Comparative Transcriptomic Analysis of Insecticide-Resistant Aedes aegypti from Puerto Rico Reveals Insecticide-Specific Patterns of Gene Expression
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area and Mosquito Collections
2.2. Insecticide Bioassays
2.3. RNA and Library Preparation Methods
2.4. Measurement of Gene Expression Using Quantitative PCR
2.5. Data Analysis
2.5.1. Quality Control Filtering and Mapping
2.5.2. Differential Gene Expression Analysis
2.5.3. Gene Ontology Annotation and Functional Enrichment Analysis
3. Results
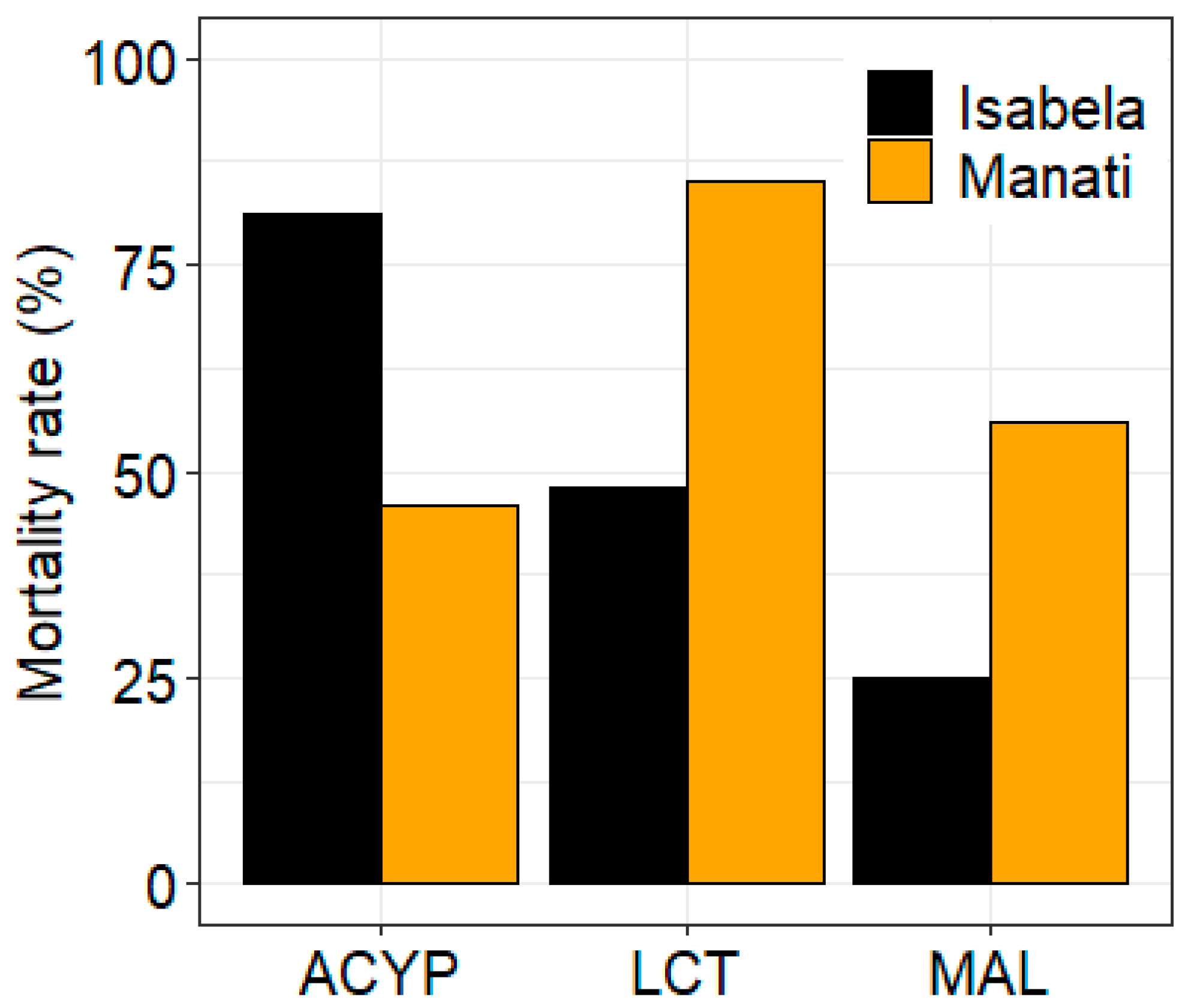
3.1. Bioassay Results
3.2. RNA Sequencing, Quality Control Filtering, and Alignment Rate
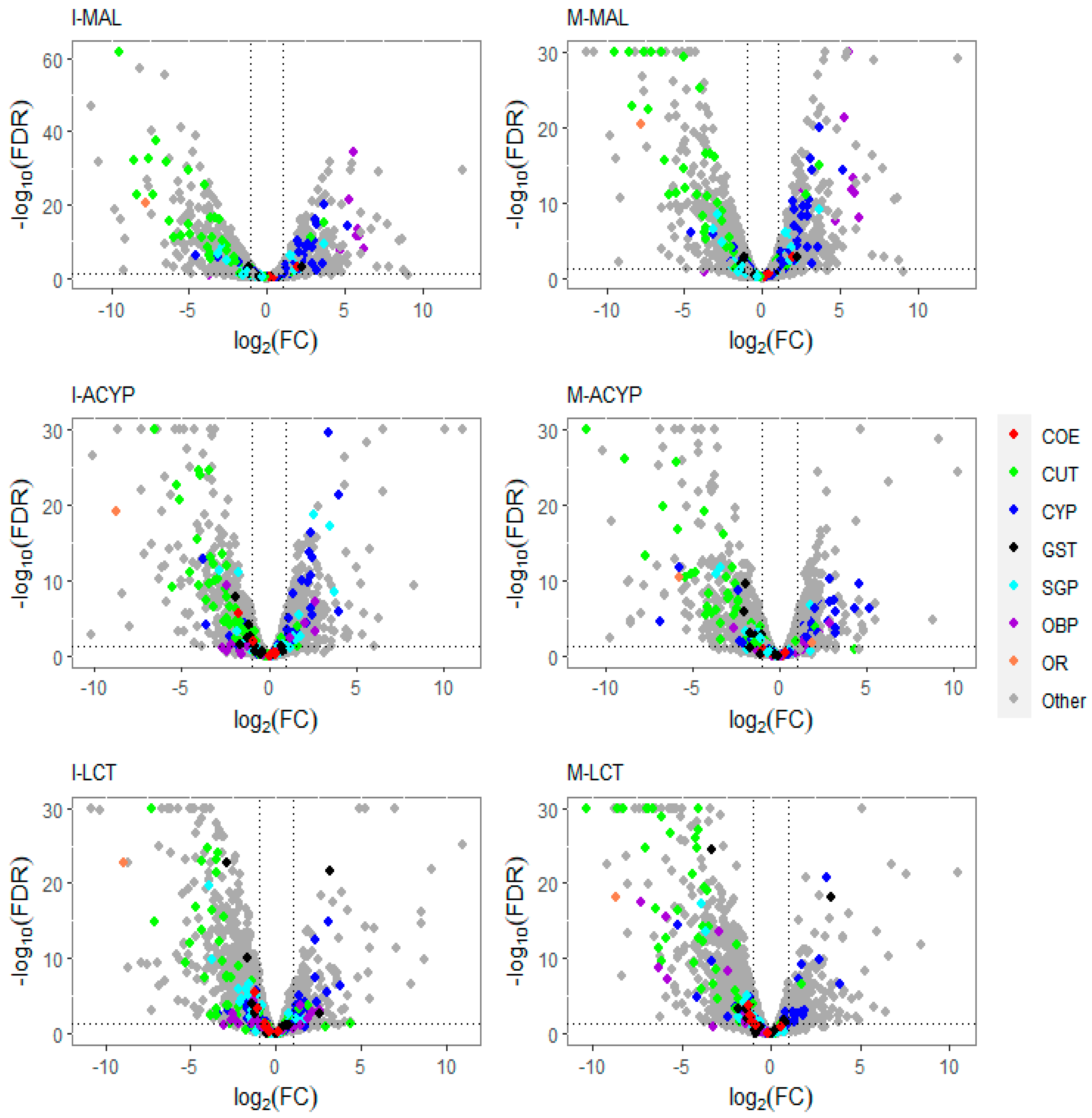
3.3. Differential Gene Expression Analysis
3.3.1. Differential Gene Expression Associated with Malathion Resistance
3.3.2. Differential Gene Expression Associated with alpha-Cypermethrin Resistance
3.3.3. Differential Gene Expression Associated with Lambda-Cyhalothrin Resistance
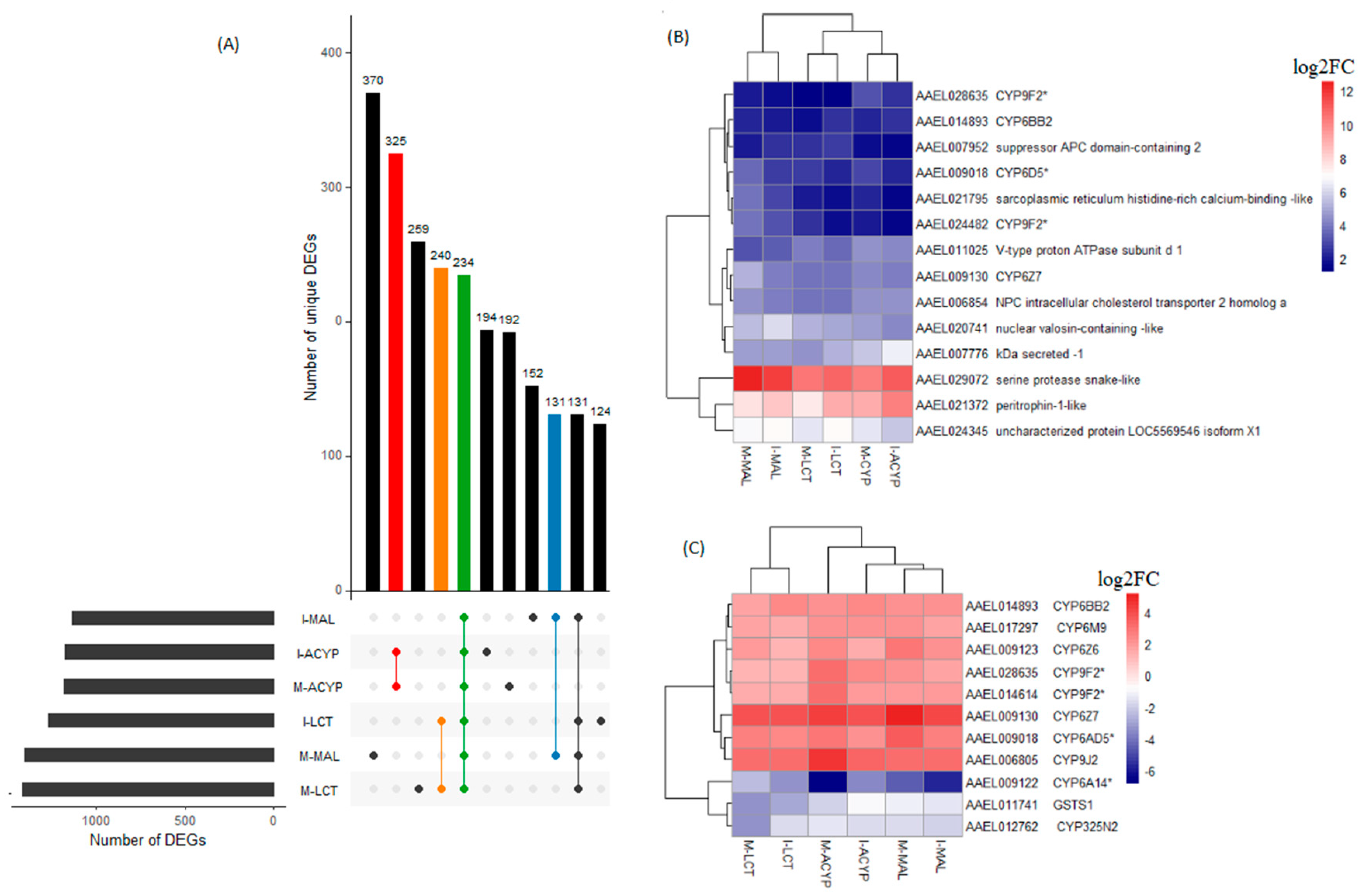
3.3.4. Genes Associated with Resistance to Multiple Insecticides
3.4. Gene Ontology Enrichment Analysis
3.5. Validation of Relative Expression Levels Estimated Using RNA-Seq with Quantitative RT-PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Code Availability
Disclaimer
References
- Sharp, T.M.; Quandelacy, T.M.; Adams, L.E.; Aponte, J.T.; Lozier, M.J.; Ryff, K.; Flores, M.; Rivera, A.; Santiago, G.A.; Muñoz-Jordán, J.L.; et al. Epidemiologic and spatiotemporal trends of Zika Virus disease during the 2016 epidemic in Puerto Rico. PLoS Negl. Trop. Dis. 2020, 14, e0008532. [Google Scholar] [CrossRef] [PubMed]
- Hemme, R.R.; Vizcaino, L.; Harris, A.F.; Felix, G.; Kavanaugh, M.; Kenney, J.L.; Nazario, N.M.; Godsey, M.S.; Barrera, R.; Miranda, J.; et al. Rapid Screening of Aedes aegypti Mosquitoes for Susceptibility to Insecticides as Part of Zika Emergency Response, Puerto Rico. Emerg. Infect. Dis. 2019, 25, 1959–1961. [Google Scholar] [CrossRef] [PubMed]
- Liu, N. insecticide resistance in mosquitoes: Impact, mechanisms and research directions. Annu. Rev. Entomol. 2015, 60, 537–559. [Google Scholar] [CrossRef]
- Hemingway, J.; Hawkes, N.J.; McCarroll, L.; Ranson, H. The molecular basis of insecticide resistance in mosquitoes. Insect. Biochem. Mol. Biol. 2004, 34, 653–665. [Google Scholar] [CrossRef]
