Changes in m6A in Steatotic Liver Disease
Abstract
:1. Introduction
2. N6-Methyladenosine (m6A)
m6A “Writers, Readers, and Erasers”
3. Methods for the Detection of m6A Modifications
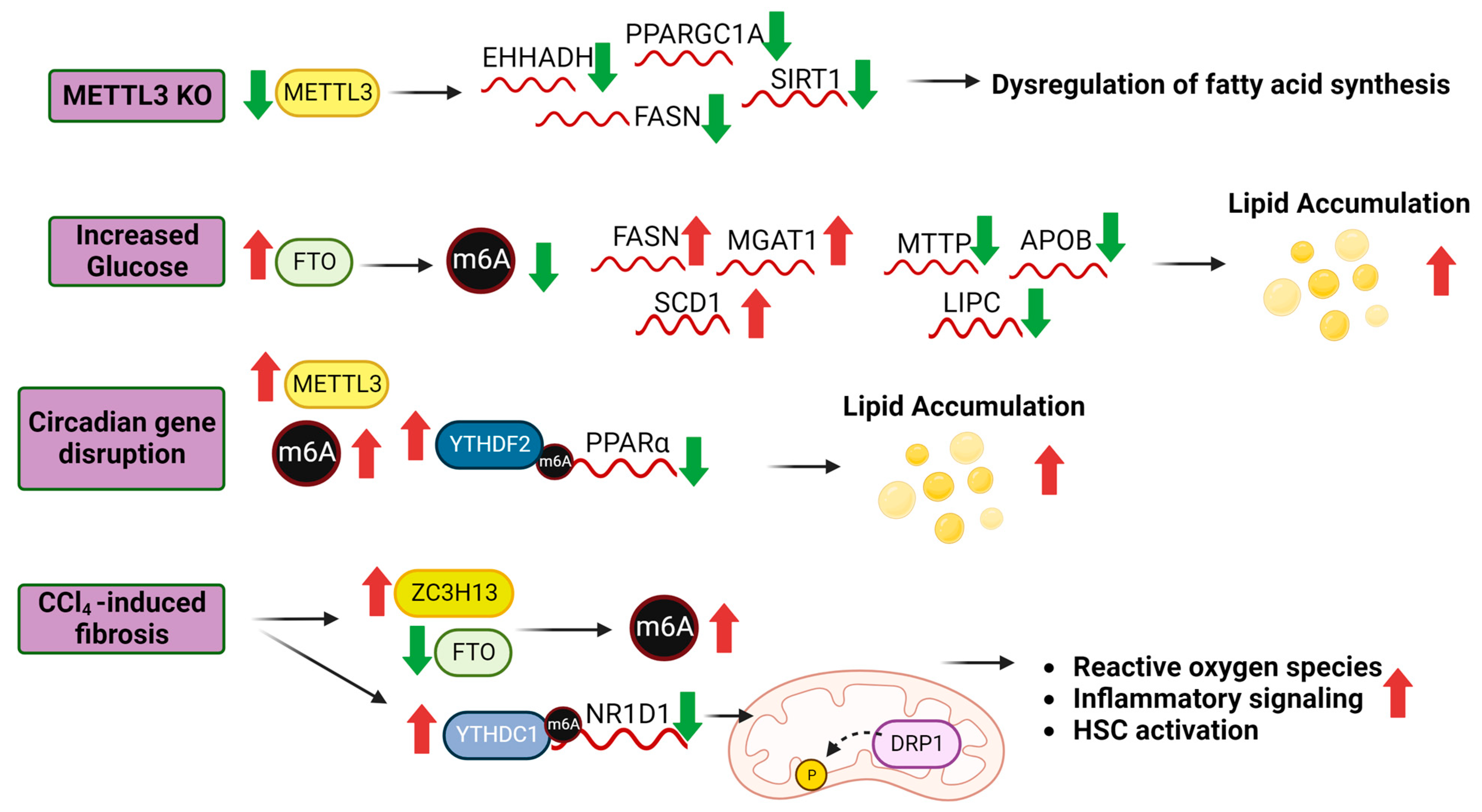
4. m6A in NAFLD
5. m6A and Liver Physiology
5.1. m6A and Lipid Metabolism
5.2. m6A and Glucose Metabolism
5.3. m6A and Hepatic Stellate Cell Activation
5.4. m6A and Hepatic Immune Response Signaling
6. Other mRNA Modifications in Liver
6.1. Pseudouridine (Ψ)
6.2. 1-Methyladenosine (m1A)
6.3. 5-Methylcytidine (m5C)
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ALD | alcohol-associated liver disease |
| ALKBH5 | alkB homolog 5 |
| AS | alternative splicing |
| ERα | estrogen receptor α |
| FTO | fat mass obesity-associated protein |
| FXR | farnesoid X receptor |
| HCC | hepatocellular carcinoma |
| HFD | high fat diet |
| HNRNP | heteronuclear family RNA binding proteins |
| HNRNPA1, HNRNPA2B1, HNRNPC, HNRNPG | |
| HSC | Hepatic stellate cells |
| IG2BP | Insulin-like growth factor 2 mRNA binding proteins |
| IGF2BP1, IGF2BP2, IGF2BP3 | |
| KC | Kupffer cells |
| lncRNAs | long non-coding RNAs |
| m6A | N-6-methyladenosine |
| MAC | m6A-METTL complex |
| MASLD | metabolic dysfunction-associated steatotic liver disease |
| MDCs | metabolism disrupting chemicals |
| METTL3 | methyltransferase like 3 |
| METTL14 | methyltransferase like 14 |
| NAFLD | non-alcoholic fatty liver disease |
| NASH | non-alcoholic steatohepatitis |
| PBXs | polychlorinated biphenyls |
| RBPs | RNA binding proteins |
| RWE | readers, writers, and erasers |
| SAM | S-adenosyl methionine |
| T2DM | type 2 diabetes mellitus |
| VIRMA | Vir like m6A methyltransferase associated |
| WTAP | Wilm’s tumor 1 (WT1)-associating protein |
| YTH | domain family proteins |
| YTHDF1, YTHDF2, YTHDF3, YTHDC1, and YTHDC2 | |
| ZC3H13 | zinc finger CCCH-type containing 13 |
References
- Loomba, R.; Friedman, S.L.; Shulman, G.I. Mechanisms and disease consequences of nonalcoholic fatty liver disease. Cell 2021, 184, 2537–2564. [Google Scholar] [CrossRef] [PubMed]
- Cotter, T.G.; Rinella, M. NAFLD 2020: The State of the Disease. Gastroenterology 2020, 158, 1851–1864. [Google Scholar] [CrossRef] [PubMed]
- Jones, G.S.; Alvarez, C.S.; Graubard, B.I.; McGlynn, K.A. Agreement between the Prevalence of Nonalcoholic Fatty Liver Disease Determined by Transient Elastography and Fatty Liver Indices. Clin. Gastroenterol. Hepatol. 2022, 20, 227–229.e2. [Google Scholar] [CrossRef] [PubMed]
- Hirsova, P.; Bamidele, A.O.; Wang, H.; Povero, D.; Revelo, X.S. Emerging Roles of T Cells in the Pathogenesis of Nonalcoholic Steatohepatitis and Hepatocellular Carcinoma. Front. Endocrinol. 2021, 12, 760860. [Google Scholar] [CrossRef]
