Abundance and Diversification of Repetitive Elements in Decapoda Genomes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Genomic Datasets
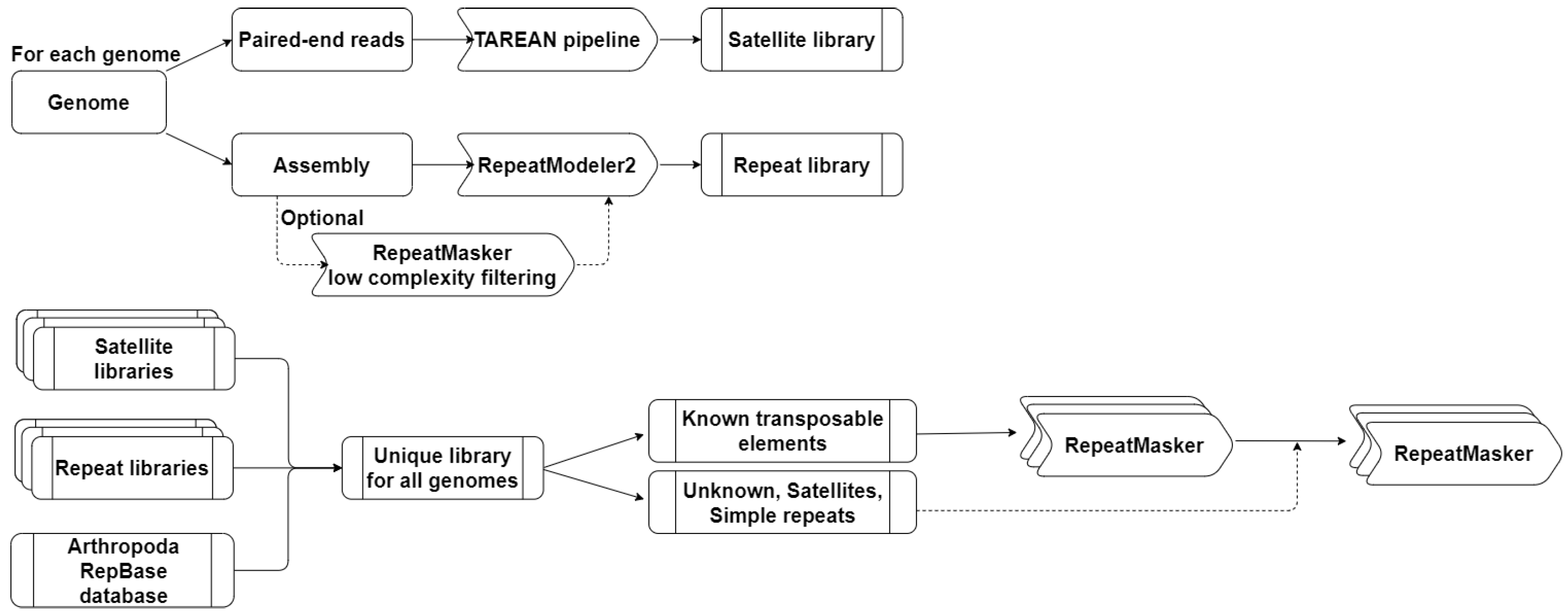
2.2. Identification and Annotation of Repetitive Elements
2.2.1. Identification of Satellite DNA Families
2.2.2. Construction of a Common Library of Repetitive Elements
2.2.3. Identification of Repetitive Elements
2.2.4. Statistical Analysis
3. Results and Discussion
3.1. Construction of Repetitive Elements Reference
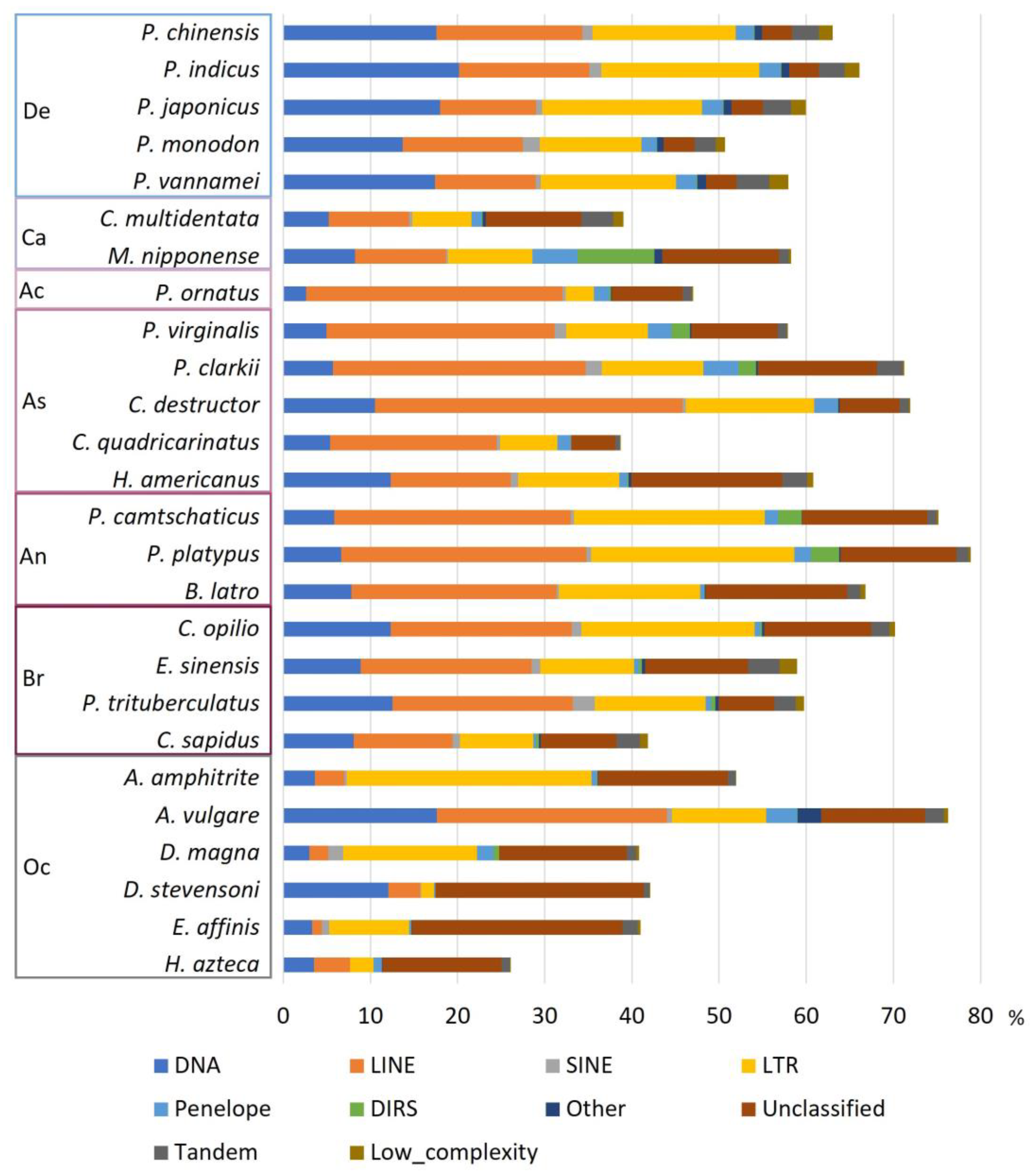
3.2. Annotation of Repetitive Elements in Decapoda Genomes
3.3. Proportion of Repetitive Elements in Decapoda Genomes
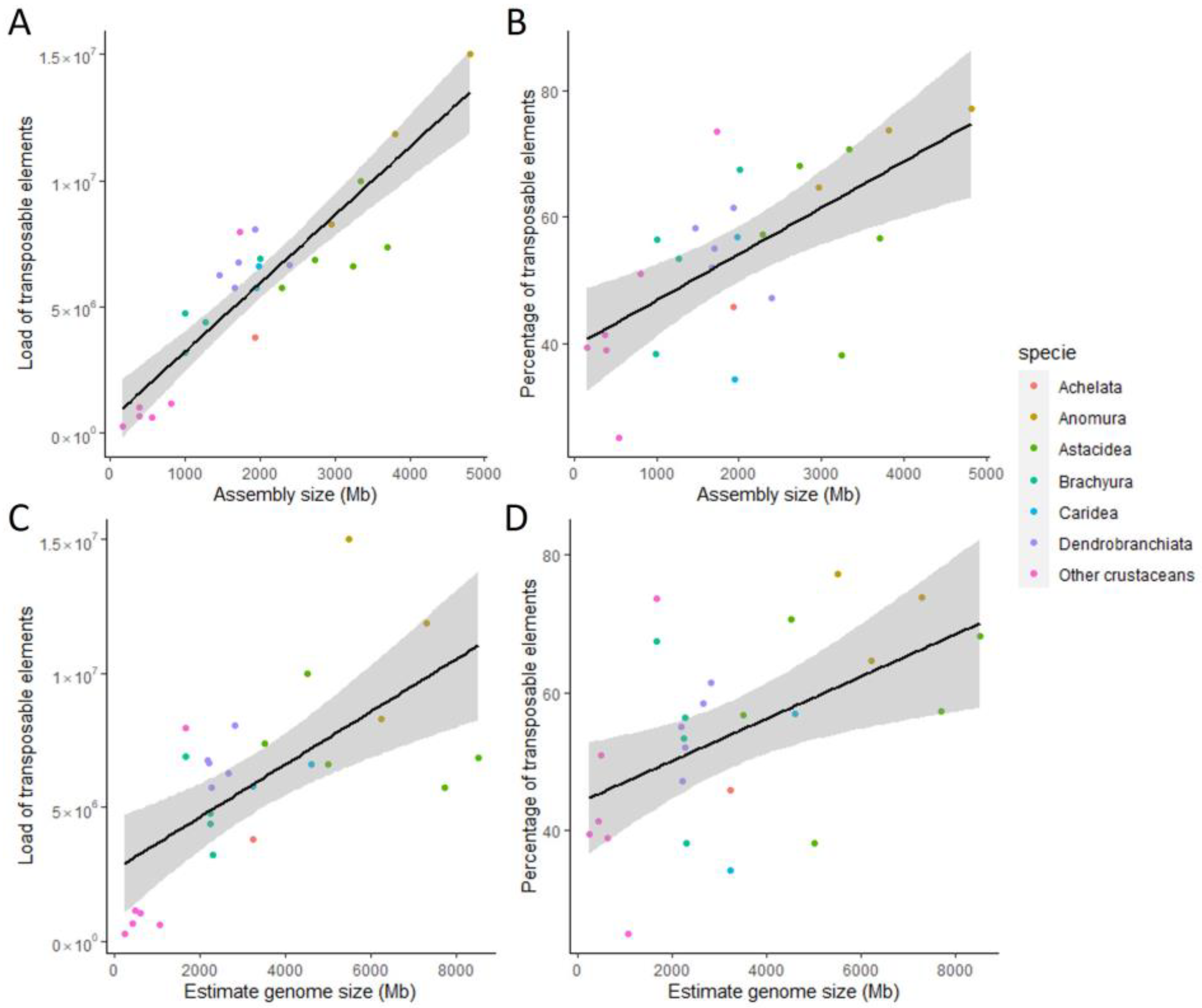
3.4. Correlation between Genome Size and Repetitive Elements
3.5. Frequency of satDNA Families Occurrence
3.6. Diversity of Repetitive Elements
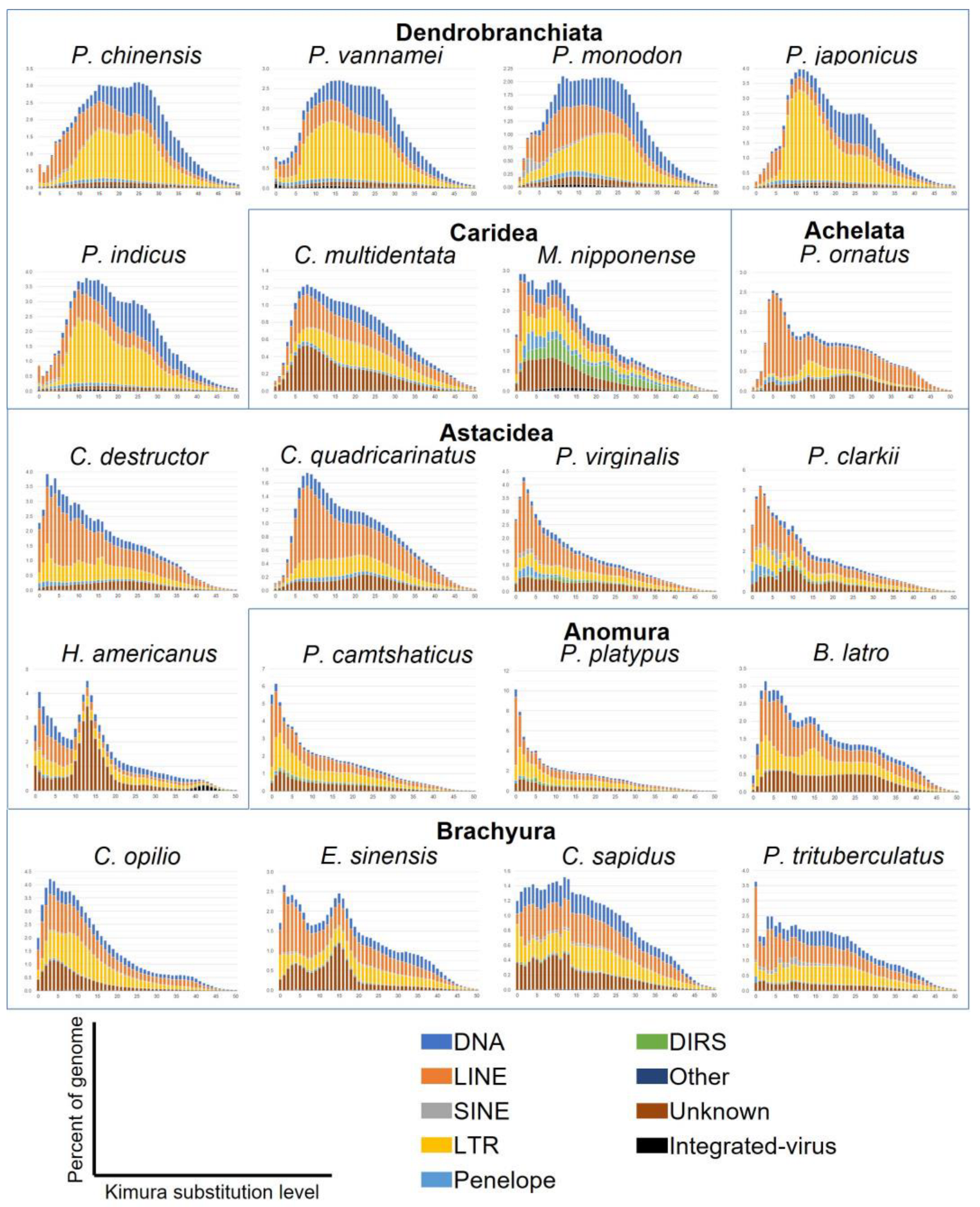
3.7. Sequence Divergence Distribution of Transposable Elements
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- De Grave, S.; Pentcheff, N.D.; Ahyong, S.T.; Chan, T.-Y.; Crandall, K.A.; Dworschak, P.C.; Felder, D.L.; Feldmann, R.M.; Fransen, C.H.; Goulding, L.Y.; et al. A Classification of Living and Fossil Genera of Decapod Crustaceans. Raffles Bull. Zool. 2009, 21, 1–109. [Google Scholar]
- Reynolds, J.; Souty-Grosset, C.; Richardson, A. Ecological Roles of Crayfish in Freshwater and Terrestrial Habitats. Freshw. Crayfish 2013, 19, 197–218. [Google Scholar]
- Souty-Grosset, C.; Holdich, D.D.M.; Noël, P.Y.; Reynolds, J.; Haffner, P. Atlas of Crayfish in Europe; Muséum national d’Histoire naturelle: Paris, France, 2006; Volume 187. [Google Scholar]
- Wolfe, J.M.; Breinholt, J.W.; Crandall, K.A.; Lemmon, A.R.; Lemmon, E.M.; Timm, L.E.; Siddall, M.E.; Bracken-Grissom, H.D. A Phylogenomic Framework, Evolutionary Timeline and Genomic Resources for Comparative Studies of Decapod Crustaceans. Proc. R. Soc. B Biol. Sci. 2019, 286, 20190079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boštjančić, L.L.; Bonassin, L.; Anušić, L.; Lovrenčić, L.; Besendorfer, V.; Maguire, I.; Grandjean, F.; Austin, C.M.; Greve, C.; Hamadou, A.B.; et al. The Pontastacus Leptodactylus (Astacidae) Repeatome Provides Insight into Genome Evolution and Reveals Remarkable Diversity of Satellite DNA. Front. Genet. 2021, 11, 611745. [Google Scholar] [CrossRef]
