Impaired Repopulating Ability of Uhrf2−/− Hematopoietic Progenitor Cells in Mice
Abstract
:1. Introduction
2. Materials and Methods
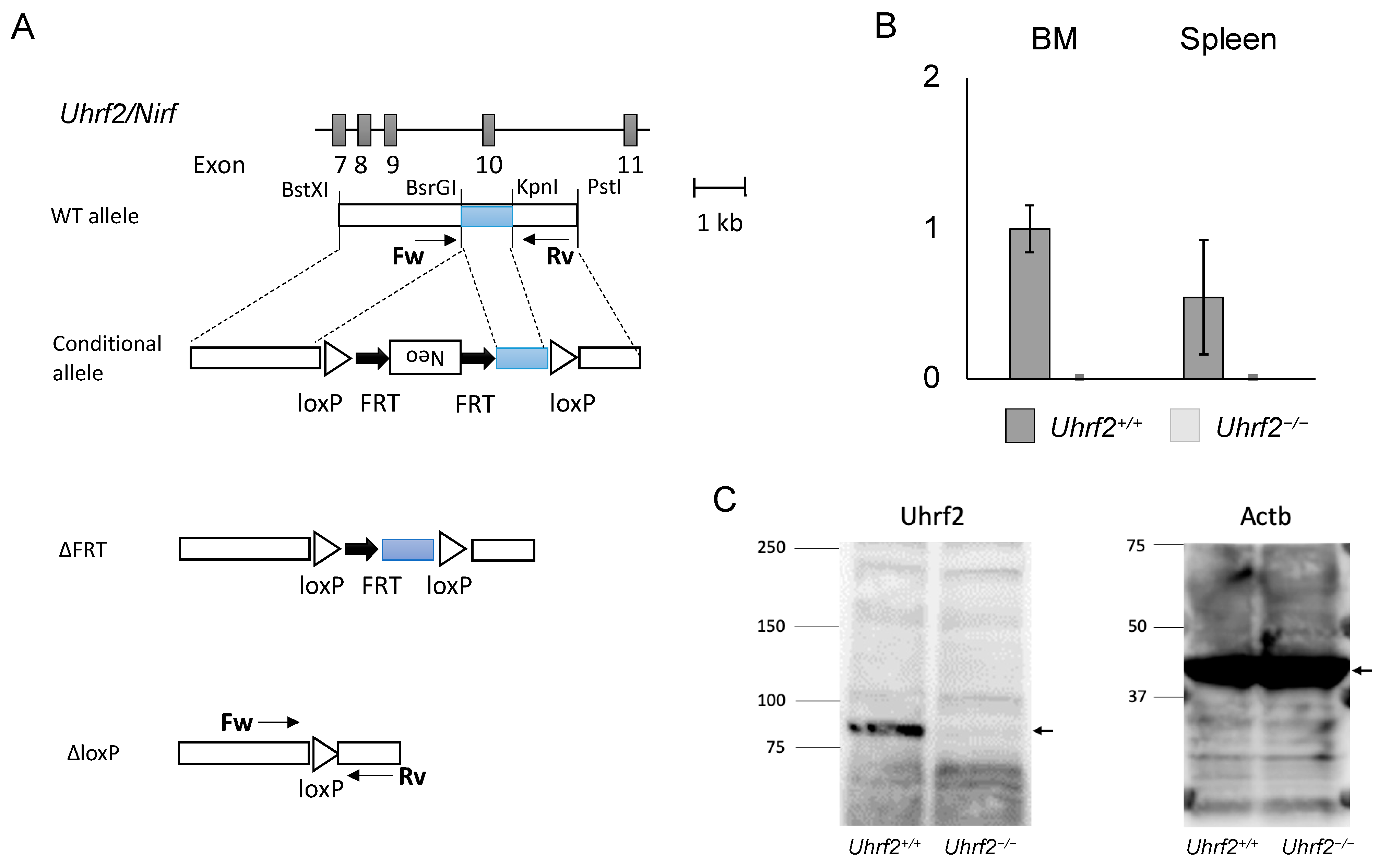
2.1. Gene Targeting and Mice
2.2. Cell Preparations and Counts
2.3. Quantitative RT-PCR
2.4. Histopathology
2.5. Flow Cytometry and Cell Sorting
2.6. Competitive Repopulation Assay with Bone Marrow Transplantation
2.7. RNA Sequencing (RNAseq)
2.8. Cleavage under Targets and Tagmentation Assay (CUT&Tag)
2.9. Statistical Analysis
3. Results
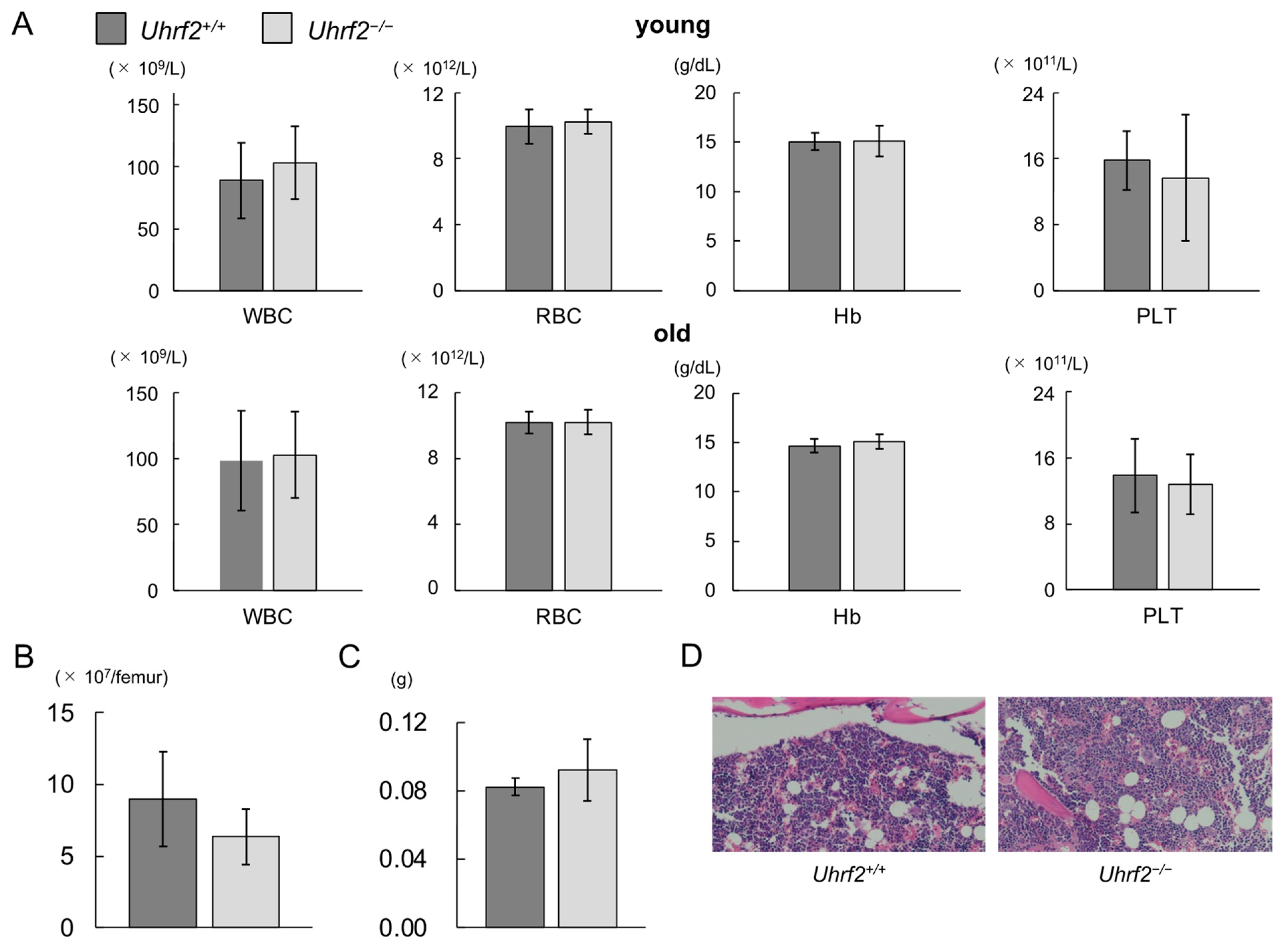
3.1. Uhrf2 KO Mice and Hematologic Findings
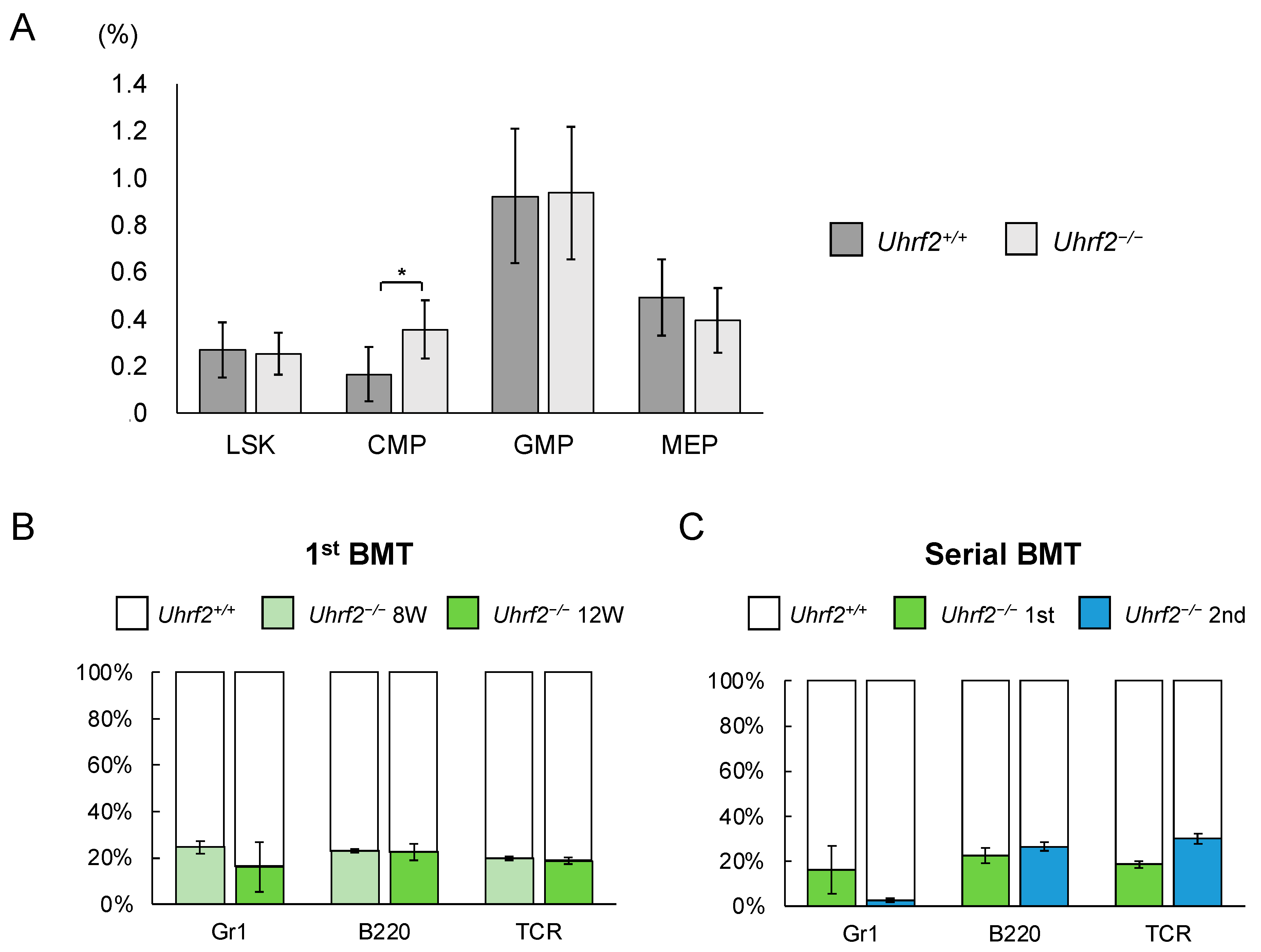
3.2. Impairment of HSPC Repopulating Capacity in Uhrf2–/– Mice
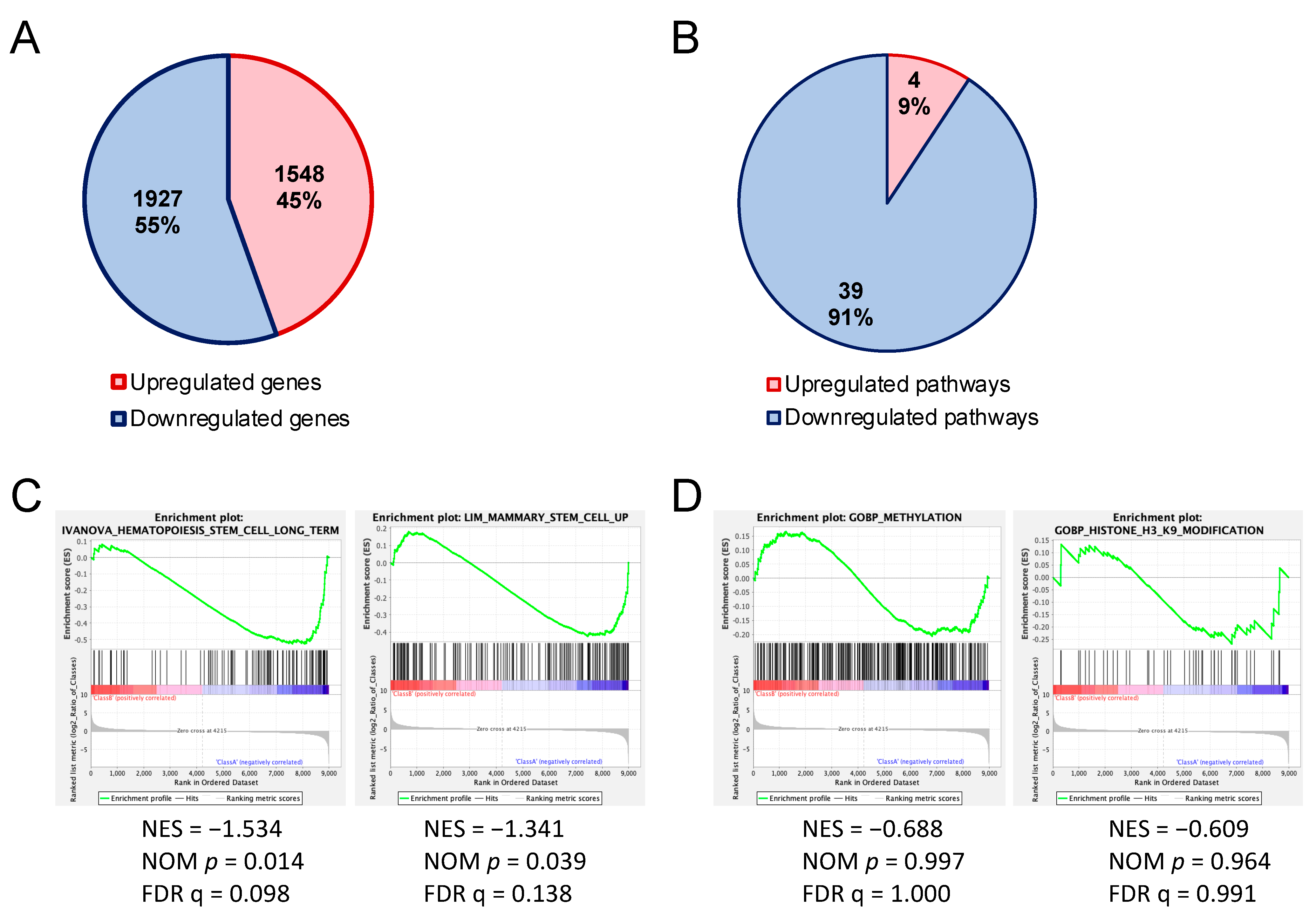
3.3. Gene Expression in HSPCs of Uhrf2–/– Mice
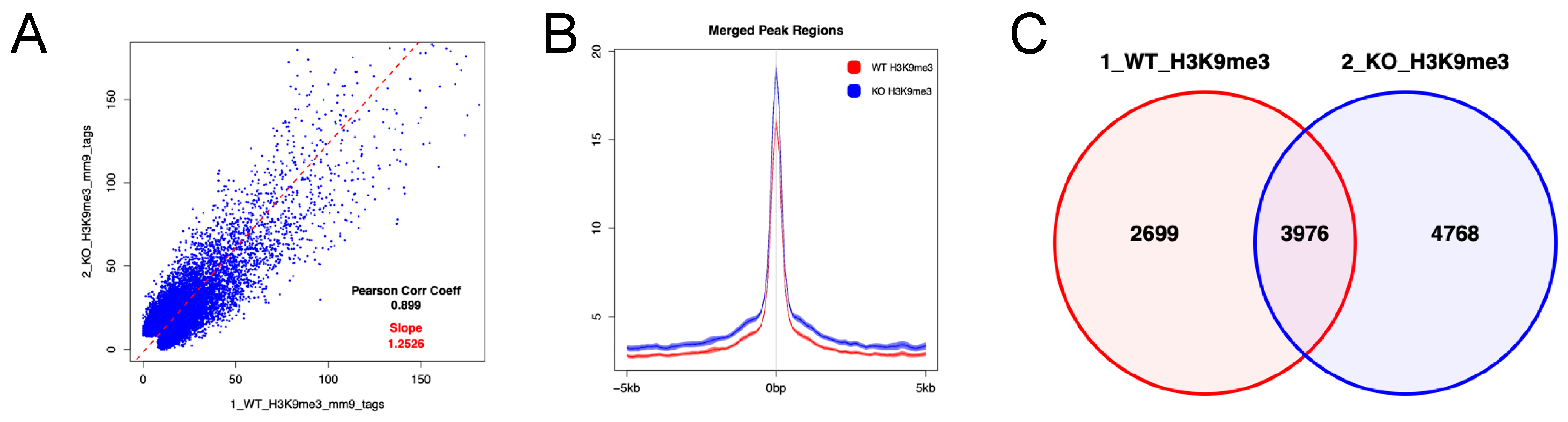
3.4. Enrichment of H3K9me3 in HSPCs of Uhrf2−/− Mice
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mori, T.; Ikeda, D.D.; Yamaguchi, Y.; Unoki, M. NIRF/UHRF2 Occupies a Central Position in the Cell Cycle Network and Allows Coupling with the Epigenetic Landscape. FEBS Lett. 2012, 586, 1570–1583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Unoki, M.; Sasaki, H. The UHRF Protein Family in Epigenetics, Development, and Carcinogenesis. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2022, 98, 401–415. [Google Scholar] [CrossRef]