- Rinella, M.E.; Lazarus, J.V.; Ratziu, V.; Francque, S.M.; Sanyal, A.J.; Kanwal, F.; Romero, D.; Abdelmalek, M.F.; Anstee, Q.M.; Arab, J.P.; et al. A multi-society Delphi consensus statement on new fatty liver disease nomenclature. J. Hepatol. 2023. online ahead of print. [Google Scholar]
- Basu, R.; Noureddin, M.; Clark, J.M. Nonalcoholic Fatty Liver Disease: Review of Management for Primary Care Providers. Mayo Clin. Proc. 2022, 97, 1700–1716. [Google Scholar] [CrossRef]
- Dolgin, E. NASH therapies head toward landmark approval. Nat. Biotechnol. 2023, 41, 587–590. [Google Scholar] [CrossRef]
- Ng, C.H.; Tang, A.S.P.; Xiao, J.; Wong, Z.Y.; Yong, J.N.; Fu, C.E.; Zeng, R.W.; Tan, C.; Wong, G.H.Z.; Teng, M.; et al. Safety and tolerability of obeticholic acid in chronic liver disease: A pooled analysis of 1878 individuals. Hepatol. Commun. 2023, 7, e0005. [Google Scholar] [CrossRef]
- Cvitanović Tomaš, T.; Urlep, Ž.; Moškon, M.; Mraz, M.; Rozman, D. LiverSex Computational Model: Sexual Aspects in Hepatic Metabolism and Abnormalities. Front. Physiol. 2018, 9, 360. [Google Scholar] [CrossRef]
- Lonardo, A.; Nascimbeni, F.; Ballestri, S.; Fairweather, D.; Win, S.; Than, T.A.; Abdelmalek, M.F.; Suzuki, A. Sex Differences in NAFLD: State of the Art and Identification of Research Gaps. Hepatology 2019, 70, 1457–1469. [Google Scholar] [CrossRef]
- DiStefano, J.K. NAFLD and NASH in Postmenopausal Women: Implications for Diagnosis and Treatment. Endocrinology 2020, 161, bqaa134. [Google Scholar] [CrossRef]
- Yang, Y.-J.; Kim, K.M.; An, J.H.; Lee, D.B.; Shim, J.H.; Lim, Y.-S.; Lee, H.C.; Lee, Y.S.; Ahn, J.-H.; Jung, K.H.; et al. Clinical significance of fatty liver disease induced by tamoxifen and toremifene in breast cancer patients. Breast 2016, 28, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Jung, E.-A.; Yoo, J.-J.; Kim, S.G.; Lee, C.-B.; Kim, Y.S.; Jeong, S.W.; Jang, J.Y.; Lee, S.H.; Kim, H.S.; et al. Prevalence, incidence and risk factors of tamoxifen-related non-alcoholic fatty liver disease: A systematic review and meta-analysis. Liver Int. 2020, 40, 1344–1355. [Google Scholar] [CrossRef] [PubMed]
- Hong, N.; Yoon, H.G.; Seo, D.H.; Park, S.; Kim, S.I.; Sohn, J.H.; Rhee, Y. Different patterns in the risk of newly developed fatty liver and lipid changes with tamoxifen versus aromatase inhibitors in postmenopausal women with early breast cancer: A propensity score–matched cohort study. Eur. J. Cancer 2017, 82, 103–114. [Google Scholar] [CrossRef]
- Lee, J.I.; Yu, J.-H.; Anh, S.G.; Lee, H.W.; Jeong, J.; Lee, K.S. Aromatase Inhibitors and Newly Developed Nonalcoholic Fatty Liver Disease in Postmenopausal Patients with Early Breast Cancer: A Propensity Score-Matched Cohort Study. Oncologist 2019, 24, e653–e661. [Google Scholar] [CrossRef]