- Lécher, P.; Defaye, D.; Noel, P. Chromosomes and Nuclear DNA of Crustacea. Invertebr. Reprod. Dev.t 1995, 27, 85–114. [Google Scholar] [CrossRef]
- González-Tizón, A.M.; Rojo, V.; Menini, E.; Torrecilla, Z.; Martínez-Lage, A. Karyological Analysis of the Shrimp Palaemon Serratus (Decapoda: Palaemonidae). J. Crustac. Biol. 2013, 33, 843–848. [Google Scholar] [CrossRef] [Green Version]
- Niiyama, H. On the Unprecedentedly Large Number of Chromosomes of the Crayfish, Astacus Trowbridgii Stimpson. Annot. Zool. Japon. 1962, 35, 229–233. [Google Scholar]
- Crandall, K.A.; De Grave, S. An Updated Classification of the Freshwater Crayfishes (Decapoda: Astacidea) of the World, with a Complete Species List. J. Crustac. Biol. 2017, 37, 615–653. [Google Scholar] [CrossRef] [Green Version]
- Gregory, T.R. Chapter 1—Genome Size Evolution in Animals. In The Evolution of the Genome; Gregory, T.R., Ed.; Academic Press: Burlington, NJ, USA, 2005; pp. 3–87. [Google Scholar]
- Tørresen, O.K.; Star, B.; Mier, P.; Andrade-Navarro, M.A.; Bateman, A.; Jarnot, P.; Gruca, A.; Grynberg, M.; Kajava, A.V.; Promponas, V.J.; et al. Tandem repeats lead to sequence assembly errors and impose multi-level challenges for genome and protein databases. Nucleic Acids Res. 2019, 47, 10994–11006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Treangen, T.J.; Salzberg, S.L. Repetitive DNA and next-generation sequencing: Computational challenges and solutions. Nat. Rev. Genet. 2012, 13, 36–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pop, M. Genome assembly reborn: Recent computational challenges. Brief. Bioinform. 2009, 10, 354–366. [Google Scholar] [CrossRef] [Green Version]
- Shapiro, J.A.; von Sternberg, R. Why repetitive DNA is essential to genome function. Biol. Rev. 2005, 80, 227–250. [Google Scholar] [CrossRef] [Green Version]
- Jurka, J.; Kapitonov, V.V.; Kohany, O.; Jurka, M.V. Repetitive Sequences in Complex Genomes: Structure and Evolution. Annu. Rev. Genom. Hum. Genet. 2007, 8, 241–259. [Google Scholar] [CrossRef] [Green Version]
