Role of Actin Dynamics and GhACTIN1 Gene in Cotton Fiber Development: A Prototypical Cell for Study
Abstract
:1. Introduction
2. Qualitative Traits of Cotton Fiber
2.1. Fiber Length
2.2. Fiber Strength
2.3. Micronaire Value
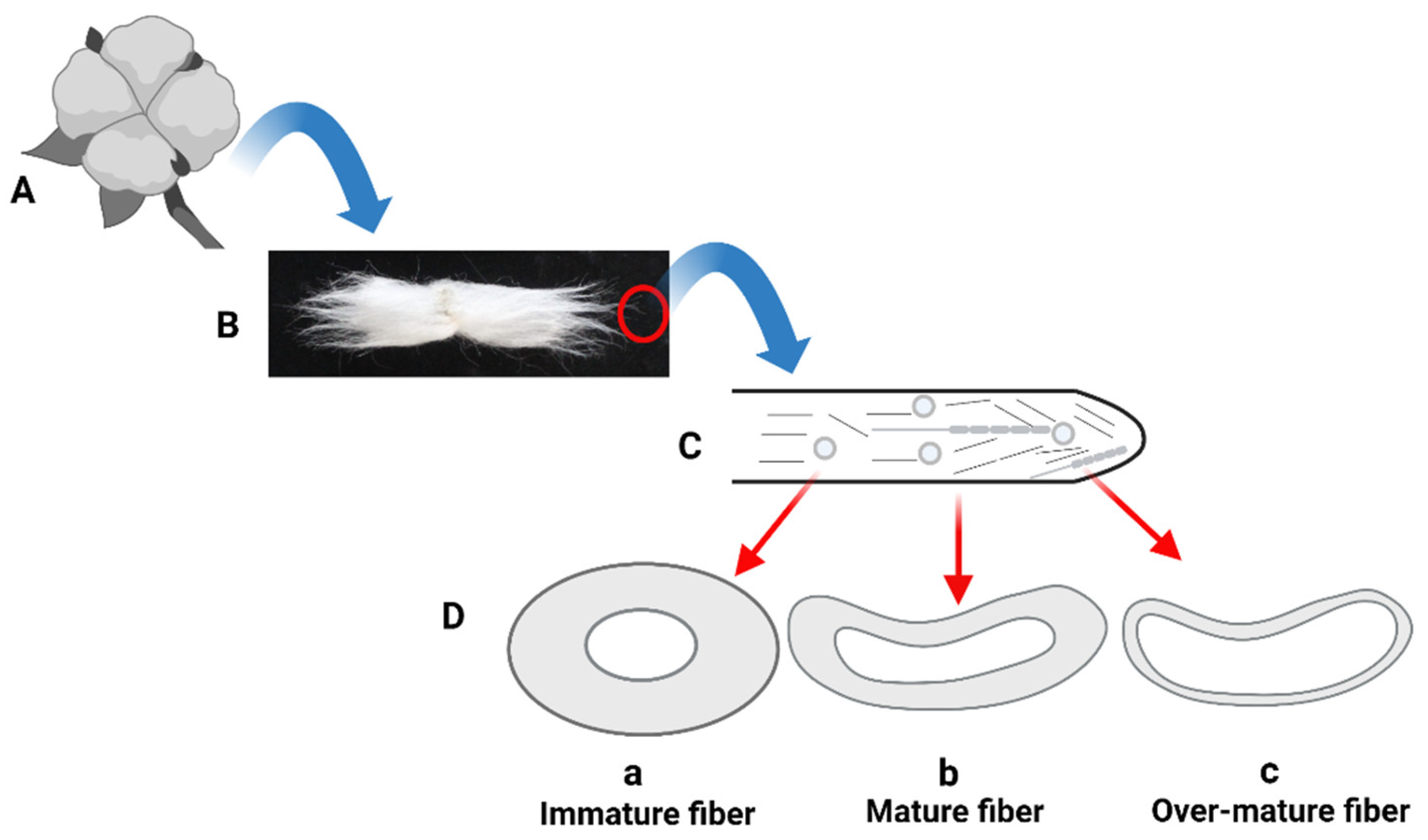
2.4. Fiber Maturity
2.5. Fiber Fineness
3. Cotton Fiber Development Stages
3.1. The Initiation Stage of Cotton Fiber
3.2. Role of Phytohormones in Fiber Initiation
3.3. Cotton Fiber Elongation Stage
3.4. Secondary Cell Wall Synthesis
3.5. Cotton Fiber Maturation Stage
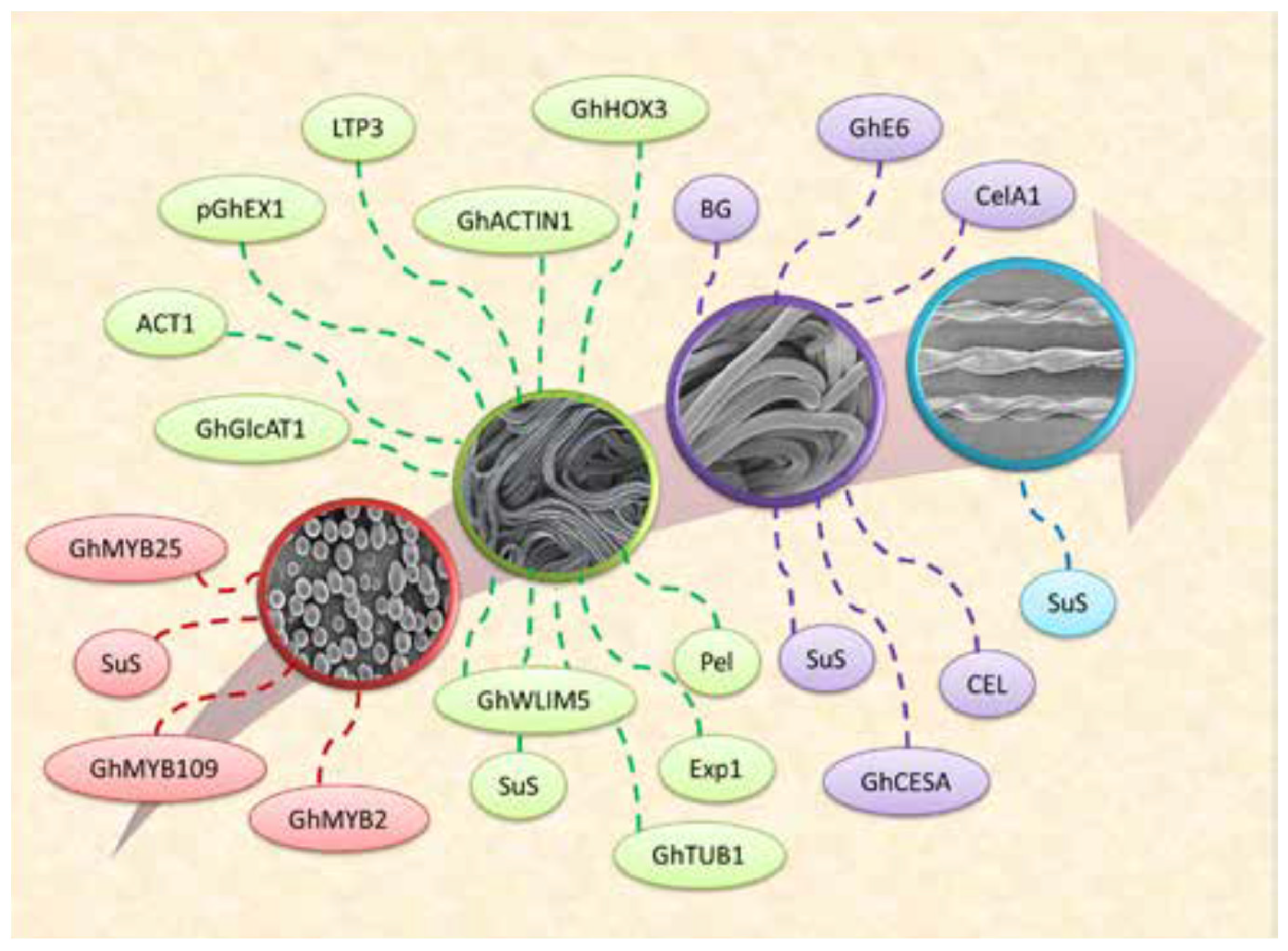
4. Role of ACTIN Genes in Cotton Fiber Development
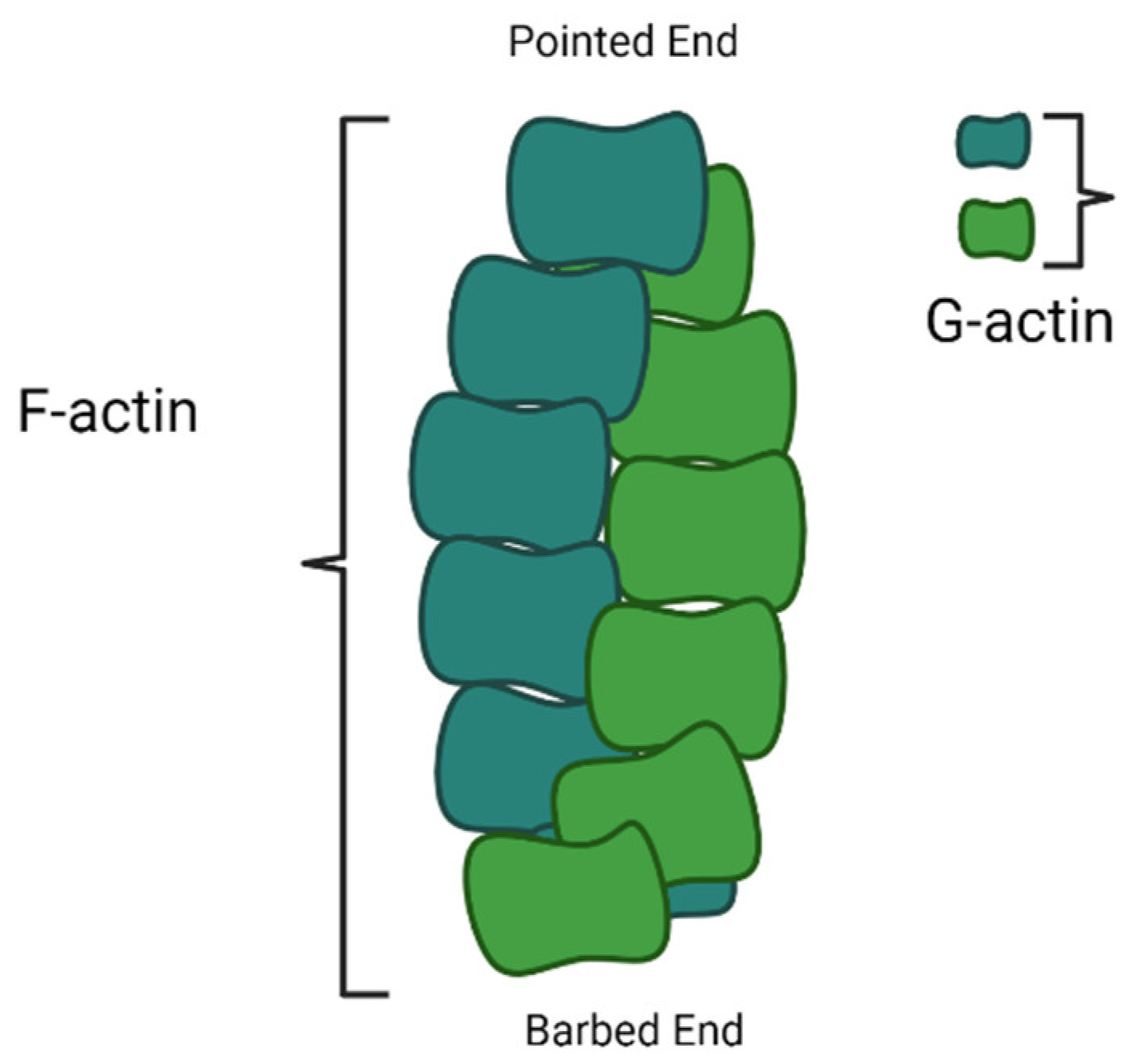
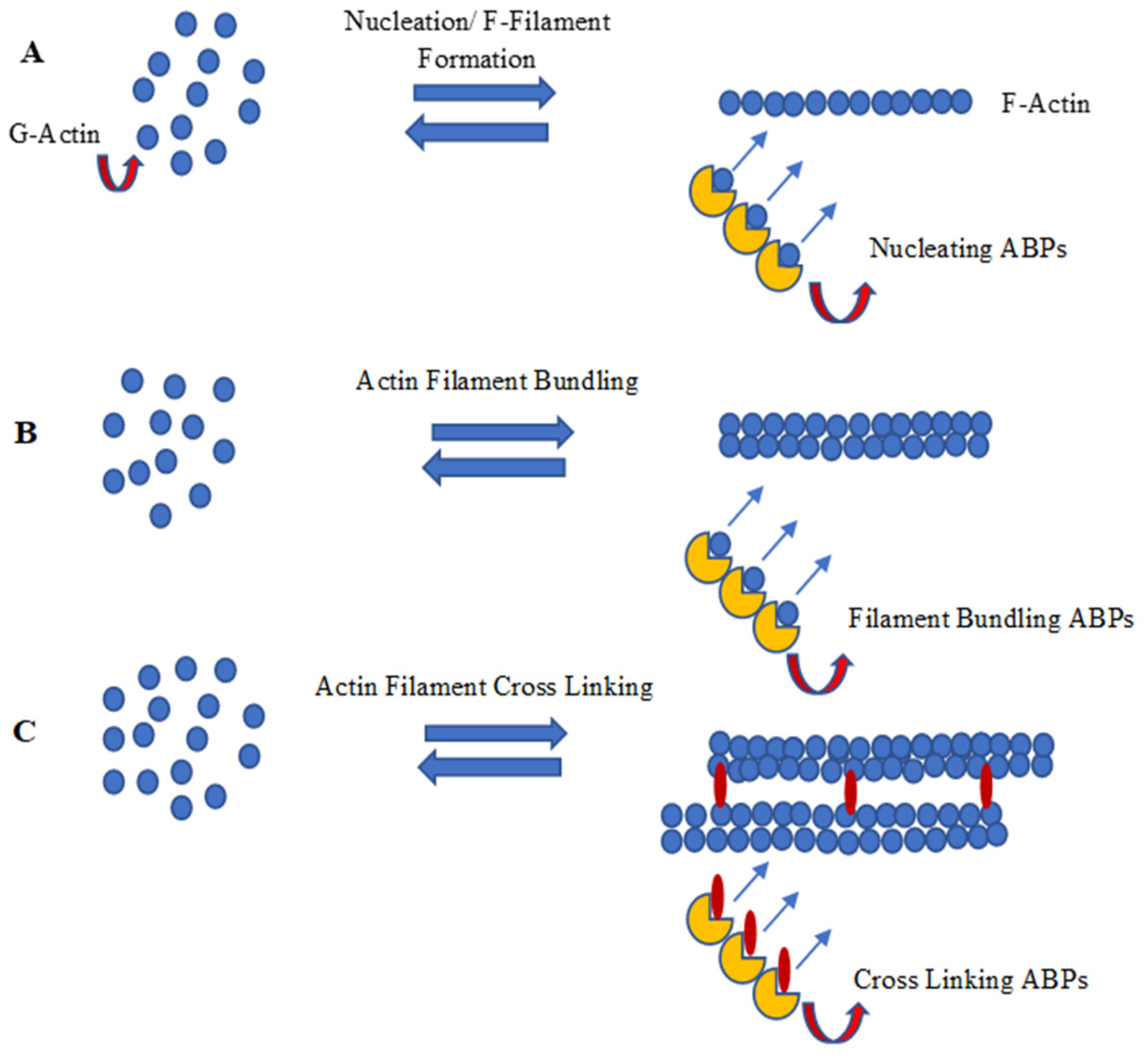
4.1. Actin Filament Development Pathway and Actin-Binding Proteins (ABPs)
4.1.1. Nucleation of Actin Filaments
4.1.2. Polymerization and Capping of Microfilament
4.1.3. F-Actin Bundling and Cross Linking
4.1.4. Plant LIM, an Actin-Bundling Protein
5. Classification and Function of Plant Actin
6. Role of GhACTIN1 Gene in Cotton Fiber Development
Regulation of Fiber Elongation by Interaction between Cotton-Annexin (Gbanx6) and ACTIN1
7. Biotechnological Approach of Genetic Transformation to Improve Cotton Fiber
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Bradow, J.M.; Hinojosa, O.; Wartelle, L.H.; Davidonis, G.; Sassenrath-Cole, G.F.; Bauer, P.J. Applications of AFIS fineness and maturity module and X-ray fluorescence spectroscopy in fiber maturity evaluation. Text. Res. J. 1996, 66, 545–554. [Google Scholar] [CrossRef]
- Haigler, C.H.; Zhang, D.; Wilkerson, C.G. Biotechnological improvement of cotton fibre maturity. Physiol. Plant. 2005, 124, 285–294. [Google Scholar] [CrossRef]
- Li, C.Q.; Song, L.; Zhao, H.H.; Wang, Q.L.; Fu, Y.Z. Identification of quantitative trait loci with main and epistatic effects for plant architecture traits in Upland cotton (Gossypium hirsutum L.). Plant Breed. 2014, 133, 390–400. [Google Scholar] [CrossRef]
- May, O.L. Quality improvement of upland cotton (Gossypium hirsutum L.). J. Crop Prod. 2002, 5, 371–394. [Google Scholar] [CrossRef]
- Gipson, J.; Joham, H. Influence of night temperature on growth and development of cotton (Gossypium hirsutum L.). III. fiber elongation 1. Crop Sci. 1969, 9, 127–129. [Google Scholar] [CrossRef]
- Behery, H.M. Short Fiber Content and Uniformity Index in Cotton; International Cotton Advisory Committee Review Article No. 4; CAB International: Wallingford, UK, 1993. [Google Scholar]
- Krifa, M. Fiber length distribution in cotton processing: A finite mixture distribution model. Text. Res. J. 2008, 78, 688–698. [Google Scholar] [CrossRef]
- Patil, N.; Singh, M. Development of medium staple high-strength cotton suitable for rotor spinning systems. In Proceedings of the Challenging the Future, World Cotton Conference I, Brisbane, Australia, 14–17 February 1994; pp. 14–17. [Google Scholar]
- Wakelyn, P.J.; Bertoniere, N.R.; French, A.D.; Thibodeaux, D.P.; Triplett, B.A.; Rousselle, M.-A.; Goynes, W.R., Jr.; Edwards, J.V.; Hunter, L.; McAlister, D.D. Cotton Fiber Chemistry and Technology; CRC Press: Boca Raton, FL, USA, 2006. [Google Scholar]
- Taylor, R. High speed measurements of strength and elongation. In Proceedings of the World Cotton Research Conference I, Brisbane, Australia, 14–17 February 1994; pp. 268–273. [Google Scholar]
- Lord, E.; Heap, S. The Origin and Assessment of Cotton Fibre Maturity; International Institute for Cotton: Manchester, UK, 1981. [Google Scholar]