- Scalzo, R.L.; Foright, R.M.; Hull, S.E.; Knaub, L.A.; Johnson-Murguia, S.; Kinanee, F.; Kaplan, J.; Houck, J.A.; Johnson, G.; Sharp, R.R.; et al. Breast Cancer Endocrine Therapy Promotes Weight Gain with Distinct Adipose Tissue Effects in Lean and Obese Female Mice. Endocrinology 2021, 162, bqab174. [Google Scholar] [CrossRef] [PubMed]
- Meda, C.; Barone, M.; Mitro, N.; Lolli, F.; Pedretti, S.; Caruso, D.; Maggi, A.; Della Torre, S. Hepatic ERα accounts for sex differences in the ability to cope with an excess of dietary lipids. Mol. Metab. 2020, 32, 97–108. [Google Scholar] [CrossRef]
- Hart-Unger, S.; Arao, Y.; Hamilton, K.J.; Lierz, S.L.; Malarkey, D.E.; Hewitt, S.C.; Freemark, M.; Korach, K.S. Hormone signaling and fatty liver in females: Analysis of estrogen receptor α mutant mice. Int. J. Obes. 2017, 41, 945–954. [Google Scholar] [CrossRef] [PubMed]
- Carlsson, B.; Lindén, D.; Brolén, G.; Liljeblad, M.; Bjursell, M.; Romeo, S.; Loomba, R. Review article: The emerging role of genetics in precision medicine for patients with non-alcoholic steatohepatitis. Aliment. Pharmacol. Ther. 2020, 51, 1305–1320. [Google Scholar] [CrossRef] [PubMed]
- Jonas, W.; Schurmann, A. Genetic and epigenetic factors determining NAFLD risk. Mol. Metab. 2021, 50, 101111. [Google Scholar] [CrossRef]
- Maude, H.; Sanchez-Cabanillas, C.; Cebola, I. Epigenetics of Hepatic Insulin Resistance. Front. Endocrinol. 2021, 12, 681356. [Google Scholar] [CrossRef]
- Wahlang, B.; Alexander, N.C.; Li, X.; Rouchka, E.C.; Kirpich, I.A.; Cave, M.C. Polychlorinated biphenyls altered gut microbiome in CAR and PXR knockout mice exhibiting toxicant-associated steatohepatitis. Toxicol. Rep. 2021, 8, 536–547. [Google Scholar] [CrossRef]
- Heindel, J.J.; Blumberg, B.; Cave, M.; Machtinger, R.; Mantovani, A.; Mendez, M.A.; Nadal, A.; Palanza, P.; Panzica, G.; Sargis, R.; et al. Metabolism disrupting chemicals and metabolic disorders. Reprod. Toxicol. 2017, 68, 3–33. [Google Scholar] [CrossRef]
- Sanoudou, D.; Gkouskou, K.K.; Eliopoulos, A.G.; Mantzoros, C.S. Epitranscriptomic challenges and promises in metabolic diseases. Metabolism 2022, 132, 155219. [Google Scholar] [CrossRef] [PubMed]
- Rose, R.E.; Pazos, M.A., 2nd; Curcio, M.J.; Fabris, D. Global Epitranscriptomics Profiling of RNA Post-Transcriptional Modifications as an Effective Tool for Investigating the Epitranscriptomics of Stress Response. Mol. Cell. Proteom. 2016, 15, 932–944. [Google Scholar] [CrossRef] [PubMed]