- Chareonviriyaphap, T.; Bangs, M.J.; Suwonkerd, W.; Kongmee, M.; Corbel, V.; Ngoen-Klan, R. Review of insecticide resistance and behavioral avoidance of vectors of human diseases in Thailand. Parasit. Vectors 2013, 6, 280. [Google Scholar] [CrossRef]
- Simma, E.A.; Dermauw, W.; Balabanidou, V.; Snoeck, S.; Bryon, A.; Clark, R.M.; Yewhalaw, D.; Vontas, J.; Duchateau, L.; Van Leeuwen, T. Genome-wide gene expression profiling reveals that cuticle alterations and P450 detoxification are associated with deltamethrin and DDT resistance in Anopheles arabiensis populations from Ethiopia. Pest. Manag. Sci. 2019, 75, 1808–1818. [Google Scholar] [CrossRef] [PubMed]
- Bass, C.; Nikou, D.; Donnelly, M.J.; Williamson, M.S.; Ranson, H.; Ball, A.; Vontas, J.; Field, L.M. Detection of knockdown resistance (kdr) mutations in Anopheles gambiae: A comparison of two new high-throughput assays with existing methods. Malar. J. 2007, 6, 111. [Google Scholar] [CrossRef] [PubMed]
- Mathias, D.K.; Ochomo, E.; Atieli, F.; Ombok, M.; Bayoh, M.N.; Olang, G.; Muhia, D.; Kamau, L.; Vulule, J.M.; Hamel, M.J.; et al. Spatial and temporal variation in the kdr allele L1014S in Anopheles gambiae s.s. and phenotypic variability in susceptibility to insecticides in Western Kenya. Malar. J. 2011, 10, 10. [Google Scholar] [CrossRef]
- Weill, M.; Malcolm, C.; Chandre, F.; Mogensen, K.; Berthomieu, A.; Marquine, M.; Raymond, M. The unique mutation in ace-1 giving high insecticide resistance is easily detectable in mosquitoes. Insect. Mol. Biol. 2004, 13, 1–7. [Google Scholar] [CrossRef]
- Li, X.; Schuler, M.A.; Berenbaum, M.R. Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu. Rev. Entomol. 2007, 52, 231–253. [Google Scholar] [CrossRef]
- Ishak, I.H.; Kamgang, B.; Ibrahim, S.S.; Riveron, J.M.; Irving, H.; Wondji, C.S. Pyrethroid Resistance in Malaysian Populations of Dengue Vector Aedes aegypti Is Mediated by CYP9 Family of Cytochrome P450 Genes. PLoS Negl. Trop. Dis. 2017, 11, e0005302. [Google Scholar] [CrossRef] [PubMed]
- Riveron, J.M.; Ibrahim, S.S.; Mulamba, C.; Djouaka, R.; Irving, H.; Wondji, M.J.; Ishak, I.H.; Wondji, C.S. Genome-Wide Transcription and Functional Analyses Reveal Heterogeneous Molecular Mechanisms Driving Pyrethroids Resistance in the Major Malaria Vector Anopheles funestus Across Africa. G3 2017, 7, 1819–1832. [Google Scholar] [CrossRef] [PubMed]