- Ramey, H. The Meaning and Assessment of Cotton Fibre Fineness; International Institute for Cotton: Manchester, UK, 1982. [Google Scholar]
- El Mogahzy, Y.E.; Gowayed, Y. Theory and Practice of Cotton Fiber Selection: Part II: Sources of Cotton Mix Variability and Critical Factors Affecting It. Text. Res. J. 1995, 65, 75–84. [Google Scholar] [CrossRef]
- Schwartz, B.M.; Smith, C.W. Genetic gain in fiber properties of upland cotton under varying plant densities. Crop Sci. 2008, 48, 1321–1327. [Google Scholar] [CrossRef]
- Kim, H.J.; Tang, Y.; Moon, H.S.; Delhom, C.D.; Fang, D.D. Functional analyses of cotton (Gossypium hirsutum L.) immature fiber (im) mutant infer that fiber cell wall development is associated with stress responses. BMC Genom. 2013, 14, 889. [Google Scholar] [CrossRef]
- Thibodeaux, D.; Rajasekaran, K. Development of new reference standards for cotton fiber maturity. J. Cotton Sci. 1999, 3, 188–193. [Google Scholar]
- Lawrence, C.A. Advances in Yarn Spinning Technology; Elsevier: Amsterdam, The Netherlands, 2010. [Google Scholar]
- Basra, A.; Saha, S.B. Growth regulation of cotton fibers. In Cotton Fibers: Developmental Biology, Quality Improvement, and Textile Processing; Basra, A.S., Ed.; The Haworth Press: New York, NY, USA, 1999; pp. 47–58. [Google Scholar]
- Kim, H.J.; Triplett, B.A. Cotton fiber growth in planta and in vitro. Models for plant cell elongation and cell wall biogenesis. Plant Physiol. 2001, 127, 1361–1366. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.J.; Woodward, A.W.; Chen, Z.J. Gene expression changes and early events in cotton fibre development. Ann. Bot. 2007, 100, 1391–1401. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Xu, X.; Wang, M.; Li, C.; Li, C.; Zhao, R.; Zhu, S.; He, Q.; Chen, J. Identification and profiling of upland cotton microRNAs at fiber initiation stage under exogenous IAA application. BMC Genom. 2019, 20, 421. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; He, P.; Yang, Z.; Huang, G.; Wang, L.; Pang, C.; Xiao, H.; Zhao, P.; Yu, J.; Xiao, G. A genome-scale analysis of the PIN gene family reveals its functions in cotton fiber development. Front. Plant Sci. 2017, 8, 461. [Google Scholar] [CrossRef] [PubMed]
- Guan, X.; Lee, J.J.; Pang, M.; Shi, X.; Stelly, D.M.; Chen, Z.J. Activation of Arabidopsis seed hair development by cotton fiber-related genes. PLoS ONE 2011, 6, e21301. [Google Scholar] [CrossRef]
- Wang, S.; Wang, J.-W.; Yu, N.; Li, C.-H.; Luo, B.; Gou, J.-Y.; Wang, L.-J.; Chen, X.-Y. Control of plant trichome development by a cotton fiber MYB gene. Plant Cell 2004, 16, 2323–2334. [Google Scholar] [CrossRef]
- Huang, Y.; Liu, X.; Tang, K.; Zuo, K. Functional analysis of the seed coat-specific gene GbMYB2 from cotton. Plant Physiol. Biochem. 2013, 73, 16–22. [Google Scholar] [CrossRef]
- Walford, S.A.; Wu, Y.; Llewellyn, D.J.; Dennis, E.S. Epidermal cell differentiation in cotton mediated by the homeodomain leucine zipper gene, GhHD-1. Plant J. 2012, 71, 464–478. [Google Scholar] [CrossRef]
- John, M.E. Characterization of a cotton (Gossypium hirsutum L.) fiber mRNA (Fb-B6). Plant Physiol. 1995, 107, 1477. [Google Scholar] [CrossRef]
- Ruan, Y.-L.; Llewellyn, D.J.; Furbank, R.T. Suppression of sucrose synthase gene expression represses cotton fiber cell initiation, elongation, and seed development. Plant Cell 2003, 15, 952–964. [Google Scholar] [CrossRef]