- Garrido-Ramos, M.A. Satellite DNA: An Evolving Topic. Genes 2017, 8, 230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macas, J.; Neumann, P.; Navrátilová, A. Repetitive DNA in the pea (Pisum sativum L.) genome: Comprehensive characterization using 454 sequencing and comparison to soybean and Medicago truncatula. BMC Genom. 2007, 8, 427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruiz-Ruano, F.J.; López-León, M.D.; Cabrero, J.; Camacho, J.P.M. High-throughput analysis of the satellitome illuminates satellite DNA evolution. Sci. Rep. 2016, 6, 28333. [Google Scholar] [CrossRef] [Green Version]
- Mravinac, B.; Plohl, M.; Ugarković, Ð. Preservation and High Sequence Conservation of Satellite DNAs Suggest Functional Constraints. J. Mol. Evol. 2005, 61, 542–550. [Google Scholar] [CrossRef]
- Miga, K.H. Completing the human genome: The progress and challenge of satellite DNA assembly. Chromosome Res. 2015, 23, 421–426. [Google Scholar] [CrossRef]
- Plohl, M.; Luchetti, A.; Meštrović, N.; Mantovani, B. Satellite DNAs between selfishness and functionality: Structure, genomics and evolution of tandem repeats in centromeric (hetero)chromatin. Gene 2008, 409, 72–82. [Google Scholar] [CrossRef]
- Plohl, M.; MešTrović, N.; Mravinac, B. Satellite DNA Evolution. Repetitive DNA 2012, 7, 126–152. [Google Scholar]
- Pezer, Ž.; Brajković, J.; Feliciello, I.; Ugarković, Đ. Satellite DNA-Mediated Effects on Genome Regulation. Genome Dyn. 2012, 7, 153–169. [Google Scholar]
- Biscotti, M.A.; Canapa, A.; Forconi, M.; Olmo, E.; Barucca, M. Transcription of tandemly repetitive DNA: Functional roles. Chromosome Res. 2015, 23, 463–477. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.Y.; Biesiot, P.M.; Skinner, D.M. Toward an Understanding of Satellite DNA Function in Crustacea. Integr. Comp. Biol. 1999, 39, 471–486. [Google Scholar] [CrossRef] [Green Version]
- Bourque, G.; Burns, K.H.; Gehring, M.; Gorbunova, V.; Seluanov, A.; Hammell, M.; Imbeault, M.; Izsvák, Z.; Levin, H.L.; Macfarlan, T.S.; et al. Ten things you should know about transposable elements. Genome Biol. 2018, 19, 199. [Google Scholar] [CrossRef]