- David, J.P.; Ismail, H.M.; Chandor-Proust, A.; Paine, M.J. Role of cytochrome P450s in insecticide resistance: Impact on the control of mosquito-borne diseases and use of insecticides on Earth. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013, 368, 20120429. [Google Scholar] [CrossRef]
- Schluep, S.M.; Buckner, E.A. Metabolic resistance in permethrin-resistant Florida Aedes aegypti (Diptera: Culicidae). Insects 2021, 12, 866. [Google Scholar] [CrossRef]
- Braga, I.A.; Lima, J.B.; Soares Sda, S.; Valle, D. Aedes aegypti resistance to temephos during 2001 in several municipalities in the states of Rio de Janeiro, Sergipe, and Alagoas, Brazil. Mem. Inst. Oswaldo Cruz 2004, 99, 199–203. [Google Scholar] [CrossRef] [PubMed]
- Vontas, J.; Kioulos, E.; Pavlidi, N.; Morou, E.; della Torre, A.; Ranson, H. Insecticide resistance in the major dengue vectors Aedes albopictus and Aedes aegypti. Pestic. Biochem. Physiol. 2012, 104, 126–131. [Google Scholar] [CrossRef]
- Amelia-Yap, Z.H.; Chen, C.D.; Sofian-Azirun, M.; Low, V.L. Pyrethroid resistance in the dengue vector Aedes aegypti in Southeast Asia: Present situation and prospects for management. Parasit. Vectors 2018, 11, 332. [Google Scholar] [CrossRef]
- Lien, N.T.K.; Ngoc, N.T.H.; Lan, N.N.; Hien, N.T.; Tung, N.V.; Ngan, N.T.T.; Hoang, N.H.; Binh, N.T.H. Transcriptome Sequencing and Analysis of Changes Associated with Insecticide Resistance in the Dengue Mosquito (Aedes aegypti) in Vietnam. Am. J. Trop. Med. Hyg. 2019, 100, 1240–1248. [Google Scholar] [CrossRef]
- David, J.P.; Faucon, F.; Chandor-Proust, A.; Poupardin, R.; Riaz, M.A.; Bonin, A.; Navratil, V.; Reynaud, S. Comparative analysis of response to selection with three insecticides in the dengue mosquito Aedes aegypti using mRNA sequencing. BMC Genom. 2014, 15, 174. [Google Scholar] [CrossRef]
- Faucon, F.; Dusfour, I.; Gaude, T.; Navratil, V.; Boyer, F.; Chandre, F.; Sirisopa, P.; Thanispong, K.; Juntarajumnong, W.; Poupardin, R.; et al. Identifying genomic changes associated with insecticide resistance in the dengue mosquito Aedes aegypti by deep targeted sequencing. Genome Res. 2015, 25, 1347–1359. [Google Scholar] [CrossRef]