- Zhang, M.; Zeng, J.-Y.; Long, H.; Xiao, Y.-H.; Yan, X.-Y.; Pei, Y. Auxin regulates cotton fiber initiation via GhPIN-mediated auxin transport. Plant Cell Physiol. 2017, 58, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Beasley, C. Hormonal regulation of growth in unfertilized cotton ovules. Science 1973, 179, 1003–1005. [Google Scholar] [CrossRef] [PubMed]
- Gialvalis, S.; Seagull, R.W. Plant hormones alter fiber initiation in unfertilized, cultured ovules of Gossypium hirsutum. J. Cotton Sci. 2001, 5, 252–258. [Google Scholar]
- Beasley, C.; Ting, I.P. The effects of plant growth substances on in vitro fiber development from fertilized cotton ovules. Am. J. Bot. 1973, 60, 130–139. [Google Scholar] [CrossRef]
- Qin, Y.; Sun, H.; Hao, P.; Wang, H.; Wang, C.; Ma, L.; Wei, H.; Yu, S. Transcriptome analysis reveals differences in the mechanisms of fiber initiation and elongation between long-and short-fiber cotton (Gossypium hirsutum L.) lines. BMC Genom. 2019, 20, 633. [Google Scholar] [CrossRef]
- Ruan, Y.-L.; Llewellyn, D.J.; Furbank, R.T. The control of single-celled cotton fiber elongation by developmentally reversible gating of plasmodesmata and coordinated expression of sucrose and K+ transporters and expansin. Plant Cell 2001, 13, 47–60. [Google Scholar]
- Kawai, M.; Aotsuka, S.; Uchimiya, H. Isolation of a cotton CAP gene: A homologue of adenylyl cyclase-associated protein highly expressed during fiber elongation. Plant Cell Physiol. 1998, 39, 1380–1383. [Google Scholar] [CrossRef]
- Xiao, G.; Zhao, P.; Zhang, Y. A pivotal role of hormones in regulating cotton fiber development. Front. Plant Sci. 2019, 10, 87. [Google Scholar] [CrossRef]
- Ahmed, M.; Iqbal, A.; Latif, A.; Din, S.U.; Sarwar, M.B.; Wang, X.; Rao, A.Q.; Husnain, T.; Shahid, A.A. Overexpression of a sucrose synthase gene indirectly improves cotton fiber quality through sucrose cleavage. Front. Plant Sci. 2020, 11, 476251. [Google Scholar] [CrossRef]
- Hu, W.; Chen, L.; Qiu, X.; Wei, J.; Lu, H.; Sun, G.; Ma, X.; Yang, Z.; Zhu, C.; Hou, Y. AKR2A participates in the regulation of cotton fibre development by modulating biosynthesis of very-long-chain fatty acids. Plant Biotechnol. J. 2020, 18, 520–539. [Google Scholar] [CrossRef]
- Tang, W.; Tu, L.; Yang, X.; Tan, J.; Deng, F.; Hao, J.; Guo, K.; Lindsey, K.; Zhang, X. The calcium sensor G h C a M 7 promotes cotton fiber elongation by modulating reactive oxygen species (ROS) production. New Phytol. 2014, 202, 509–520. [Google Scholar] [CrossRef]
- Iqbal, A.; Latif, A.; Galbraith, D.W.; Jabbar, B.; Ali, M.A.; Ahmed, M.; Gul, A.; Rao, A.Q.; Shahid, A.A.; Husnain, T. Structure-based prediction of protein–protein interactions between GhWlim5 Domain1 and GhACTIN-1 proteins: A practical evidence with improved fibre strength. J. Plant Biochem. Biotechnol. 2021, 30, 373–386. [Google Scholar] [CrossRef]
- Li, Y.; Jiang, J.; Li, L.; Wang, X.-L.; Wang, N.-N.; Li, D.-D.; Li, X.-B. A cotton LIM domain-containing protein (GhWLIM5) is involved in bundling actin filaments. Plant Physiol. Biochem. 2013, 66, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Lv, F.; Wang, H.; Wang, X.; Han, L.; Ma, Y.; Wang, S.; Feng, Z.; Niu, X.; Cai, C.; Kong, Z. GhCFE1A, a dynamic linker between the ER network and actin cytoskeleton, plays an important role in cotton fibre cell initiation and elongation. J. Exp. Bot. 2015, 66, 1877–1889. [Google Scholar] [CrossRef]
- Shan, C.-M.; Shangguan, X.-X.; Zhao, B.; Zhang, X.-F.; Chao, L.-m.; Yang, C.-Q.; Wang, L.-J.; Zhu, H.-Y.; Zeng, Y.-D.; Guo, W.-Z. Control of cotton fibre elongation by a homeodomain transcription factor GhHOX3. Nat. Commun. 2014, 5, 5519. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-R.; Wang, L.; Ruan, Y.-L. Developmental and molecular physiological evidence for the role of phospho enol pyruvate carboxylase in rapid cotton fibre elongation. J. Exp. Bot. 2010, 61, 287–295. [Google Scholar] [CrossRef]