- Destefanis, E.; Avşar, G.; Groza, P.; Romitelli, A.; Torrini, S.; Pir, P.; Conticello, S.G.; Aguilo, F.; Dassi, E. A mark of disease: How mRNA modifications shape genetic and acquired pathologies. RNA 2021, 27, 367–389. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.; Pham, P.; Dedon, P.C.; Begley, T.J. Lifestyle modifications: Coordinating the tRNA epitranscriptome with codon bias to adapt translation during stress responses. Genome Biol. 2018, 19, 228. [Google Scholar] [CrossRef]
- Sarkar, A.; Gasperi, W.; Begley, U.; Nevins, S.; Huber, S.M.; Dedon, P.C.; Begley, T.J. Detecting the epitranscriptome. WIREs RNA 2021, 12, e1663. [Google Scholar] [CrossRef]
- Zaccara, S.; Ries, R.J.; Jaffrey, S.R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 608–624. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.; Wang, R.; Xiong, F.; Krakowiak, J.; Liao, Z.; Nguyen, P.T.; Moroz-Omori, E.V.; Shao, J.; Zhu, X.; Bolt, M.J.; et al. Enhancer RNA m6A methylation facilitates transcriptional condensate formation and gene activation. Mol. Cell 2021, 81, 3368–3385.e3369. [Google Scholar] [CrossRef]
- Jenjaroenpun, P.; Wongsurawat, T.; Wadley, T.D.; Wassenaar, T.M.; Liu, J.; Dai, Q.; Wanchai, V.; Akel, N.S.; Jamshidi-Parsian, A.; Franco, A.T.; et al. Decoding the epitranscriptional landscape from native RNA sequences. Nucleic Acids Res. 2020, 49, e7. [Google Scholar] [CrossRef]
- Licht, K.; Jantsch, M.F. Rapid and dynamic transcriptome regulation by RNA editing and RNA modifications. J. Cell Biol. 2016, 213, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Knuckles, P.; Bühler, M. Adenosine methylation as a molecular imprint defining the fate of RNA. FEBS Lett. 2018, 592, 2845–2859. [Google Scholar] [CrossRef] [PubMed]
- Cayir, A.; Byun, H.-M.; Barrow, T.M. Environmental epitranscriptomics. Environ. Res. 2020, 189, 109885. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Meng, J.; Su, R.; Zhang, J.; Chen, J.; Ma, X.; Xia, Q. Epitranscriptomics in liver disease: Basic concepts and therapeutic potential. J. Hepatol. 2020, 73, 664–679. [Google Scholar] [CrossRef]
- Roundtree, I.A.; Evans, M.E.; Pan, T.; He, C. Dynamic RNA Modifications in Gene Expression Regulation. Cell 2017, 169, 1187–1200. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, Y.; Toth, J.I.; Petroski, M.D.; Zhang, Z.; Zhao, J.C. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 2014, 16, 191–198. [Google Scholar] [CrossRef]
- Huang, M.; Xu, S.; Liu, L.; Zhang, M.; Guo, J.; Yuan, Y.; Xu, J.; Chen, X.; Zou, J. m6A Methylation Regulates Osteoblastic Differentiation and Bone Remodeling. Front. Cell Dev. Biol. 2021, 9, 783322. [Google Scholar] [CrossRef]
- Li, J.; Zhang, X.; Wang, X.; Sun, C.; Zheng, J.; Li, J.; Yi, G.; Yang, N. The m6A methylation regulates gonadal sex differentiation in chicken embryo. J. Anim. Sci. Biotechnol. 2022, 13, 52. [Google Scholar] [CrossRef]