- Strode, C.; Wondji, C.S.; David, J.-P.; Hawkes, N.J.; Lumjuan, N.; Nelson, D.R.; Drane, D.R.; Karunaratne, S.H.P.P.; Hemingway, J.; Black, W.C.; et al. Genomic analysis of detoxification genes in the mosquito Aedes aegypti. Insect Biochem. Mol. Biol. 2008, 38, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Lima, E.P.; Paiva, M.H.; de Araújo, A.P.; da Silva, E.V.; da Silva, U.M.; de Oliveira, L.N.; Santana, A.E.; Barbosa, C.N.; de Paiva Neto, C.C.; Goulart, M.O.; et al. Insecticide resistance in Aedes aegypti populations from Ceará, Brazil. Parasit. Vectors 2011, 4, 5. [Google Scholar] [CrossRef] [PubMed]
- Aponte, A.; Penilla, R.P.; Rodríguez, A.D.; Ocampo, C.B. Mechanisms of pyrethroid resistance in Aedes (Stegomyia) aegypti from Colombia. Acta Tropica 2019, 191, 146–154. [Google Scholar] [CrossRef]
- Badolo, A.; Sombié, A.; Pignatelli, P.M.; Sanon, A.; Yaméogo, F.; Wangrawa, D.W.; Sanon, A.; Kanuka, H.; McCall, P.J.; Weetman, D. Insecticide resistance levels and mechanisms in Aedes aegypti populations in and around Ouagadougou, Burkina Faso. PLoS Negl. Trop. Dis. 2019, 13, e0007439. [Google Scholar] [CrossRef]
- Saavedra-Rodriguez, K.; Campbell, C.L.; Lozano, S.; Penilla-Navarro, P.; Lopez-Solis, A.; Solis-Santoyo, F.; Rodriguez, A.D.; Perera, R.; Black Iv, W.C. Permethrin resistance in Aedes aegypti: Genomic variants that confer knockdown resistance, recovery, and death. PLoS Genet. 2021, 17, e1009606. [Google Scholar] [CrossRef] [PubMed]
- Sene, N.M.; Mavridis, K.; Ndiaye, E.H.; Diagne, C.T.; Gaye, A.; Ngom, E.H.M.; Ba, Y.; Diallo, D.; Vontas, J.; Dia, I.; et al. Insecticide resistance status and mechanisms in Aedes aegypti populations from Senegal. PLoS Negl. Trop. Dis. 2021, 15, e0009393. [Google Scholar] [CrossRef]
- Samal, R.R.; Panmei, K.; Lanbiliu, P.; Kumar, S. Metabolic detoxification and ace-1 target site mutations associated with acetamiprid resistance in Aedes aegypti L. Front. Physiol. 2022, 13, 988907. [Google Scholar] [CrossRef]
- Lertkiatmongkol, P.; Pethuan, S.; Jirakanjanakit, N.; Rongnoparut, P. Transcription analysis of differentially expressed genes in insecticide-resistant Aedes aegypti mosquitoes after deltamethrin exposure. J. Vector Ecol. 2010, 35, 197–203. [Google Scholar] [CrossRef]
- Brogdon, W.; Chan, A. Guideline for Evaluating Insecticide Resistance in Vectors Using the CDC Bottle Bioassay; CDC: Atlanta, GA, USA, 2010. [Google Scholar]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Version 0.11.2. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 3 September 2021).