- Li, X.-B.; Fan, X.-P.; Wang, X.-L.; Cai, L.; Yang, W.-C. The cotton ACTIN1 gene is functionally expressed in fibers and participates in fiber elongation. Plant Cell 2005, 17, 859–875. [Google Scholar] [CrossRef]
- Gokani, S.; Thaker, V. Physiological and biochemical changes associated with cotton fiber development. VIII. Wall components. Acta Physiol. Plant. 2000, 22, 403–408. [Google Scholar] [CrossRef]
- Qin, Y.-M.; Hu, C.-Y.; Pang, Y.; Kastaniotis, A.J.; Hiltunen, J.K.; Zhu, Y.-X. Saturated very-long-chain fatty acids promote cotton fiber and Arabidopsis cell elongation by activating ethylene biosynthesis. Plant Cell 2007, 19, 3692–3704. [Google Scholar] [CrossRef]
- Niu, Q.; Tan, K.; Zang, Z.; Xiao, Z.; Chen, K.; Hu, M.; Luo, M. Modification of phytosterol composition influences cotton fiber cell elongation and secondary cell wall deposition. BMC Plant Biol. 2019, 19, 208. [Google Scholar] [CrossRef]
- Han, L.-B.; Li, Y.-B.; Wang, H.-Y.; Wu, X.-M.; Li, C.-L.; Luo, M.; Wu, S.-J.; Kong, Z.-S.; Pei, Y.; Jiao, G.-L. The dual functions of WLIM1a in cell elongation and secondary wall formation in developing cotton fibers. Plant Cell 2013, 25, 4421–4439. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.Y.; Wang, J.; Gao, P.; Jiao, G.L.; Zhao, P.M.; Li, Y.; Wang, G.L.; Xia, G.X. Down-regulation of GhADF1 gene expression affects cotton fibre properties. Plant Biotechnol. J. 2009, 7, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, H.-Y.; Zhao, P.-M.; Han, L.-B.; Jiao, G.-L.; Zheng, Y.-Y.; Huang, S.-J.; Xia, G.-X. Overexpression of a profilin (GhPFN2) promotes the progression of developmental phases in cotton fibers. Plant Cell Physiol. 2010, 51, 1276–1290. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Lv, J.; Zhao, L.; Tong, X.; Zhou, B.; Zhang, T.; Guo, W. Molecular evolution and phylogenetic analysis of genes related to cotton fibers development from wild and domesticated cotton species in Gossypium. Mol. Phylogenetics 2012, 63, 589–597. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.-L.; Wang, X.-L.; Wang, H.; Li, X.-B. Molecular characterization and expression analysis of nine cotton GhEF1A genes encoding translation elongation factor 1A. Gene 2007, 389, 27–35. [Google Scholar] [CrossRef]
- Gwathmey, C.O.; Bange, M.P.; Brodrick, R. Cotton crop maturity: A compendium of measures and predictors. Field Crops Res. 2016, 191, 41–53. [Google Scholar] [CrossRef]
- Su, J.; Wang, C.; Hao, F.; Ma, Q.; Wang, J.; Li, J.; Ning, X. Genetic Detection of Lint Percentage Applying Single-Locus and Multi-Locus Genome-Wide Association Studies in Chinese Early-Maturity Upland Cotton. Front. Plant Sci. 2019, 10, 964. [Google Scholar] [CrossRef]
- Latif, A.; Ahmed, M.; Akhtar, S.; Ahad, A.; Iqbal, A.; Yaqoob, A.; Imran, A.; Usmaan, M.; Shahid, N.; Azam, S.; et al. Cotton Fibre Quality Management for Sustainable Textile Industry. ICAC Rec. 2019, XXXVII, 22–27. [Google Scholar]
- Hussey, P.J.; Ketelaar, T.; Deeks, M.J. Control of the actin cytoskeleton in plant cell growth. Annu. Rev. Plant Biol. 2006, 57, 109–125. [Google Scholar] [CrossRef]
- Komis, G.; Luptovciak, I.; Doskocilova, A.; Samaj, J. Biotechnological aspects of cytoskeletal regulation in plants. Biotechnol. Adv. 2015, 33, 1043–1062. [Google Scholar] [CrossRef]