- Engel, M.; Eggert, C.; Kaplick, P.M.; Eder, M.; Roh, S.; Tietze, L.; Namendorf, C.; Arloth, J.; Weber, P.; Rex-Haffner, M.; et al. The Role of m(6)A/m-RNA Methylation in Stress Response Regulation. Neuron 2018, 99, 389–403.e9. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Zhang, Y.; Zhang, J.; Wang, J.; Li, Q. The alteration of N6-methyladenosine (m6A) modification at the transcriptome-wide level in response of heat stress in bovine mammary epithelial cells. BMC Genom. 2022, 23, 829. [Google Scholar] [CrossRef]
- Ling, Z.; Chen, L.; Zhao, J. m6A-dependent up-regulation of DRG1 by METTL3 and ELAVL1 promotes growth, migration, and colony formation in osteosarcoma. Biosci. Rep. 2020, 40, BSR20200282. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Ji, J. N6-methyladenosine (m6A) RNA modification in cancer stem cells. Stem Cells 2020, 38, 1511–1519. [Google Scholar] [CrossRef] [PubMed]
- Jing, J.J.; Zhao, X.; Li, H.; Sun, L.P.; Yuan, Y. Expression profiles and prognostic roles of m6A writers, erasers and readers in gastric cancer. Future Oncol. 2021, 17, 2605–2620. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Zhang, X.; Yang, P.; Zhang, X.; Peng, Y.; Li, D.; Yu, Y.; Wu, Y.; Wang, Y.; Zhang, J.; et al. RNA m6A methylation orchestrates cancer growth and metastasis via macrophage reprogramming. Nat. Commun. 2021, 12, 1394. [Google Scholar] [CrossRef]
- Chen, M.; Wong, C.M. The emerging roles of N6-methyladenosine (m6A) deregulation in liver carcinogenesis. Mol. Cancer 2020, 19, 44. [Google Scholar] [CrossRef]
- Fan, C.; Ma, Y.; Chen, S.; Zhou, Q.; Jiang, H.; Zhang, J.; Wu, F. Comprehensive Analysis of the Transcriptome-Wide m6A Methylation Modification Difference in Liver Fibrosis Mice by High-Throughput m6A Sequencing. Front. Cell Dev. Biol. 2021, 9, 767051. [Google Scholar] [CrossRef]
- Malovic, E.; Ealy, A.; Kanthasamy, A.; Kanthasamy, A.G. Emerging Roles of N6-Methyladenosine (m6A) Epitranscriptomics in Toxicology. Toxicol. Sci. 2021, 181, 13–22. [Google Scholar] [CrossRef]
- Chen, L.; Xia, S.; Wang, F.; Zhou, Y.; Wang, S.; Yang, T.; Li, Y.; Xu, M.; Zhou, Y.; Kong, D.; et al. m(6)A methylation-induced NR1D1 ablation disrupts the HSC circadian clock and promotes hepatic fibrosis. Pharmacol. Res. 2023, 189, 106704. [Google Scholar] [CrossRef]
- Winkler, R.; Gillis, E.; Lasman, L.; Safra, M.; Geula, S.; Soyris, C.; Nachshon, A.; Tai-Schmiedel, J.; Friedman, N.; Le-Trilling, V.T.K.; et al. m(6)A modification controls the innate immune response to infection by targeting type I interferons. Nat. Immunol. 2019, 20, 173–182. [Google Scholar] [CrossRef]
- Huang, X.T.; Li, J.H.; Zhu, X.X.; Huang, C.S.; Gao, Z.X.; Xu, Q.C.; Zhao, W.; Yin, X.Y. HNRNPC impedes m(6)A-dependent anti-metastatic alternative splicing events in pancreatic ductal adenocarcinoma. Cancer Lett. 2021, 518, 196–206. [Google Scholar] [CrossRef] [PubMed]