- Reardon, E.S.; Shukla, V.; Xi, S.; Gara, S.K.; Liu, Y.; Straughan, D.; Zhang, M.; Hong, J.A.; Payabyab, E.C.; Kumari, A.; et al. UHRF1 Is a Novel Druggable Epigenetic Target in Malignant Pleural Mesothelioma. J. Thorac. Oncol. 2021, 16, 89–103. [Google Scholar] [CrossRef]
- Kofunato, Y.; Kumamoto, K.; Saitou, K.; Hayase, S.; Okayama, H.; Miyamoto, K.; Sato, Y.; Katakura, K.; Nakamura, I.; Ohki, S.; et al. UHRF1 Expression Is Upregulated and Associated with Cellular Proliferation in Colorectal Cancer. Oncol. Rep. 2012, 28, 1997–2002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, H.; Bhoopatiraju, S.; Wang, H.; Schmitz, N.P.; Wang, X.; Freeman, M.J.; Forster, C.L.; Verneris, M.R.; Linden, M.A.; Hallstrom, T.C. Loss of UHRF2 Expression Is Associated with Human Neoplasia, Promoter Hypermethylation, Decreased 5-Hydroxymethylcytosine, and High Proliferative Activity. Oncotarget 2016, 7, 76047–76061. [Google Scholar] [CrossRef] [Green Version]
- Jin, C.; Xiong, D.; Li, H.R.; Jiang, J.H.; Qi, J.C.; Ding, J.Y. Loss of UHRF2 Is Associated with Non-Small Cell Lung Carcinoma Progression. J. Cancer 2018, 9, 2994–3005. [Google Scholar] [CrossRef]