- Paavilainen, V.O.; Bertling, E.; Falck, S.; Lappalainen, P. Regulation of cytoskeletal dynamics by actin-monomer-binding proteins. Trends Cell Biol. 2004, 14, 386–394. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, M.; Zhang, C.; Szymanski, D.B. ARP2/3-dependent growth in the plant kingdom: SCARs for life. Front. Plant Sci. 2013, 4, 166. [Google Scholar] [CrossRef] [PubMed]
- Winder, S.J.; Ayscough, K.R. Actin-binding proteins. J. Cell Sci. 2005, 118, 651–654. [Google Scholar] [CrossRef] [PubMed]
- Goode, B.L.; Eck, M.J. Mechanism and function of formins in the control of actin assembly. Annu. Rev. Biochem. 2007, 76, 593–627. [Google Scholar] [CrossRef]
- Allingham, J.S.; Miles, C.O.; Rayment, I. A structural basis for regulation of actin polymerization by pectenotoxins. J. Mol. Biol. 2007, 371, 959–970. [Google Scholar] [CrossRef]
- Winder, S.J.; Jess, T.; Ayscough, K.R. SCP1 encodes an actin-bundling protein in yeast. Biochem. J. 2003, 375, 287–295. [Google Scholar] [CrossRef]
- Thomas, C.; Moreau, F.; Dieterle, M.; Hoffmann, C.; Gatti, S.; Hofmann, C.; Van Troys, M.; Ampe, C.; Steinmetz, A. The LIM domains of WLIM1 define a new class of actin bundling modules. J. Biol. Chem. 2007, 282, 33599–33608. [Google Scholar] [CrossRef]
- Han, L.; Li, Y.; Sun, Y.; Wang, H.; Kong, Z.; Xia, G. The two domains of cotton WLIM1a protein are functionally divergent. Sci. China Life Sci. 2016, 59, 206–212. [Google Scholar] [CrossRef]
- Zheng, Q.; Zhao, Y. The diverse biofunctions of LIM domain proteins: Determined by subcellular localization and protein—Protein interaction. Biol. Cell 2007, 99, 489–502. [Google Scholar] [CrossRef]
- Li, L.; Li, Y.; Wang, N.N.; Li, Y.; Lu, R.; Li, X.B. Cotton LIM domain-containing protein Gh PLIM 1 is specifically expressed in anthers and participates in modulating F-actin. Plant Biol. 2015, 17, 528–534. [Google Scholar] [CrossRef]
- Kadrmas, J.L.; Beckerle, M.C. The LIM domain: From the cytoskeleton to the nucleus. Nat. Rev. Mol. Cell Biol. 2004, 5, 920. [Google Scholar] [CrossRef] [PubMed]
- Arnaud, D.; Dejardin, A.; Leple, J.C.; Lesage-Descauses, M.C.; Boizot, N.; Villar, M.; Benedetti, H.; Pilate, G. Expression analysis of LIM gene family in poplar, toward an updated phylogenetic classification. BMC Res. Notes 2012, 5, 102. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.-H.; Xia, G.-X.; Hong, Y.; Ramachandran, S.; Kost, B.; Chua, N.-H. ADF proteins are involved in the control of flowering and regulate F-actin organization, cell expansion, and organ growth in Arabidopsis. Plant Cell 2001, 13, 1333–1346. [Google Scholar] [CrossRef] [PubMed]
- Kost, B.; Mathur, J.; Chua, N.-H. Cytoskeleton in plant development. Curr. Opin. Plant Biol. 1999, 2, 462–470. [Google Scholar] [CrossRef]
- Sahu, M.; Dehury, B.; Sarmah, R.; Sahoo, S.; Sahu, J.; Sarma, K.; Sen, P.; Modi, M.K.; Barooah, M. In silico prediction and characterization of three-dimensional structure of Actin-1 of Arabidopsis thaliana. BioTechnol. J. Biotechnol. Comput. Biol. Bionanotechnol. 2013, 94, 432–443. [Google Scholar] [CrossRef]
- Kandasamy, M.K.; McKinney, E.C.; Meagher, R.B. Functional nonequivalency of actin isovariants in Arabidopsis. Mol. Biol. Cell 2002, 13, 251–261. [Google Scholar] [CrossRef]
- McDowell, J.M.; Huang, S.; McKinney, E.C.; An, Y.-Q.; Meagher, R.B. Structure and evolution of the actin gene family in Arabidopsis thaliana. Genetics 1996, 142, 587–602. [Google Scholar] [CrossRef]
- Huang, S.; An, Y.Q.; McDowell, J.M.; McKinney, E.C.; Meagher, R.B. The Arabidopsis thaliana ACT4/ACT12 actin gene subclass is strongly expressed throughout pollen development. Plant J. 1996, 10, 189–202. [Google Scholar] [CrossRef]