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Giraldo-Calderón, G.I.; Emrich, S.J.; MacCallum, R.M.; Maslen, G.; Dialynas, E.; Topalis, P.; Ho, N.; Gesing, S.; Madey, G.; Collins, F.H.; et al. VectorBase: An updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res. 2015, 43, D707–D713. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. The Subread aligner: Fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013, 41, e108. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Subgroup, G.P.D.P. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Chen, Y.; McCarthy, D.; Ritchie, M.; Robinson, M.; Smyth, G.; Hall, E. edgeR: Differential Analysis of Sequence Read Count Data User’s Guide. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2796818/ (accessed on 20 September 2021).
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Matthews, B.J.; Dudchenko, O.; Kingan, S.B.; Koren, S.; Antoshechkin, I.; Crawford, J.E.; Glassford, W.J.; Herre, M.; Redmond, S.N.; Rose, N.H.; et al. Improved reference genome of Aedes aegypti informs arbovirus vector control. Nature 2018, 563, 501–507. [Google Scholar] [CrossRef]
- Hunter, S.; Apweiler, R.; Attwood, T.K.; Bairoch, A.; Bateman, A.; Binns, D.; Bork, P.; Das, U.; Daugherty, L.; Duquenne, L.; et al. InterPro: The integrative protein signature database. Nucleic Acids Res. 2008, 37, D211–D215. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef]
- Klopfenstein, D.V.; Zhang, L.; Pedersen, B.S.; Ramírez, F.; Warwick Vesztrocy, A.; Naldi, A.; Mungall, C.J.; Yunes, J.M.; Botvinnik, O.; Weigel, M.; et al. GOATOOLS: A Python library for Gene Ontology analyses. Sci. Rep. 2018, 8, 10872. [Google Scholar] [CrossRef]
- Balabanidou, V.; Grigoraki, L.; Vontas, J. Insect cuticle: A critical determinant of insecticide resistance. Curr. Opin. Insect. Sci. 2018, 27, 68–74. [Google Scholar] [CrossRef]
- Wood, O.; Hanrahan, S.; Coetzee, M.; Koekemoer, L.; Brooke, B. Cuticle thickening associated with pyrethroid resistance in the major malaria vector Anopheles funestus. Parasit. Vectors 2010, 3, 67. [Google Scholar] [CrossRef] [PubMed]
- Balabanidou, V.; Kampouraki, A.; MacLean, M.; Blomquist, G.J.; Tittiger, C.; Juarez, M.P.; Mijailovsky, S.J.; Chalepakis, G.; Anthousi, A.; Lynd, A.; et al. Cytochrome P450 associated with insecticide resistance catalyzes cuticular hydrocarbon production in Anopheles gambiae. Proc. Natl. Acad. Sci. USA 2016, 113, 9268–9273. [Google Scholar] [CrossRef] [PubMed]
- Vontas, J.; Blass, C.; Koutsos, A.C.; David, J.P.; Kafatos, F.C.; Louis, C.; Hemingway, J.; Christophides, G.K.; Ranson, H. Gene expression in insecticide resistant and susceptible Anopheles gambiae strains constitutively or after insecticide exposure. Insect. Mol. Biol. 2005, 14, 509–521. [Google Scholar] [CrossRef] [PubMed]
- Gong, M.; Shen, B.; Gu, Y.; Tian, H.; Ma, L.; Li, X.; Yang, M.; Hu, Y.; Sun, Y.; Hu, X.; et al. Serine proteinase over-expression in relation to deltamethrin resistance in Culex pipiens pallens. Arch. Biochem. Biophys. 2005, 438, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-C.; Wang, Q.-H.; Bhowmick, B.; Li, Y.-X.; Han, Q. Functional characterization of two clip domain serine proteases in innate immune responses of Aedes aegypti. Parasites Vectors 2021, 14, 584. [Google Scholar] [CrossRef]
- Shan, T.; Wang, Y.; Dittmer, N.T.; Kanost, M.R.; Jiang, H. Serine Protease Networks Mediate Immune Responses in Extra-Embryonic Tissues of Eggs in the Tobacco Hornworm, Manduca sexta. J. Innate Immun. 2022, 15, 365–379. [Google Scholar] [CrossRef] [PubMed]
- El Moussawi, L.; Nakhleh, J.; Kamareddine, L.; Osta, M.A. The mosquito melanization response requires hierarchical activation of non-catalytic clip domain serine protease homologs. PLoS Pathog. 2019, 15, e1008194. [Google Scholar] [CrossRef]