- Gokhale, N.S.; McIntyre, A.B.R.; Mattocks, M.D.; Holley, C.L.; Lazear, H.M.; Mason, C.E.; Horner, S.M. Altered m(6)A Modification of Specific Cellular Transcripts Affects Flaviviridae Infection. Mol. Cell 2020, 77, 542–555.e8. [Google Scholar] [CrossRef] [PubMed]
- Flamand, M.N.; Meyer, K.D. m6A and YTHDF proteins contribute to the localization of select neuronal mRNAs. Nucleic Acids Res. 2022, 50, 4464–4483. [Google Scholar] [CrossRef] [PubMed]
- Roundtree, I.A.; Luo, G.Z.; Zhang, Z.; Wang, X.; Zhou, T.; Cui, Y.; Sha, J.; Huang, X.; Guerrero, L.; Xie, P.; et al. YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. eLife 2017, 6, e31311. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Chen, M.; Huang, H.; Zhu, J.; Song, H.; Zhu, J.; Park, J.; Ji, S.J. Dynamic m6A modification regulates local translation of mRNA in axons. Nucleic Acids Res. 2018, 46, 1412–1423. [Google Scholar] [CrossRef]
- Mao, Y.; Dong, L.; Liu, X.M.; Guo, J.; Ma, H.; Shen, B.; Qian, S.B. m(6)A in mRNA coding regions promotes translation via the RNA helicase-containing YTHDC2. Nat. Commun. 2019, 10, 5332. [Google Scholar] [CrossRef]
- Chen, X.Y.; Liang, R.; Yi, Y.C.; Fan, H.N.; Chen, M.; Zhang, J.; Zhu, J.S. The m(6)A Reader YTHDF1 Facilitates the Tumorigenesis and Metastasis of Gastric Cancer via USP14 Translation in an m(6)A-Dependent Manner. Front. Cell Dev. Biol. 2021, 9, 647702. [Google Scholar] [CrossRef]
- Yang, Y.; Hsu, P.J.; Chen, Y.S.; Yang, Y.G. Dynamic transcriptomic m(6)A decoration: Writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018, 28, 616–624. [Google Scholar] [CrossRef]
- Liu, J.; Yue, Y.; Han, D.; Wang, X.; Fu, Y.; Zhang, L.; Jia, G.; Yu, M.; Lu, Z.; Deng, X.; et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014, 10, 93–95. [Google Scholar] [CrossRef]
- Narayan, P.; Rottman, F.M. An in vitro system for accurate methylation of internal adenosine residues in messenger RNA. Science 1988, 242, 1159–1162. [Google Scholar] [CrossRef]
- Bokar, J.A.; Shambaugh, M.E.; Polayes, D.; Matera, A.G.; Rottman, F.M. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 1997, 3, 1233–1247. [Google Scholar]
- Wei, C.M.; Moss, B. Nucleotide sequences at the N6-methyladenosine sites of HeLa cell messenger ribonucleic acid. Biochemistry 1977, 16, 1672–1676. [Google Scholar] [CrossRef]
- Harper, J.E.; Miceli, S.M.; Roberts, R.J.; Manley, J.L. Sequence specificity of the human mRNA N6-adenosine methylase in vitro. Nucleic Acids Res. 1990, 18, 5735–5741. [Google Scholar] [CrossRef] [PubMed]
- Ping, X.L.; Sun, B.F.; Wang, L.; Xiao, W.; Yang, X.; Wang, W.J.; Adhikari, S.; Shi, Y.; Lv, Y.; Chen, Y.S.; et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014, 24, 177–189. [Google Scholar] [CrossRef]