- Dixon, D.C.; Meredith, J.; William, R.; Triplett, B.A. An Assessment ofα-Tubulin Isotype Modification in Developing Cotton Fiber. Int. J. Plant Sci. 2000, 161, 63–67. [Google Scholar] [CrossRef]
- Ji, S.; Lu, Y.; Li, J.; Wei, G.; Liang, X.; Zhu, Y. A β-tubulin-like cDNA expressed specifically in elongating cotton fibers induces longitudinal growth of fission yeast. Biochem. Biophys. Res. Commun. 2002, 296, 1245–1250. [Google Scholar] [CrossRef]
- Pei, W.; Du, F.; Zhang, Y.; He, T.; Ren, H. Control of the actin cytoskeleton in root hair development. Plant Sci. 2012, 187, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Boustead, C.M.; Smallwood, M.; Small, H.; Bowles, D.J.; Walker, J.H. Identification of calcium-dependent phospholipid-binding proteins in higher plant cells. Febs Lett. 1989, 244, 456–460. [Google Scholar] [CrossRef]
- Creutz, C.E.; Pazoles, C.J.; Pollard, H.B.J. Identification and purification of an adrenal medullary protein (synexin) that causes calcium-dependent aggregation of isolated chromaffin granules. J. Biol. Chem. 1978, 253, 2858–2866. [Google Scholar] [CrossRef] [PubMed]
- Laohavisit, A.; Mortimer, J.C.; Demidchik, V.; Coxon, K.M.; Stancombe, M.A.; Macpherson, N.; Brownlee, C.; Hofmann, A.; Webb, A.A.; Miedema, H. Zea mays annexins modulate cytosolic free Ca2+ and generate a Ca2+-permeable conductance. Plant Cell 2009, 21, 479–493. [Google Scholar] [CrossRef] [PubMed]
- Blackbourn, H.D.; Barker, P.J.; Huskisson, N.S.; Battey, N.H. Properties and partial protein sequence of plant annexins. Plant Physiol. 1992, 99, 864–871. [Google Scholar] [CrossRef]
- Kwon, H.-K.; Yokoyama, R.; Nishitani, K. A proteomic approach to apoplastic proteins involved in cell wall regeneration in protoplasts of Arabidopsis suspension-cultured cells. Plant Cell Physiol. 2005, 46, 843–857. [Google Scholar] [CrossRef]
- Morel, E.; Gruenberg, J. Annexin A2 binding to endosomes and functions in endosomal transport are regulated by tyrosine 23 phosphorylation. J. Biol. Chem. 2009, 284, 1604–1611. [Google Scholar] [CrossRef]
- Reddy, V.S.; Reddy, A.S.P. Proteomics of calcium-signaling components in plants. Phytochemistry 2004, 65, 1745–1776. [Google Scholar] [CrossRef]
- Bassani, M.; Neumann, P.M.; Gepstein, S. Differential expression profiles of growth-related genes in the elongation zone of maize primary roots. Plant Mol. Biol. 2004, 56, 367–380. [Google Scholar] [CrossRef]
- Dai, S.; Li, L.; Chen, T.; Chong, K.; Xue, Y.; Wang, T. Proteomic analyses of Oryza sativa mature pollen reveal novel proteins associated with pollen germination and tube growth. Proteomics 2006, 6, 2504–2529. [Google Scholar] [CrossRef]
- Andrawis, A.; Solomon, M.; Delmer, D.P. Cotton fiber annexins: A potential role in the regulation of callose synthase. Plant J. 1993, 3, 763–772. [Google Scholar] [CrossRef] [PubMed]
- Dhaliwal, G.; Jindal, V.; Mohindru, B. Crop losses due to insect pests: Global and Indian scenario. Indian J. Entomol. 2015, 77, 165–168. [Google Scholar] [CrossRef]
- Iqbal, A.; Ali, M.A.; Ahmed, S.; Hassan, S.; Shahid, N.; Azam, S.; Rao, A.Q.; Ali, Q.; Shahid, A.A. Engineered resistance and risk assessment associated with insecticidal and weeds resistant transgenic cotton using wister rat model. Sci. Rep. 2022, 12, 2518. [Google Scholar] [CrossRef]
- Latif, A.; Rao, A.Q.; Khan, M.A.; Shahid, N.; Bajwa, K.S.; Ashraf, M.A.; Abbas, M.A.; Azam, M.; Shahid, A.A.; Nasir, I.A.; et al. Herbicide-resistant cotton (Gossypium hirsutum) plants: An alternative way of manual weed removal. BMC Res. Notes 2015, 8, 453. [Google Scholar] [CrossRef] [PubMed]