- Iguchi, T.; Ueda, M.; Masuda, T.; Nambara, S.; Kidogami, S.; Komatsu, H.; Sato, K.; Tobo, T.; Ogawa, Y.; Hu, Q.; et al. Identification of UHRF2 as a Negative Regulator of Epithelial-Mesenchymal Transition and Its Clinical Significance in Esophageal Squamous Cell Carcinoma. Oncology 2018, 95, 179–187. [Google Scholar] [CrossRef]
- Hu, C.-M.; Peng, J.; Lv, L.; Wang, X.-H.; Huo, J.-R.; Liu, D.-L. MiR-196a Promotes the Proliferation and Migration of Esophageal Cancer via the UHRF2/TET2 Axis. Mol. Cell. Biochem. 2022, 477, 537–547. [Google Scholar] [CrossRef]
- Amiri, V.; Mohammadi, M.H.; Rafiee, M.; Ghezelbash, B.; Salari, S.; Allahbakhshian Farsani, M. Transcription Analysis of a Histones Modifiers Panel Coupled with Critical Tumor Suppressor Genes Displayed Frequent Changes in Patients with AML. Curr. Res. Transl. Med. 2021, 69, 103311. [Google Scholar] [CrossRef]
- Li, L.; Duan, Q.; Zeng, Z.; Zhao, J.; Lu, J.; Sun, J.; Zhang, J.; Siwko, S.; Wong, J.; Shi, T.; et al. UHRF2 Promotes Intestinal Tumorigenesis through Stabilization of TCF4 Mediated Wnt/β-Catenin Signaling. Int. J. Cancer 2020, 147, 2239–2252. [Google Scholar] [CrossRef]