- Bennetzen, J.L.; Wang, H. The Contributions of Transposable Elements to the Structure, Function, and Evolution of Plant Genomes. Annu. Rev. Plant Biol. 2014, 65, 505–530. [Google Scholar] [CrossRef]
- Deininger, P.L.; Moran, J.V.; Batzer, M.A.; Kazazian, H.H. Mobile elements and mammalian genome evolution. Curr. Opin. Genet. Dev. 2003, 13, 651–658. [Google Scholar] [CrossRef]
- Craig, N.L.; Lambowitz, A.; Gragie, R.; Gellert, M. Mobile DNA II; ASM Press: Washington, DC, USA, 2002. [Google Scholar]
- Kim, Y.-J.; Lee, J.; Han, K. Transposable Elements: No More “Junk DNA”. Genom. Inform. 2012, 10, 226–233. [Google Scholar] [CrossRef]
- Barrón, M.G.; Fiston-Lavier, A.-S.; Petrov, D.A.; González, J. Population Genomics of Transposable Elements in Drosophila. Annu. Rev. Genet. 2014, 48, 561–581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burns, K.H.; Boeke, J.D. Human Transposon Tectonics. Cell 2012, 149, 740–752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lanciano, S.; Mirouze, M. Transposable elements: All mobile, all different, some stress responsive, some adaptive? Curr. Opin. Genet. Dev. 2018, 49, 106–114. [Google Scholar] [CrossRef]
- Slotkin, R.K.; Martienssen, R. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 2007, 8, 272–285. [Google Scholar] [CrossRef]
- Wicker, T.; Sabot, F.; Hua-Van, A.; Bennetzen, J.L.; Capy, P.; Chalhoub, B.; Flavell, A.; Leroy, P.; Morgante, M.; Panaud, O.; et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007, 8, 973–982. [Google Scholar] [CrossRef] [PubMed]
- Di Stefano, L. All Quiet on the TE Front? The Role of Chromatin in Transposable Element Silencing. Cells 2022, 11, 2501. [Google Scholar] [CrossRef] [PubMed]
- Kojima, K.K. Structural and sequence diversity of eukaryotic transposable elements. Genes Genet. Syst. 2019, 94, 233–252. [Google Scholar] [CrossRef] [Green Version]
- Flynn, J.M.; Hubley, R.; Goubert, C.; Rosen, J.; Clark, A.G.; Feschotte, C.; Smit, A.F. RepeatModeler2 for automated genomic discovery of transposable element families. Proc. Natl. Acad. Sci. USA 2020, 117, 9451–9457. [Google Scholar] [CrossRef] [PubMed]
- Smit, A.F.A.; Hubley, R.; Green, P. RepeatMasker Open-4.0 2013–2015. Available online: http://www.repeatmasker.org (accessed on 12 May 2021).