- Farooq, J.; Farooq, A.; Rizwan, M.; Petrescu-Mag, I.V.; Ali, M.A.; Mahmood, K.; Batool, A. Cotton fibers: Attributes of specialized cells and factors affecting them. Adv. Environ. Sci. 2015, 7, 369–382. [Google Scholar]
- Shahid, M.R.; Farooq, J.; Mahmood, A.; Ilahi, F.; Riaz, M.; Shakeel, A.; Petrescu-Mag, I.V.; Farooq, A. Seasonal occurrence of sucking insect pest in cotton ecosystem of Punjab, Pakistan. Adv. Agric. 2012, 4, 26–30. [Google Scholar]
- Khan, A.; Tan, D.K.Y.; Afridi, M.Z.; Luo, H.; Tung, S.A.; Ajab, M.; Fahad, S. Nitrogen fertility and abiotic stresses management in cotton crop: A review. Environ. Sci. Pollut. Res. 2017, 24, 14551–14566. [Google Scholar] [CrossRef]
- Hemphill, J.K.; Basal, H.; Smith, W. Screening method for salt tolerance in cotton. Am. J. Plant Pathol. 2006, 1, 107–112. [Google Scholar]
- Barrick, B.; Steiner, R.; Picchioni, G.; Ulery, A.; Zhang, J. Salinity responses of selected introgressed cotton lines grown in two soils from organic and conventional cotton production. J. Cotton Sci. 2015, 19, 268–278. [Google Scholar]
- Alghabari, F.; Ihsan, M.Z.; Khaliq, A.; Hussain, S.; Daur, I.; Fahad, S.; Nasim, W. Gibberellin-sensitive Rht alleles confer tolerance to heat and drought stresses in wheat at booting stage. J. Cereal Sci. 2016, 70, 72–78. [Google Scholar] [CrossRef]
- Noman, A.; Fahad, S.; Aqeel, M.; Ali, U.; Anwar, S.; Baloch, S.K.; Zainab, M. miRNAs: Major modulators for crop growth and development under abiotic stresses. Biotechnol. Lett. 2017, 39, 685–700. [Google Scholar] [CrossRef] [PubMed]
- Snider, J.L.; Oosterhuis, D.M.; Skulman, B.W.; Kawakami, E.M. Heat stress-induced limitations to reproductive success in Gossypium hirsutum. Physiol. Plant. 2009, 137, 125–138. [Google Scholar] [CrossRef] [PubMed]
- Fahad, S.; Hussain, S.; Saud, S.; Khan, F.; Hassan, S.; Nasim, W.; Arif, M.; Wang, F.; Huang, J. Exogenously applied plant growth regulators affect heat-stressed rice pollens. J. Agron. Crop Sci. 2016, 202, 139–150. [Google Scholar] [CrossRef]
- Fahad, S.; Hussain, S.; Saud, S.; Tanveer, M.; Bajwa, A.A.; Hassan, S.; Shah, A.N.; Ullah, A.; Wu, C.; Khan, F.A. A biochar application protects rice pollen from high-temperature stress. Plant Physiol. Biochem. 2015, 96, 281–287. [Google Scholar] [CrossRef]
- Pettigrew, W. The effect of higher temperatures on cotton lint yield production and fiber quality. Crop Sci. 2008, 48, 278–285. [Google Scholar] [CrossRef]
- Conaty, W.C.; Mahan, J.R.; Neilsen, J.E.; Tan, D.K.; Yeates, S.J.; Sutton, B.G. The relationship between cotton canopy temperature and yield, fibre quality and water-use efficiency. Field Crops Res. 2015, 183, 329–341. [Google Scholar] [CrossRef]
- Ahmed, M.; Shahid, A.A.; Din, S.U.; Akhtar, S.; Ahad, A.; Rao, A.Q.; Bajwa, K.S.; Khan, M.A.U.; Sarwar, M.B.; Husnain, T. An overview of genetic and hormonal control of cotton fiber development. Pak. J. Bot 2018, 50, 433–443. [Google Scholar]
- Zhang, M.; Han, L.B.; Wang, W.Y.; Wu, S.J.; Jiao, G.L.; Su, L.; Xia, G.X.; Wang, H.Y. Overexpression of GhFIM2 propels cotton fiber development by enhancing actin bundle formation. J. Integr. Plant Biol. 2017, 59, 531–534. [Google Scholar] [CrossRef]
- Su, H.; Zhu, J.; Cai, C.; Pei, W.; Wang, J.; Dong, H.; Ren, H. FIMBRIN1 is involved in lily pollen tube growth by stabilizing the actin fringe. Plant Cell 2012, 24, 4539–4554. [Google Scholar] [CrossRef]