- Sharif, J.; Muto, M.; Takebayashi, S.I.; Suetake, I.; Iwamatsu, A.; Endo, T.A.; Shinga, J.; Mizutani-Koseki, Y.; Toyoda, T.; Okamura, K.; et al. The SRA Protein Np95 Mediates Epigenetic Inheritance by Recruiting Dnmt1 to Methylated DNA. Nature 2007, 450, 908–912. [Google Scholar] [CrossRef] [PubMed]
- Ueda, K.; Ikeda, K.; Ikezoe, T.; Harada-Shirado, K.; Ogawa, K.; Hashimoto, Y.; Sano, T.; Ohkawara, H.; Kimura, S.; Shichishima-Nakamura, A.; et al. Hmga2 Collaborates with JAK2V617F in the Development of Myeloproliferative Neoplasms. Blood Adv. 2017, 1, 1001–1015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minakawa, K.; Yokokawa, T.; Ueda, K.; Nakajima, O.; Misaka, T.; Kimishima, Y.; Wada, K.; Tomita, Y.; Miura, S.; Sato, Y.; et al. Myeloproliferative Neoplasm-Driving Calr Frameshift Promotes the Development of Pulmonary Hypertension in Mice. J. Hematol. Oncol. 2021, 14, 52. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, K.; Mason, P.J.; Bessler, M. 3′UTR-Truncated Hmga2 CDNA Causes MPN-like Hematopoiesis by Conferring a Clonal Growth Advantage at the Level of HSC in Mice. Blood 2011, 117, 5860–5869. [Google Scholar] [CrossRef] [Green Version]