- Bao, Z.; Eddy, S.R. Automated De Novo Identification of Repeat Sequence Families in Sequenced Genomes. Genome Res. 2002, 12, 1269–1276. [Google Scholar] [CrossRef] [Green Version]
- Price, A.L.; Jones, N.C.; Pevzner, P.A. De novo identification of repeat families in large genomes. Bioinformatics 2005, 21 (Suppl. S1), i351–i358. [Google Scholar] [CrossRef] [Green Version]
- Ou, S.; Jiang, N. LTR_retriever: A Highly Accurate and Sensitive Program for Identification of Long Terminal Repeat Retrotransposons. Plant Physiol. 2018, 176, 1410–1422. [Google Scholar] [CrossRef] [Green Version]
- Flutre, T.; Duprat, E.; Feuillet, C.; Quesneville, H. Considering Transposable Element Diversification in De Novo Annotation Approaches. PLoS ONE. 2011, 6, e16526. [Google Scholar] [CrossRef]
- Novák, P.; Neumann, P.; Pech, J.; Steinhaisl, J.; Macas, J. RepeatExplorer: A Galaxy-based web server for genome-wide characterization of eukaryotic repetitive elements from next-generation sequence reads. Bioinformatics 2013, 29, 792–793. [Google Scholar] [CrossRef] [Green Version]
- Holt, C.; Campbell, M.; Keays, D.A.; Edelman, N.; Kapusta, A.; Maclary, E.; Domyan, E.T.; Suh, A.; Warren, W.C.; Yandell, M.; et al. Improved Genome Assembly and Annotation for the Rock Pigeon (Columba livia). G3 Genes Genomes Genet. 2018, 8, 1391–1398. [Google Scholar] [CrossRef] [Green Version]
- Meng, X.; Fu, Q.; Luan, S.; Luo, K.; Sui, J.; Kong, J. Genome Survey and High-Resolution Genetic Map Provide Valuable Genetic Resources for Fenneropenaeus Chinensis. Sci. Rep. 2021, 11, 7533. [Google Scholar] [CrossRef] [PubMed]
- Swathi, A.; Shekhar, M.S.; Katneni, V.K.; Vijayan, K.K. Genome Size Estimation of Brackishwater Fishes and Penaeid Shrimps by Flow Cytometry. Mol. Biol. Rep. 2018, 45, 951–960. [Google Scholar] [CrossRef]
- Kawato, S.; Nishitsuji, K.; Arimoto, A.; Hisata, K.; Kawamitsu, M.; Nozaki, R.; Kondo, H.; Shinzato, C.; Ohira, T.; Satoh, N.; et al. Genome and Transcriptome Assemblies of the Kuruma Shrimp, Marsupenaeus Japonicus. G3 Genes Genomes Genet. 2021, 11, jkab268. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Bian, C.; Jiang, S.; Han, K.; Xiong, Y.; Zhang, W.; Shi, C.; Qiao, H.; Gao, Z.; Li, R.; et al. A Chromosome-Level Genome Assembly of the Oriental River Prawn, Macrobrachium Nipponense. GigaScience 2021, 10, giaa160. [Google Scholar] [CrossRef] [PubMed]
- Veldsman, W.P.; Ma, K.Y.; Hui, J.H.L.; Chan, T.F.; Baeza, J.A.; Qin, J.; Chu, K.H. Comparative Genomics of the Coconut Crab and Other Decapod Crustaceans: Exploring the Molecular Basis of Terrestrial Adaptation. BMC Genom. 2021, 22, 313. [Google Scholar] [CrossRef]
- Gutekunst, J.; Andriantsoa, R.; Falckenhayn, C.; Hanna, K.; Stein, W.; Rasamy, J.; Lyko, F. Clonal Genome Evolution and Rapid Invasive Spread of the Marbled Crayfish. Nat. Ecol. Evol. 2018, 2, 567–573. [Google Scholar] [CrossRef] [Green Version]
- Shi, L.; Yi, S.; Li, Y. Genome Survey Sequencing of Red Swamp Crayfish Procambarus Clarkii. Mol. Biol. Rep. 2018, 45, 799–806. [Google Scholar] [CrossRef]
- Austin, C.M.; Croft, L.J.; Grandjean, F.; Gan, H.M. The NGS Magic Pudding: A Nanopore-Led Long-Read Genome Assembly for the Commercial Australian Freshwater Crayfish, Cherax Destructor. Front. Genet. 2022, 12, 695763. [Google Scholar] [CrossRef]
- Tan, M.H.; Gan, H.M.; Lee, Y.P.; Grandjean, F.; Croft, L.J.; Austin, C.M. A Giant Genome for a Giant Crayfish (Cherax Quadricarinatus) With Insights Into Cox1 Pseudogenes in Decapod Genomes. Front. Genet. 2020, 11, 201. [Google Scholar] [CrossRef] [Green Version]
- Polinski, J.M.; Zimin, A.V.; Clark, K.F.; Kohn, A.B.; Sadowski, N.; Timp, W.; Ptitsyn, A.; Khanna, P.; Romanova, D.Y.; Williams, P.; et al. The American Lobster Genome Reveals Insights on Longevity, Neural, and Immune Adaptations. Sci. Adv. 2021, 7, eabe8290. [Google Scholar] [CrossRef]
- Tang, B.; Wang, Z.; Liu, Q.; Wang, Z.; Ren, Y.; Guo, H.; Qi, T.; Li, Y.; Zhang, H.; Jiang, S.; et al. Chromosome-level Genome Assembly of Paralithodes platypus Provides Insights into Evolution and Adaptation of King Crabs. Mol. Ecol. Resour. 2021, 21, 511–525. [Google Scholar] [CrossRef]
- Liu, L.; Cui, Z.; Song, C.; Liu, Y.; Hui, M.; Wang, C. Flow Cytometric Analysis of DNA Content for Four Commercially Important Crabs in China. Acta Oceanol. Sin. 2016, 35, 7–11. [Google Scholar] [CrossRef]
- Jimenez, A.G.; Kinsey, S.T.; Dillaman, R.M.; Kapraun, D.F. Nuclear DNA Content Variation Associated with Muscle Fiber Hypertrophic Growth in Decapod Crustaceans. Genome 2010, 53, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-H.; Kim, H.; Kim, H.; Chan, B.; Kang, S.; Kim, W. Draft Genome Assembly of a Fouling Barnacle, Amphibalanus Amphitrite (Darwin, 1854): The First Reference Genome for Thecostraca. Front. Ecol. Evol. 2019, 7, 465. [Google Scholar] [CrossRef] [Green Version]
- Chebbi, M.A.; Becking, T.; Moumen, B.; Giraud, I.; Gilbert, C.; Peccoud, J.; Cordaux, R. The Genome of Armadillidium vulgare (Crustacea, Isopoda) Provides Insights into Sex Chromosome Evolution in the Context of Cytoplasmic Sex Determination. Mol. Biol. Evol. 2019, 36, 727–741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Routtu, J.; Hall, M.D.; Albere, B.; Beisel, C.; Bergeron, R.D.; Chaturvedi, A.; Choi, J.-H.; Colbourne, J.; De Meester, L.; Stephens, M.T.; et al. An SNP-Based Second-Generation Genetic Map of Daphnia magna and Its Application to QTL Analysis of Phenotypic Traits. BMC Genom. 2014, 15, 1033. [Google Scholar] [CrossRef] [Green Version]
