplay to Shape the Major Histocompatibility Complex Spatial Patterns in the Striped Hamster in Inner Mongolia: Suggestive of Broad-Scale Local Adaptation" Genes 14, no. 7: 1500. https://doi.org/10.3390/genes14071500
APA StyleLiu, P., Li, G., Zhao, N., Song, X., Wang, J., Shi, X., Wang, B., Zhang, L., Dong, L., Li, Q., Liu, Q., & Lu, L. (2023). Neutral Forces and Balancing Selection Interplay to Shape the Major Histocompatibility Complex Spatial Patterns in the Striped Hamster in Inner Mongolia: Suggestive of Broad-Scale Local Adaptation. Genes, 14(7), 1500. https://doi.org/10.3390/genes14071500