- Kimishima, Y.; Misaka, T.; Yokokawa, T.; Wada, K.; Ueda, K.; Sugimoto, K.; Minakawa, K.; Nakazato, K.; Ishida, T.; Oshima, M.; et al. Clonal Hematopoiesis with JAK2V617F Promotes Pulmonary Hypertension with ALK1 Upregulation in Lung Neutrophils. Nat. Commun. 2021, 12, 6177. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Ginnard, S.M.; Winkler, A.E.; Mellado Fritz, C.; Bluhm, T.; Kemmer, R.; Gilliam, M.; Butkevich, N.; Abdrabbo, S.; Bricker, K.; Feiler, J.; et al. Molecular Investigation of the Tandem Tudor Domain and Plant Homeodomain Histone Binding Domains of the Epigenetic Regulator UHRF2. Proteins Struct. Funct. Bioinform. 2022, 90, 835–847. [Google Scholar] [CrossRef]
- Pichler, G.; Wolf, P.; Schmidt, C.S.; Meilinger, D.; Schneider, K.; Frauer, C.; Fellinger, K.; Rottach, A.; Leonhardt, H. Cooperative DNA and Histone Binding by Uhrf2 Links the Two Major Repressive Epigenetic Pathways. J. Cell. Biochem. 2011, 112, 2585–2593. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Gao, Q.; Li, P.; Liu, X.; Jia, Y.; Wu, W.; Li, J.; Dong, S.; Koseki, H.; Wong, J. S Phase-Dependent Interaction with DNMT1 Dictates the Role of UHRF1 but Not UHRF2 in DNA Methylation Maintenance. Cell Res. 2011, 21, 1723–1739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Zhang, B.; Meng, X.; Korn, M.J.; Parent, J.M.; Lu, L.Y.; Yu, X. UHRF2 Regulates Local 5-Methylcytosine and Suppresses Spontaneous Seizures. Epigenetics 2017, 12, 551–560. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Zhang, Q.; Duan, X.; York, P.; Chen, G.D.; Yin, P.; Zhu, H.; Xu, M.; Chen, P.; Wu, Q.; et al. The 5-Hydroxymethylcytosine (5hmC) Reader UHRF2 Is Required for Normal Levels of 5hmC in Mouse Adult Brain and Spatial Learning and Memory. J. Biol. Chem. 2017, 292, 4533–4543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Sarver, A.L.; Han, Q.; Seiler, C.L.; Xie, C.; Lu, H.; Forster, C.L.; Tretyakova, N.Y.; Hallstrom, T.C. UHRF2 Regulates Cell Cycle, Epigenetics and Gene Expression to Control the Timing of Retinal Progenitor and Ganglion Cell Differentiation. Development 2022, 149, dev195644. [Google Scholar] [CrossRef]
- Zhao, J.; Chen, X.; Song, G.; Zhang, J.; Liu, H.; Liu, X. Uhrf1 Controls the Self-Renewal versus Differentiation of Hematopoietic Stem Cells by Epigenetically Regulating the Cell-Division Modes. Proc. Natl. Acad. Sci. USA 2017, 114, E142–E151. [Google Scholar] [CrossRef]
- Shooshtarizadeh, P.; Helness, A.; Vadnais, C.; Brouwer, N.; Beauchemin, H.; Chen, R.; Bagci, H.; Staal, F.J.T.; Coté, J.F.; Möröy, T. Gfi1b Regulates the Level of Wnt/β-Catenin Signaling in Hematopoietic Stem Cells and Megakaryocytes. Nat. Commun. 2019, 10, 1270. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Patnana, P.K.; Xie, X.; Frank, D.; Nimmagadda, S.C.; Su, M.; Zhang, D.; Koenig, T.; Rosenbauer, F.; Liebmann, M.; et al. GFI1B Acts as a Metabolic Regulator in Hematopoiesis and Acute Myeloid Leukemia. Leukemia 2022, 36, 2196–2207. [Google Scholar] [CrossRef]
- Sugiyama, T.; Kohara, H.; Noda, M.; Nagasawa, T. Maintenance of the Hematopoietic Stem Cell Pool by CXCL12-CXCR4 Chemokine Signaling in Bone Marrow Stromal Cell Niches. Immunity 2006, 25, 977–988. [Google Scholar] [CrossRef] [Green Version]
- Takubo, K.; Goda, N.; Yamada, W.; Iriuchishima, H.; Ikeda, E.; Kubota, Y.; Shima, H.; Johnson, R.S.; Hirao, A.; Suematsu, M.; et al. Regulation of the HIF-1α Level Is Essential for Hematopoietic Stem Cells. Cell Stem Cell 2010, 7, 391–402. [Google Scholar] [CrossRef] [Green Version]
- Tosato, G. Ephrin Ligands and Eph Receptors Contribution to Hematopoiesis. Cell. Mol. Life Sci. 2017, 74, 3377–3394. [Google Scholar] [CrossRef] [PubMed]
- Hanaki, S.; Habara, M.; Shimada, M. UV-Induced Activation of ATR Is Mediated by UHRF2. Genes Cells 2021, 26, 447–454. [Google Scholar] [CrossRef]
- Kohli, L.; Passegué, E. Surviving Change: The Metabolic Journey of Hematopoietic Stem Cells. Trends Cell Biol. 2014, 24, 479–487. [Google Scholar] [CrossRef] [Green Version]