- Tran Van, P.; Anselmetti, Y.; Bast, J.; Dumas, Z.; Galtier, N.; Jaron, K.S.; Martens, K.; Parker, D.J.; Robinson-Rechavi, M.; Schwander, T.; et al. First Annotated Draft Genomes of Nonmarine Ostracods (Ostracoda, Crustacea) with Different Reproductive Modes. G3 Genes Genomes Genet. 2021, 11, jkab043. [Google Scholar] [CrossRef] [PubMed]
- Rasch, E.; Lee, C.; Wyngaard, G. DNA-Feulgen Cytophotometric Determination of Genome Size for the Freshwater-Invading Copepod Eurytemora Affinis. Genome/Natl. Res. Counc. Can. 2004, 47, 559–564. [Google Scholar] [CrossRef] [Green Version]
- Poynton, H.; Hasenbein, S.; Benoit, J.; Sepulveda, M.; Poelchau, M.; Hughes, D.; Murali, S.; Chen, S.; Glastad, K.; Goodisman, M.; et al. The Toxicogenome of Hyalella azteca: A Model for Sediment Ecotoxicology and Evolutionary Toxicology. Environ. Sci. Technol. 2018, 52, 6009–6022. [Google Scholar] [CrossRef]
- Chikhi, R.; Medvedev, P. Informed and Automated K-Mer Size Selection for Genome Assembly. Bioinformatics 2014, 30, 31–37. [Google Scholar] [CrossRef] [Green Version]
- Novák, P.; Ávila Robledillo, L.; Koblížková, A.; Vrbová, I.; Neumann, P.; Macas, J. TAREAN: A Computational Tool for Identification and Characterization of Satellite DNA from Unassembled Short Reads. Nucleic Acids Res. 2017, 45, e111. [Google Scholar] [CrossRef]
- Bao, W.; Kojima, K.K.; Kohany, O. Repbase Update, a Database of Repetitive Elements in Eukaryotic Genomes. Mob. DNA 2015, 6, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Demšar, J.; Curk, T.; Erjavec, A.; Gorup, Č.; Hočevar, T.; Milutinovič, M.; Možina, M.; Polajnar, M.; Toplak, M.; Starič, A.; et al. Orange: Data mining toolbox in python. J. Mach. Learn. Res. 2013, 14, 2349–2353. [Google Scholar]
- Silva, B.S.M.L.; Picorelli, A.C.R.; Kuhn, G.C.S. In Silico Identification and Characterization of Satellite DNAs in 23 Drosophila Species from the Montium Group. Genes 2023, 14, 300. [Google Scholar] [CrossRef]
- Pita, S.; Panzera, F.; Mora, P.; Vela, J.; Cuadrado, Á.; Sánchez, A.; Palomeque, T.; Lorite, P. Comparative Repeatome Analysis on Triatoma Infestans Andean and Non-Andean Lineages, Main Vector of Chagas Disease. PLoS ONE 2017, 12, e0181635. [Google Scholar] [CrossRef] [Green Version]
- Palacios-Gimenez, O.M.; Koelman, J.; Palmada-Flores, M.; Bradford, T.M.; Jones, K.K.; Cooper, S.J.B.; Kawakami, T.; Suh, A. Comparative Analysis of Morabine Grasshopper Genomes Reveals Highly Abundant Transposable Elements and Rapidly Proliferating Satellite DNA Repeats. BMC Biol. 2020, 18, 199. [Google Scholar] [CrossRef]
- Utsunomia, R.; Silva, D.M.Z.d.A.; Ruiz-Ruano, F.J.; Goes, C.A.G.; Melo, S.; Ramos, L.P.; Oliveira, C.; Porto-Foresti, F.; Foresti, F.; Hashimoto, D.T. Satellitome Landscape Analysis of Megaleporinus Macrocephalus (Teleostei, Anostomidae) Reveals Intense Accumulation of Satellite Sequences on the Heteromorphic Sex Chromosome. Sci. Rep. 2019, 9, 5856. [Google Scholar] [PubMed] [Green Version]
- Sproul, J.S.; Hotaling, S.; Heckenhauer, J.; Powell, A.; Larracuente, A.M.; Kelley, J.L.; Pauls, S.U.; Frandsen, P.B. Repetitive Elements in the Era of Biodiversity Genomics: Insights from 600+ Insect Genomes. bioRxiv 2022. [Google Scholar]
- Logsdon, G.A.; Vollger, M.R.; Eichler, E.E. Long-Read Human Genome Sequencing and Its Applications. Nat. Rev. Genet. 2020, 21, 597–614. [Google Scholar] [CrossRef]
- Paajanen, P.; Kettleborough, G.; López-Girona, E.; Giolai, M.; Heavens, D.; Baker, D.; Lister, A.; Cugliandolo, F.; Wilde, G.; Hein, I.; et al. A Critical Comparison of Technologies for a Plant Genome Sequencing Project. GigaScience 2019, 8, giy163. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Gao, T.; Xu, Y.; Li, X.; Li, J.; Lin, H.; Yan, W.; Pan, J.; Tang, J. A chromosome-level reference genome of red swamp crayfish Procambarus clarkii provides insights into the gene families regarding growth or development in crustaceans. Genomics 2021, 113, 3274–3284. [Google Scholar] [CrossRef]
- Wang, Q.; Ren, X.; Liu, P.; Li, J.; Lv, J.; Wang, J.; Zhang, H.; Wei, W.; Zhou, Y.; He, Y.; et al. Improved genome assembly of Chinese shrimp (Fenneropenaeus chinensis) suggests adaptation to the environment during evolution and domestication. Mol. Ecol. Res. 2022, 334–344. [Google Scholar] [CrossRef] [PubMed]
- Katneni, V.K.; Shekhar, M.S.; Jangam, A.K.; Krishnan, K.; Prabhudas, S.K.; Kaikkolante, N.; Baghel, D.S.; Koyadan, V.K.; Jena, J.; Mohapatra, T. A Superior Contiguous Whole Genome Assembly for Shrimp (Penaeus indicus). Front. Mar. Sci. 2022, 8, 808354. [Google Scholar] [CrossRef]
- Uengwetwanit, T.; Pootakham, W.; Nookaew, I.; Sonthirod, C.; Angthong, P.; Sittikankaew, K.; Rungrassamee, W.; Arayamethakorn, S.; Wongsurawat, T.; Jenjaroenpun, P.; et al. A chromosome-level assembly of the black tiger shrimp (Penaeus monodon) genome facilitates the identification of growth-associated genes. Mol. Ecol. Resour. 2021, 21, 1620–1640. [Google Scholar] [CrossRef]