- Kasai, S.; Komagata, O.; Itokawa, K.; Shono, T.; Ng, L.C.; Kobayashi, M.; Tomita, T. Mechanisms of pyrethroid resistance in the dengue mosquito vector, Aedes aegypti: Target site insensitivity, penetration, and metabolism. PLoS Negl. Trop. Dis. 2014, 8, e2948. [Google Scholar] [CrossRef]
- Dusfour, I.; Zorrilla, P.; Guidez, A.; Issaly, J.; Girod, R.; Guillaumot, L.; Robello, C.; Strode, C. Deltamethrin Resistance Mechanisms in Aedes aegypti Populations from Three French Overseas Territories Worldwide. PLoS Negl. Trop. Dis. 2015, 9, e0004226. [Google Scholar] [CrossRef]
- Bariami, V.; Jones, C.M.; Poupardin, R.; Vontas, J.; Ranson, H. Gene amplification, ABC transporters and cytochrome P450s: Unraveling the molecular basis of pyrethroid resistance in the dengue vector, Aedes aegypti. PLoS Negl. Trop. Dis. 2012, 6, e1692. [Google Scholar] [CrossRef]
- Reid, W.R.; Thornton, A.; Pridgeon, J.W.; Becnel, J.J.; Tang, F.; Estep, A.; Clark, G.G.; Allan, S.; Liu, N. Transcriptional analysis of four family 4 P450s in a Puerto Rico strain of Aedes aegypti (Diptera: Culicidae) compared with an Orlando strain and their possible functional roles in permethrin resistance. J. Med. Entomol. 2014, 51, 605–615. [Google Scholar] [CrossRef]
- Moyes, C.L.; Vontas, J.; Martins, A.J.; Ng, L.C.; Koou, S.Y.; Dusfour, I.; Raghavendra, K.; Pinto, J.; Corbel, V.; David, J.P.; et al. Contemporary status of insecticide resistance in the major Aedes vectors of arboviruses infecting humans. PLoS Negl. Trop. Dis. 2017, 11, e0005625. [Google Scholar] [CrossRef] [PubMed]
- Epelboin, Y.; Wang, L.; Giai Gianetto, Q.; Choumet, V.; Gaborit, P.; Issaly, J.; Guidez, A.; Douche, T.; Chaze, T.; Matondo, M.; et al. CYP450 core involvement in multiple resistance strains of Aedes aegypti from French Guiana highlighted by proteomics, molecular and biochemical studies. PLoS ONE 2021, 16, e0243992. [Google Scholar] [CrossRef]
- Marcombe, S.; Poupardin, R.; Darriet, F.; Reynaud, S.; Bonnet, J.; Strode, C.; Brengues, C.; Yébakima, A.; Ranson, H.; Corbel, V.; et al. Exploring the molecular basis of insecticide resistance in the dengue vector Aedes aegypti: A case study in Martinique Island (French West Indies). BMC Genom. 2009, 10, 494. [Google Scholar] [CrossRef] [PubMed]
- Saavedra-Rodriguez, K.; Suarez, A.F.; Salas, I.F.; Strode, C.; Ranson, H.; Hemingway, J.; Black, W.C.t. Transcription of detoxification genes after permethrin selection in the mosquito Aedes aegypti. Insect. Mol. Biol. 2012, 21, 61–77. [Google Scholar] [CrossRef] [PubMed]
- Morgan, J.; Salcedo-Sora, J.E.; Triana-Chavez, O.; Strode, C. Expansive and Diverse Phenotypic Landscape of Field Aedes aegypti (Diptera: Culicidae) Larvae with Differential Susceptibility to Temephos: Beyond Metabolic Detoxification. J. Med. Entomol. 2022, 59, 192–212. [Google Scholar] [CrossRef]
- Ranson, H.; Rossiter, L.; Ortelli, F.; Jensen, B.; Wang, X.; Roth, C.W.; Collins, F.H.; Hemingway, J. Identification of a novel class of insect glutathione S-transferases involved in resistance to DDT in the malaria vector Anopheles gambiae. Biochem. J. 2001, 359, 295–304. [Google Scholar] [CrossRef]
- Tang, A.H.; Tu, C.P. Biochemical characterization of Drosophila glutathione S-transferases D1 and D21. J. Biol. Chem. 1994, 269, 27876–27884. [Google Scholar] [CrossRef]
- Faucon, F.; Gaude, T.; Dusfour, I.; Navratil, V.; Corbel, V.; Juntarajumnong, W.; Girod, R.; Poupardin, R.; Boyer, F.; Reynaud, S.; et al. In the hunt for genomic markers of metabolic resistance to pyrethroids in the mosquito Aedes aegypti: An integrated next-generation sequencing approach. PLoS Negl. Trop. Dis. 2017, 11, e0005526. [Google Scholar] [CrossRef]
- Eldefrawi, M.E.; Eldefrawi, A.T. Insecticide Actions on Gaba Receptors and Voltage-Dependent Chloride Channels. 1989. Available online: https://api.semanticscholar.org/CorpusID:92680854 (accessed on 3 September 2021).