- Wu, Y.; Yan, J.; Zhang, R.; Qu, X.; Ren, S.; Chen, N.; Huang, S. Arabidopsis FIMBRIN5, an actin bundling factor, is required for pollen germination and pollen tube growth. Plant Cell 2010, 22, 3745–3763. [Google Scholar] [CrossRef]
- Haigler, C.H.; Singh, B.; Zhang, D.; Hwang, S.; Wu, C.; Cai, W.X.; Hozain, M.; Kang, W.; Kiedaisch, B.; Strauss, R.E. Transgenic cotton over-producing spinach sucrose phosphate synthase showed enhanced leaf sucrose synthesis and improved fiber quality under controlled environmental conditions. Plant Mol. Biol. 2007, 63, 815–832. [Google Scholar] [CrossRef] [PubMed]
- Haigler, C.H.; Singh, B.; Wang, G.; Zhang, D. Genomics of cotton fiber secondary wall deposition and cellulose biogenesis. In Genetics and Genomics of Cotton; Springer: Berlin/Heidelberg, Germany, 2009; pp. 385–417. [Google Scholar]
- Hou, L.; Liu, H.; Li, J.; Yang, X.; Xiao, Y.; Luo, M.; Song, S.; Yang, G.; Pei, Y. SCFP, a novel fiber-specific promoter in cotton. Chin. Sci. Bull. 2008, 53, 2639–2645. [Google Scholar] [CrossRef]
- Li, M.; Wang, S.; Liu, Y.; Zhang, Y.; Ren, M.; Liu, L.; Lu, T.; Wei, H.; Wei, Z. Overexpression of PsnSuSy1, 2 genes enhances secondary cell wall thickening, vegetative growth, and mechanical strength in transgenic tobacco. Plant Mol. Biol. 2019, 100, 215–230. [Google Scholar] [CrossRef] [PubMed]
- Manivannan, A.; Cheeran Amal, T. Deciphering the complex cotton genome for improving fiber traits and abiotic stress resilience in sustainable agriculture. Mol. Biol. Rep. 2023, 50, 6937–6953. [Google Scholar] [CrossRef] [PubMed]
| Gene | Function | Reference |
|---|---|---|
| GhPIN3a | Fiber initiation | [22] |
| GhMYB2 | Fiber initiation | [23] |
| GaMYB2 | Fiber initiation | [24] |
| GhMYB2 | Fiber initiation | [25] |
| GhHD1 | Fiber initiation | [26] |
| GhE6 | Fiber initiation | [27] |
| GhSuS | Fiber initiation and elongation | [28] |
| GhPIN1a_Dt, GhPIN6_At and GhPIN8_At | Fiber initiation and elongation | [29] |
| Gene | Function | Reference |
|---|---|---|
| SuS | Fiber elongation | [37] |
| AKR2A | Fiber elongation | [38] |
| GhCaM7 | Fiber elongation | [39] |
| WLIM5 | Fiber elongation | [40,41] |
| GhCFE1A | Fiber elongation | [42] |
| GhHOX3 | Fiber elongation | [43] |
| GhPEPC1, GhPEPC2 | Fiber elongation | [44] |
| GhACTIN1 | Fiber elongation | [40,45] |
| GhEXP1 | Fiber elongation | [34] |
| Gene | Function | Reference |
|---|---|---|
| GhSMT2–1 | Secondary wall synthesis | [48] |
| WLIM1a | Fiber elongation and secondary wall synthesis | [49] |
| GhADF1 | Fiber elongation and secondary wall synthesis | [50] |
| GhPFN2 | Fiber elongation and secondary wall synthesis | [51] |
| CelA1 | Fiber elongation and secondary wall synthesis | [52] |
| GhEF1A | Fiber elongation and secondary wall synthesis | [53] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iqbal, A.; Aslam, S.; Ahmed, M.; Khan, F.; Ali, Q.; Han, S. Role of Actin Dynamics and GhACTIN1 Gene in Cotton Fiber Development: A Prototypical Cell for Study. Genes 2023, 14, 1642. https://doi.org/10.3390/genes14081642
Iqbal A, Aslam S, Ahmed M, Khan F, Ali Q, Han S. Role of Actin Dynamics and GhACTIN1 Gene in Cotton Fiber Development: A Prototypical Cell for Study. Genes. 2023; 14(8):1642. https://doi.org/10.3390/genes14081642
Chicago/Turabian StyleIqbal, Adnan, Sibgha Aslam, Mukhtar Ahmed, Fahad Khan, Qurban Ali, and Shiming Han. 2023. "Role of Actin Dynamics and GhACTIN1 Gene in Cotton Fiber Development: A Prototypical Cell for Study" Genes 14, no. 8: 1642. https://doi.org/10.3390/genes14081642