- Yuan, J.; Zhang, X.; Li, F.; Xiang, J. Genome Sequencing and Assembly Strategies and a Comparative Analysis of the Genomic Characteristics in Penaeid Shrimp Species. Front. Genet. 2021, 12, 658619. [Google Scholar] [CrossRef]
- Zhang, X.; Yuan, J.; Sun, Y.; Li, S.; Gao, Y.; Yu, Y.; Liu, C.; Wang, Q.; Lv, X.; Zhang, X.; et al. Penaeid shrimp genome provides insights into benthic adaptation and frequent molting. Nat. Commun. 2019, 10, 356. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Ge, S.; Bhandari, S.; Fan, C.; Jiao, Y.; Gai, C.; Wang, Y.; Liu, H. Genome characterization and comparative analysis among three swimming crab species. Front. Mar. Sci. 2022, 9, 895119. [Google Scholar] [CrossRef]
- Tang, B.; Zhang, D.; Li, H.; Jiang, S.; Zhang, H.; Xuan, F.; Ge, B.; Wang, Z.; Liu, Y.; Sha, Z.; et al. Chromosome-level genome assembly reveals the unique genome evolution of the swimming crab (Portunus trituberculatus). GigaScience 2020, 9, giz161. [Google Scholar] [CrossRef] [PubMed]
- Bachvaroff, T.R.; McDonald, R.C.; Plough, L.V.; Chung, J.S. Chromosome-level genome assembly of the blue crab, Callinectes sapidus. G3 Genes Genomes Genet. 2021, 11, jkab212. [Google Scholar] [CrossRef]
- Tang, B.; Wang, Z.; Liu, Q.; Zhang, H.; Jiang, S.; Li, X.; Wang, Z.; Sun, Y.; Sha, Z.; Jiang, H.; et al. High-Quality Genome Assembly of Eriocheir japonica sinensis Reveals Its Unique Genome Evolution. Front. Genet. 2020, 10, 1340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petersen, M.; Armisén, D.; Gibbs, R.A.; Hering, L.; Khila, A.; Mayer, G.; Richards, S.; Niehuis, O.; Misof, B. Diversity and Evolution of the Transposable Element Repertoire in Arthropods with Particular Reference to Insects. BMC Ecol. Evol. 2019, 19, 11. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.; Lu, J. Diversification of Transposable Elements in Arthropods and Its Impact on Genome Evolution. Genes 2019, 10, 338. [Google Scholar] [CrossRef] [Green Version]
- Shao, C.; Sun, S.; Liu, K.; Wang, J.; Li, S.; Liu, Q.; Deagle, B.E.; Seim, I.; Biscontin, A.; Wang, Q.; et al. The Enormous Repetitive Antarctic Krill Genome Reveals Environmental Adaptations and Population Insights. Cell 2023, 186, 1279–1294.e19. [Google Scholar] [CrossRef] [PubMed]
- Lyon, M.F. Do LINEs Have a Role in X-Chromosome Inactivation? J. Biomed. Biotechnol. 2006, 2006, 59746. [Google Scholar] [CrossRef]
- Dodsworth, S.; Chase, M.W.; Kelly, L.J.; Leitch, I.J.; Macas, J.; Novák, P.; Piednoël, M.; Weiss-Schneeweiss, H.; Leitch, A.R. Genomic Repeat Abundances Contain Phylogenetic Signal. Syst. Biol. 2015, 64, 112–126. [Google Scholar] [CrossRef] [Green Version]
- Dodsworth, S.; Jang, T.-S.; Struebig, M.; Chase, M.W.; Weiss-Schneeweiss, H.; Leitch, A.R. Genome-Wide Repeat Dynamics Reflect Phylogenetic Distance in Closely Related Allotetraploid Nicotiana (Solanaceae). Plant Syst. Evol. 2017, 303, 1013–1020. [Google Scholar] [CrossRef]
- Zhu, L.; Swergold, G.D.; Seldin, M.F. Examination of Sequence Homology between Human Chromosome 20 and the Mouse Genome: Intense Conservation of Many Genomic Elements. Hum. Genet. 2003, 113, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.C.; Shabalina, S.A.; Harris, D.G.; Spouge, J.L.; Kondrashovi, A.S. Conserved Fragments of Transposable Elements in Intergenic Regions: Evidence for Widespread Recruitment of MIR- and L2-Derived Sequences within the Mouse and Human Genomes. Genet Res 2003, 82, 1–18. [Google Scholar] [CrossRef]
- Vitales, D.; Garcia, S.; Dodsworth, S. Reconstructing Phylogenetic Relationships Based on Repeat Sequence Similarities. Mol. Phylogenet. Evol. 2020, 147, 106766. [Google Scholar] [CrossRef]
- Bao, W.; Tang, K.F.J.; Alcivar-Warren, A. The Complete Genome of an Endogenous Nimavirus (Nimav-1_LVa) From the Pacific Whiteleg Shrimp Penaeus (Litopenaeus) vannamei. Genes 2020, 11, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cawthorn, R.J. Diseases of American Lobsters (Homarus Americanus): A Review. J. Invertebr. Pathol. 2011, 106, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Clark, K.F.; Greenwood, S.J.; Acorn, A.R.; Byrne, P.J. Molecular Immune Response of the American Lobster (Homarus Americanus) to the White Spot Syndrome Virus. J. Invertebr. Pathol. 2013, 114, 298–308. [Google Scholar] [CrossRef] [PubMed]
- Sotero-Caio, C.G.; Platt, R.N., II; Suh, A.; Ray, D.A. Evolution and Diversity of Transposable Elements in Vertebrate Genomes. Genome Biol. Evol. 2017, 9, 161–177. [Google Scholar] [CrossRef] [Green Version]


| Suborder/Infraorder | Species | Assembly Access ID | Assembly Size (Mb) | BUSCO Completeness (%) | Paired-End Illumina Reads SRA Access ID | Estimate Genome Size (Mb) | Estimate Genome Size Reference |
|---|---|---|---|---|---|---|---|
| Dendrobranchiata | Penaeus chinensis | GCF019202785.1 | 1466 | 90.7 | SRR13452153 | 2660 | [46] |
| Penaeus indicus | GCA018983055.1 | 1936 | 88.5 | SRR12969543 | 2810 | [47] | |
| Penaeus japonicus | GCF017312705.1 | 1705 | 96.6 | DRR278744 | 2170 | [47] | |
| Penaeus monodon | GCF015228065.1 | 2394 | 83.9 | SRR11278066 | 2200 | [47] | |
| Penaeus vannamei | GCF003789085.1 | 1664 | 84.8 | SRR13661692 | 2270 | [47] | |
| Caridea | Caridina multidentata | GCA002091895.1 | 1949 | 25.2 | DRR054559 | 3230 | [48] |
| Macrobrachium nipponense | GCA015104395.1 | 1985 | 41 | SRR9026393 | 4600 | [49] | |