- World Health Organization. The World Health Report: 1998: Life in the 21st Century: A Vision for All: Report of the Director-General; World Health Organization: Geneva, Switzerland, 1998. [Google Scholar]



| # Of Genes Tested | DE Genes(|log2FC| > 1 and FDR < 0.05) | DE Genes(|log2FC| > 1 and FDR < 0.01) | |||||
|---|---|---|---|---|---|---|---|
| Insecticide | Sites | Comparisons | Up | Down | Up | Down | |
| Malathion | Isabela | I-MAL vs. Rock (R–S) | 10,153 | 699 | 939 | 441 | 691 |
| I-MAL vs. I-U1 (R–C) | 10,161 | 120 | 186 | 33 | 86 | ||
| I-U1 vs. Rock (C–S) | 10,088 | 337 | 403 | 256 | 307 | ||
| Manatí | M-MAL vs. Rock (R–S) | 10,573 | 961 | 792 | 739 | 657 | |
| M-MAL vs. M-U (R–C) | 10,885 | 322 | 98 | 174 | 41 | ||
| M-U vs. Rock (C–S) | 10,039 | 333 | 634 | 197 | 456 | ||
| alpha-cypermethrin | Isabela | I-ACYP vs. Rock (R–S) | 9825 | 711 | 625 | 642 | 529 |
| I-U vs. Rock (R–C) | 9711 | 230 | 426 | 158 | 322 | ||
| I-ACYP vs. I-U (R–C) | 9308 | 127 | 426 | 87 | 346 | ||
| Manatí | M-ACYP vs. Rock (R–S) | 9599 | 686 | 692 | 605 | 570 | |
| M-ACYP vs. M-U (R–C) | 7835 | 237 | 561 | 154 | 448 | ||
| M-U vs. Rock (C–S) | 9561 | 301 | 758 | 222 | 589 | ||
| Lambda-cyhalothrin | Isabela | I-LCT vs. R vs. Rock (R–S) | 9916 | 429 | 1008 | 341 | 920 |
| I-U vs. Rock (C–S) | 9711 | 230 | 426 | 158 | 322 | ||
| I-LCT vs. I-U (R–C) | 9550 | 51 | 366 | 30 | 256 | ||
| Manatí | M-LCT vs. Rock (R–S) | 9710 | 461 | 1163 | 362 | 1051 | |
| M-U2 vs. Rock (C–S) | 9561 | 301 | 758 | 222 | 589 | ||
| M-LCT vs. M-U (R–C) | 8618 | 126 | 271 | 67 | 197 | ||
| Gene ID | Description | I-MAL vs. Rock (R–S) | I-U1 vs. Rock (C–S) | M-MAL vs. Rock (R–S) | M-U1 vs. Rock (C–S) | I-ACYP vs. Rock (R–S) | I-LCT-R vs. Rock (R–S) | I-U2 vs. Rock (C–S) | M-ACYP vs. Rock (R–S) | M-LCT vs. Rock (R–S) | M-U2 vs. Rock (C–S) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cytochrome P450s monooxygenases | |||||||||||
| AAEL009130 | CYP6Z7 | 17.19 | 17.45 | 36.13 | 14.77 | 15.5 | 13.63 | 14.92 | 19.44 | 14.69 | |
| AAEL006805 | CYP9J2 | 9.25 | 8.16 | 8.8 | 11.56 | 10.58 | 8.29 | 7.15 | 23.33 | 8.87 | 8.96 |
| AAEL009121 | CYP6N9 | 5.57 | 3.17 | 8.42 | 4.5 | 5.16 | 7.28 | ||||
| AAEL006044 | CYP325Q1 | 5.18 | 3.23 | 9.48 | 3.23 | 2.77 | 3.55 | ||||
| AAEL014893 | CYP6BB2 | 4.27 | 7.01 | 4.37 | 6.73 | 5.48 | 5.34 | 6.01 | 4.71 | 3.37 | 3.75 |
| AAEL009123 | CYP6Z6 | 4.22 | 7.4 | 2.81 | 2.31 | 5.14 | 3.69 | ||||
| AAEL010154 | CYP4AR2 | 3.62 | 2.84 | 2.54 | 2.24 | 2.35 | |||||
| AAEL014605 | CYP9J9 | 3.43 | 2.47 | 2.91 | 3.32 | 2.57 | 2.2 | 6.79 | 2.62 | ||
| AAEL017297 | CYP6M9 | 3.35 | 6.91 | 4.36 | 4.67 | 4.84 | 2.68 | 5.39 | 4.15 | 3.41 | 3.38 |
| AAEL009127 | CYP6M11 | 3.34 | 5.13 | 3.69 | 2.23 | 2.96 | |||||
| AAEL012772 | CYP325G3 | 3.19 | 7.72 | ||||||||
| AAEL009124 | CYP6N12 | 3.18 | 5.06 | 4.71 | 4.18 | 5.56 | 2.86 | 3.91 | 3.45 | ||
| AAEL014619 | CYP9J22 | 3.13 | 2.44 | 3.09 | 5.27 | 2.39 | 2.27 | 8.75 | |||
| AAEL014615 | CYP9J23 | 3.08 | 3.31 | 9.06 | 0.03 | 0.3 | |||||
| AAEL014019 | CYP4J16 | 2.86 | 2.54 | ||||||||
| AAEL017136 | CYP325V1 | 2.79 | 2.95 | 6.14 | 2.94 | 2.68 | |||||
| AAEL007808 | CYP4D39 | 2.38 | 2.91 | ||||||||
| AAEL004054 | CYP4G36 | 2.37 | 2.13 | 2.54 | 2.18 | 2.57 | |||||
| AAEL009125 | CYP6M10 | 0.46 | 0.4 | ||||||||
| AAEL009132 | CYP6Y3 | 0.39 | 0.3 | 0.41 | 0.34 | 0.45 | 0.38 | 0.38 | 0.34 | ||
| AAEL003748 | CYP9AE1 | 0.38 | |||||||||
| AAEL012762 | CYP325N2 | 0.28 | 0.29 | 0.22 | 0.31 | 0.3 | 0.34 | 0.1 | 0.22 | ||
| AAEL009762 | CYP307A1 | 2.73 | 2.71 | 2.38 | |||||||
| AAEL014609 | CYP9J26 | 2.73 | 3.04 | 2.23 | 2.77 | 2.02 | |||||
| AAEL004941 | CYP6AK1 | 0.38 | 0.47 | ||||||||
| AAEL007010 | CYP6AG4 | 0.34 | |||||||||
| AAEL009120 | CYP6S3 | 0.28 | 0.31 | 0.33 | 0.27 | 0.22 | |||||
| AAEL007812 | CYP4H32 | 0.12 | 0.11 | 0.05 | 0.08 | 0.1 | 0.02 | 0.26 | 0.12 | ||