- Zhang, X.; Li, M.J.; Xia, L.; Zhang, H. The biological function of m6A methyltransferase KIAA1429 and its role in human disease. PeerJ 2022, 10, e14334. [Google Scholar] [CrossRef] [PubMed]
- Rong, J.; Jie, Y.; Zhao, H. m6A ‘writer’ KIAA1429 regulates the proliferation and migration of endothelial cells in atherosclerosis. Mol. Biotechnol. 2022, 65, 1198–1206. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, S.; Mumbach, M.R.; Jovanovic, M.; Wang, T.; Maciag, K.; Bushkin, G.G.; Mertins, P.; Ter-Ovanesyan, D.; Habib, N.; Cacchiarelli, D.; et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5’ sites. Cell Rep. 2014, 8, 284–296. [Google Scholar] [CrossRef] [PubMed]
- Patil, D.P.; Chen, C.K.; Pickering, B.F.; Chow, A.; Jackson, C.; Guttman, M.; Jaffrey, S.R. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 2016, 537, 369–373. [Google Scholar] [CrossRef]
- Li, T.; Wang, T.; Jing, J.; Sun, L. Expression Pattern and Clinical Value of Key m6A RNA Modification Regulators in Abdominal Aortic Aneurysm. J. Inflamm. Res. 2021, 14, 4245–4258. [Google Scholar] [CrossRef]
- Wen, J.; Lv, R.; Ma, H.; Shen, H.; He, C.; Wang, J.; Jiao, F.; Liu, H.; Yang, P.; Tan, L.; et al. Zc3h13 Regulates Nuclear RNA m(6)A Methylation and Mouse Embryonic Stem Cell Self-Renewal. Mol. Cell 2018, 69, 1028–1038.e6. [Google Scholar] [CrossRef]
- Knuckles, P.; Lence, T.; Haussmann, I.U.; Jacob, D.; Kreim, N.; Carl, S.H.; Masiello, I.; Hares, T.; Villasenor, R.; Hess, D.; et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m(6)A machinery component Wtap/Fl(2)d. Genes Dev. 2018, 32, 415–429. [Google Scholar] [CrossRef]
- Meng, Y.; Zhang, Q.; Wang, K.; Zhang, X.; Yang, R.; Bi, K.; Chen, W.; Diao, H. RBM15-mediated N6-methyladenosine modification affects COVID-19 severity by regulating the expression of multitarget genes. Cell Death Dis. 2021, 12, 732. [Google Scholar] [CrossRef]
- Cheng, X.; Li, M.; Rao, X.; Zhang, W.; Li, X.; Wang, L.; Huang, G. KIAA1429 regulates the migration and invasion of hepatocellular carcinoma by altering m6A modification of ID2 mRNA. OncoTargets Ther. 2019, 12, 3421–3428. [Google Scholar] [CrossRef] [PubMed]
- Petri, B.J.; Klinge, C.M. m6A readers, writers, erasers, and the m6A epitranscriptome in breast cancer. J. Mol. Endocrinol. 2023, 70, e220110. [Google Scholar] [CrossRef]
- Lee, Y.; Choe, J.; Park, O.H.; Kim, Y.K. Molecular Mechanisms Driving mRNA Degradation by m(6)A Modification. Trends Genet. 2020, 36, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Rauch, S.; He, C.; Dickinson, B.C. Targeted m(6)A Reader Proteins To Study Epitranscriptomic Regulation of Single RNAs. J. Am. Chem. Soc. 2018, 140, 11974–11981. [Google Scholar] [CrossRef]