| Achelata | Panulirus ornatus | GCA018397875.1 | 1926 | 70 | SSR13822589 | 3230 | [50] |
| Astacidea | Procambarus virginalis | GCA020271785.1 | 3701 | 67 | SRR12901906 | 3500 | [51] |
| Procambarus clarkii | GCF020424385.1 | 2735 | 94.3 | SRR14457195 | 8500 | [52] | |
| Cherax destructor | GCA009830355.1 | 3337 | 81.7 | SRR10467055 | 4500 | [53] | |
| Cherax quadricarinatus | GCA009761615.1 | 3237 | 69.9 | SRR10484712 | 5000 | [54] | |
| Homarus americanus | GCF018991925.1 | 2292 | 93 | SRR12699166 | 7700 | [55] | |
| Anomura | Paralithodes camtschaticus | GCA018397895.1 | 3810 | 44.2 | SRR13805857 | 7290 | [50] |
| Paralithodes platypus | GCA013283005.1 | 4805 | 71.7 | SRR1145749 | 5490 | [56] | |
| Birgus latro | GCA018397915.1 | 2959 | 57.7 | SRR13816158 | 6220 | [50] | |
| Brachyura | Chionoecetes opilio | GCA016584305.1 | 2003 | 91 | SRR11278230 | 1655 | |
| Eriocheir sinensis | GCA013436485.1 | 1272 | 92.6 | SRR11971329 | 2230 | [57] | |
| Portunus trituberculatus | GCF017591435.1 | 1005 | 93.5 | SRR9964028 | 2250 | [57] | |
| Callinectes sapidus | GCA020233015.1 | 998 | 90.4 | SRR15834103 | 2290 | [58] | |
| Other Crustacea | Amphibalanus Amphitrite (Cirripedia) | GCA019059575.1 | 808 | 93.9 | SRR9595623 | 481 | [59] |
| Armadillidium vulgare (Isopoda) | GCA004104545.1 | 1725 | 84.5 | SRR8156178 | 1660 | [60] | |
| Daphnia magna (Phyllopoda) | GCA020631705.2 | 161 | 98.6 | SRR15012074 | 238 | [61] | |
| Darwinula stevensoni (Podocopida) | GCA905338385.1 | 382 | 90.3 | SRR8695251 | 437 | [62] | |
| Eurytemora affinis (Copepoda) | GCA000591075.2 | 389 | 91 | SRR2452640 | 616 | [63] | |
| Hyalella Azteca (Amphipoda) | GCA000764305.4 | 551 | 93.8 | SRR1556043 | 1050 | [64] |
| Suborder/Infraorder | Species | Ab Initio satDNA Families Identified | Number of Families Annotated Using RMo Species-Specific and Repbase as Library for Each Species | Number of Families Annotated Using Merged Libraries of RMo and Tp Libraries for All Species and Repbase | |||||
|---|---|---|---|---|---|---|---|---|---|
| RMo | Tp | All RE Families | Percentage of Unknown | satDNA Only | All RE Families | Percentage of Unknown | Satdna Only | ||
| Dendrobranchiata | P. chinensis | 1 | 7 | 7547 | 12.38% | 24 | 22,702 | 3.44% | 56 |
| P. indicus | 1 | 2 | 8252 | 7.72% | 30 | 24,237 | 3.40% | 57 | |
| P. japonicus | 3 | 5 | 7693 | 7.25% | 29 | 22,611 | 3.61% | 59 | |
| P. monodon | 0 | 4 | 8647 | 9.28% | 28 | 25,183 | 3.57% | 57 | |
| P. vannamei | 0 | 3 | 7621 | 8.85% | 30 | 23,240 | 3.49% | 55 | |
| Caridea | C. multidentata | 1 | 6 | 11,104 | 11.93% | 38 | 28,065 | 11% | 74 |
| M. nipponense | 2 | 0 | 10,455 | 19.68% | 38 | 26,021 | 13.42% | 57 | |
| Achelata | P. ornatus | 1 | 6 | 8850 | 21.13% | 35 | 25,995 | 8.12% | 60 |
| Astacidea | P. virginalis | 1 | 31 | 9213 | 28.26% | 33 | 26,483 | 9.95% | 96 |
| P. clarkii | 2 | 39 | 8838 | 22.52% | 34 | 26,051 | 13.67% | 97 | |
| C. destructor | 4 | 24 | 10,391 | 14.10% | 40 | 29,970 | 6.88% | 92 | |
| C. quadricarinatus | 1 | 43 | 10,411 | 14.33% | 35 | 26,966 | 4.99% | 96 | |
| H. americanus | 1 | 2 | 9557 | 24.16% | 35 | 27,873 | 17.29% | 61 | |
| Anomura | P. camtschaticus | 2 | 19 | 11,431 | 24.95% | 33 | 30,169 | 14.36% | 95 |
| P. platypus | 0 | 36 | 11,332 | 32.76% | 34 | 31,798 | 13.27% | 109 | |
| B. latro | 1 | 2 | 11,053 | 25.48% | 37 | 31,207 | 16.30% | 59 | |
| Brachyura | C. opilio | 0 | 0 | 10,400 | 22.89% | 29 | 26,561 | 12.26% | 52 |
| E. sinensis | 1 | 0 | 8486 | 20.74% | 29 | 23,937 | 11.82% | 49 | |
| P. trituberculatus | 0 | 0 | 7399 | 12.28% | 20 | 21,070 | 6.42% | 39 | |
| C. sapidus | 0 | 2 | 6911 | 13.68% | 18 | 19,041 | 8.68% | 31 | |
| Other Crustacea | A. Amphitrite (Cirripedia) | 1 | 1 | 6717 | 27.06% | 14 | 11,969 | 14.90% | 22 |
| A. vulgare (Isopoda) | 0 | 13 | 9431 | 17.40% | 27 | 19,098 | 11.91% | 47 | |
| D. magna (Phyllopoda) | 2 | 3 | 3643 | 17.90% | 10 | 6805 | 14.63% | 11 | |
| D. stevensoni (Podocopida) | 1 | 2 | 9762 | 25.59% | 22 | 17,339 | 23.89% | 38 | |
| E. affinis (Copepoda) | 1 | 8 | 6069 | 33.37% | 32 | 13,334 | 24.15% | 46 | |
| H. Azteca (Amphipoda) | 1 | 10 | 6851 | 16.21% | 28 | 14,424 | 13.69% | 46 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rutz, C.; Bonassin, L.; Kress, A.; Francesconi, C.; Boštjančić, L.L.; Merlat, D.; Theissinger, K.; Lecompte, O. Abundance and Diversification of Repetitive Elements in Decapoda Genomes. Genes 2023, 14, 1627. https://doi.org/10.3390/genes14081627
Rutz C, Bonassin L, Kress A, Francesconi C, Boštjančić LL, Merlat D, Theissinger K, Lecompte O. Abundance and Diversification of Repetitive Elements in Decapoda Genomes. Genes. 2023; 14(8):1627. https://doi.org/10.3390/genes14081627
Chicago/Turabian StyleRutz, Christelle, Lena Bonassin, Arnaud Kress, Caterina Francesconi, Ljudevit Luka Boštjančić, Dorine Merlat, Kathrin Theissinger, and Odile Lecompte. 2023. "Abundance and Diversification of Repetitive Elements in Decapoda Genomes" Genes 14, no. 8: 1627. https://doi.org/10.3390/genes14081627
APA StyleRutz, C., Bonassin, L., Kress, A., Francesconi, C., Boštjančić, L. L., Merlat, D., Theissinger, K., & Lecompte, O. (2023). Abundance and Diversification of Repetitive Elements in Decapoda Genomes. Genes, 14(8), 1627. https://doi.org/10.3390/genes14081627