| AAEL006989 | CYP6AG7 | 3.24 | 3.24 | ||||||||
| AAEL009126 | CYP6N6 | 3.49 | 4.87 | 4.3 | 3.65 | ||||||
| AAEL010151 | CYP6N16 | 2.31 | |||||||||
| AAEL014891 | CYP6P12 | 0.44 | 0.45 | ||||||||
| AAEL007830 | CYP4H29 | 0.32 | 0.38 | ||||||||
| AAEL009131 | CYP6Z8 | 0.3 | |||||||||
| AAEL006815 | CYP9J16 | 3.58 | |||||||||
| AAEL009129 | CYP6Z9 | 2.13 | |||||||||
| AAEL002633 | CYP9J31 | 2.27 | |||||||||
| AAEL007024 | CYP6AG3 | 2.88 | |||||||||
| AAEL026582 | CYP6AA5 | 2.17 | 2.08 | ||||||||
| AAEL001960 | CYP12F5 | 0.37 | 0.42 | 0.49 | 0.4 | 0.38 | 0.47 | ||||
| AAEL014604 | CYP9f2* | 10.73 | 12.18 | 7.65 | 14.15 | 16.08 | 8.03 | 12.05 | 36.68 | ||
| AAEL009018 | CYP6d5* | 6.24 | 3.07 | 12.91 | 4.33 | 4.81 | 4.94 | 3.56 | 7.26 | 6.66 | 5.05 |
| AAEL019504 | CYP9f2* | 5.12 | 4.14 | 5.81 | 6.98 | 4.16 | 3.89 | 4.04 | 3.43 | ||
| AAEL014614 | CYP9f2* | 3.81 | 4 | 3.96 | 5.69 | 3.72 | 2.54 | 3.05 | 9.18 | 2.92 | 4.28 |
| AAEL028635 | CYP9f2* | 3.16 | 3.98 | 4.18 | 5.02 | 5.34 | 2.3 | 3.15 | 9.43 | 2.45 | 4.78 |
| AAEL003890 | CYP28a5* | 2.39 | 2.82 | 2.67 | |||||||
| AAEL009122 | CYP6a14* | 0.02 | 0.07 | 0.04 | 0.09 | 0.08 | 0.1 | 0.01 | 0.01 | 0.19 | 0.11 |
| AAEL021861 | CYP28a5* | 2.89 | 2.4 | 5.03 | 3.85 | 3.02 | 2.37 | 2.32 | |||
| AAEL014830 | CYP4ac1* | 0.47 | 0.29 | 0.37 | 0.19 | 0.33 | |||||
| AAEL000340 | CYP4C1-like | 0.24 | 0.15 | 0.06 | 0.19 | ||||||
| AAEL019603 | CYP9f2* | 2.05 | |||||||||
| AAEL014924 | CYP6d5* | 0.21 | |||||||||
| AAEL017539 | CYP6BY1* | 17.22 | 12.48 | ||||||||
| Glutathione S-transferases | |||||||||||
| AAEL010591 | GSTD6 | 7.03 | 5.58 | 4.81 | 3.98 | 8.87 | 5.23 | 10.56 | |||
| AAEL010582 | GSTD11 | 0.43 | 0.33 | 0.41 | 0.37 | 0.41 | 0.3 | 0.38 | |||
| AAEL000092 | GSTX1 | 0.4 | 0.42 | 0.42 | 0.39 | 0.39 | |||||
| AAEL007962 | GSTE4 | 0.37 | 0.27 | 0.23 | 0.26 | 0.31 | 0.24 | 0.24 | 0.21 | ||
| AAEL011741 | GSTS1 | 0.35 | 0.44 | 0.37 | 0.49 | 0.13 | 0.26 | 0.1 | 0.29 | ||
| AAEL007946 | GSTE6 | 0.24 | 0.41 | 0.28 | 0.18 | ||||||
| AAEL001061 | GSTD1 | 0.44 | 0.48 | ||||||||
| AAEL001054 | GSTD4 | 5.97 | 8.34 | ||||||||
| Carboxylesterases | |||||||||||
| AAEL004022 | CES5A | 3.19 | 4.07 | ||||||||
| AAEL005199 | CES6-like | 0.36 | 0.33 | 0.3 | 0.48 | 0.24 | |||||
| AAEL005200 | CES6-like | 0.48 | |||||||||
| AAEL012886 | CES-6 | 0.41 | 0.43 | ||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Derilus, D.; Impoinvil, L.M.; Muturi, E.J.; McAllister, J.; Kenney, J.; Massey, S.E.; Hemme, R.; Kothera, L.; Lenhart, A. Comparative Transcriptomic Analysis of Insecticide-Resistant Aedes aegypti from Puerto Rico Reveals Insecticide-Specific Patterns of Gene Expression. Genes 2023, 14, 1626. https://doi.org/10.3390/genes14081626
Derilus D, Impoinvil LM, Muturi EJ, McAllister J, Kenney J, Massey SE, Hemme R, Kothera L, Lenhart A. Comparative Transcriptomic Analysis of Insecticide-Resistant Aedes aegypti from Puerto Rico Reveals Insecticide-Specific Patterns of Gene Expression. Genes. 2023; 14(8):1626. https://doi.org/10.3390/genes14081626
Chicago/Turabian StyleDerilus, Dieunel, Lucy Mackenzie Impoinvil, Ephantus J. Muturi, Janet McAllister, Joan Kenney, Steven E. Massey, Ryan Hemme, Linda Kothera, and Audrey Lenhart. 2023. "Comparative Transcriptomic Analysis of Insecticide-Resistant Aedes aegypti from Puerto Rico Reveals Insecticide-Specific Patterns of Gene Expression" Genes 14, no. 8: 1626. https://doi.org/10.3390/genes14081626
APA StyleDerilus, D., Impoinvil, L. M., Muturi, E. J., McAllister, J., Kenney, J., Massey, S. E., Hemme, R., Kothera, L., & Lenhart, A. (2023). Comparative Transcriptomic Analysis of Insecticide-Resistant Aedes aegypti from Puerto Rico Reveals Insecticide-Specific Patterns of Gene Expression. Genes, 14(8), 1626. https://doi.org/10.3390/genes14081626