- Ito, K.; Bonora, M.; Ito, K. Metabolism as Master of Hematopoietic Stem Cell Fate. Int. J. Hematol. 2019, 109, 18–27. [Google Scholar] [CrossRef] [Green Version]
- Oguro, H.; Ding, L.; Morrison, S.J. SLAM Family Markers Resolve Functionally Distinct Subpopulations of Hematopoietic Stem Cells and Multipotent Progenitors. Cell Stem Cell 2013, 13, 102–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sano, T.; Ueda, K.; Minakawa, K.; Mori, T.; Hashimoto, Y.; Koseki, H.; Takeishi, Y.; Ikeda, K.; Ikezoe, T. Impaired Repopulating Ability of Uhrf2−/− Hematopoietic Progenitor Cells in Mice. Genes 2023, 14, 1531. https://doi.org/10.3390/genes14081531
Sano T, Ueda K, Minakawa K, Mori T, Hashimoto Y, Koseki H, Takeishi Y, Ikeda K, Ikezoe T. Impaired Repopulating Ability of Uhrf2−/− Hematopoietic Progenitor Cells in Mice. Genes. 2023; 14(8):1531. https://doi.org/10.3390/genes14081531
Chicago/Turabian StyleSano, Takahiro, Koki Ueda, Keiji Minakawa, Tsutomu Mori, Yuko Hashimoto, Haruhiko Koseki, Yasuchika Takeishi, Kazuhiko Ikeda, and Takayuki Ikezoe. 2023. "Impaired Repopulating Ability of Uhrf2−/− Hematopoietic Progenitor Cells in Mice" Genes 14, no. 8: 1531. https://doi.org/10.3390/genes14081531