- Alarcon, C.R.; Goodarzi, H.; Lee, H.; Liu, X.; Tavazoie, S.; Tavazoie, S.F. HNRNPA2B1 Is a Mediator of m(6)A-Dependent Nuclear RNA Processing Events. Cell 2015, 162, 1299–1308. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, S.; Xiao, W.; Zhao, Y.L.; Yang, Y.G. m(6)A: Signaling for mRNA splicing. RNA Biol. 2016, 13, 756–759. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Weng, H.; Sun, W.; Qin, X.; Shi, H.; Wu, H.; Zhao, B.S.; Mesquita, A.; Liu, C.; Yuan, C.L. Recognition of RNA N 6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2018, 20, 285–295. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Chen, Y.S.; Ping, X.L.; Yang, X.; Xiao, W.; Yang, Y.; Sun, H.Y.; Zhu, Q.; Baidya, P.; Wang, X.; et al. Cytoplasmic m(6)A reader YTHDF3 promotes mRNA translation. Cell Res. 2017, 27, 444–447. [Google Scholar] [CrossRef]
- Patil, D.P.; Pickering, B.F.; Jaffrey, S.R. Reading m(6)A in the Transcriptome: m(6)A-Binding Proteins. Trends Cell Biol. 2018, 28, 113–127. [Google Scholar] [CrossRef]
- Zhao, Y.; Shi, Y.; Shen, H.; Xie, W. m(6)A-binding proteins: The emerging crucial performers in epigenetics. J. Hematol. Oncol. 2020, 13, 35. [Google Scholar] [CrossRef]
- Liao, J.; Wei, Y.; Liang, J.; Wen, J.; Chen, X.; Zhang, B.; Chu, L. Insight into the structure, physiological function, and role in cancer of m6A readers-YTH domain-containing proteins. Cell Death Discov. 2022, 8, 137. [Google Scholar] [CrossRef] [PubMed]
- Liao, S.; Sun, H.; Xu, C. YTH Domain: A Family of N(6)-methyladenosine (m(6)A) Readers. Genom. Proteom. Bioinform. 2018, 16, 99–107. [Google Scholar] [CrossRef]
- Kasowitz, S.D.; Ma, J.; Anderson, S.J.; Leu, N.A.; Xu, Y.; Gregory, B.D.; Schultz, R.M.; Wang, P.J. Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. PLoS Genet. 2018, 14, e1007412. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Sun, H.; Chen, K.; Gu, X.; Chen, H.; Jiang, L.; Chen, L.; Zhang, S.; Liu, Y.; Shi, D.; et al. Depletion of m(6) A reader protein YTHDC1 induces dilated cardiomyopathy by abnormal splicing of Titin. J. Cell. Mol. Med. 2021, 25, 10879–10891. [Google Scholar] [CrossRef] [PubMed]
- Widagdo, J.; Anggono, V.; Wong, J.J. The multifaceted effects of YTHDC1-mediated nuclear m(6)A recognition. Trends Genet. 2022, 38, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.S.; Zhu, Y.Q.; Xu, Q.C.; Chen, S.; Huang, Y.; Zhao, G.; Ni, X.; Liu, B.; Zhao, W.; Yin, X.Y. YTHDF2 promotes intrahepatic cholangiocarcinoma progression and desensitises cisplatin treatment by increasing CDKN1B mRNA degradation. Clin. Transl. Med. 2022, 12, e848. [Google Scholar] [CrossRef]
- Shi, H.; Wang, X.; Lu, Z.; Zhao, B.S.; Ma, H.; Hsu, P.J.; Liu, C.; He, C. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 2017, 27, 315–328. [Google Scholar] [CrossRef]
- Du, H.; Zhao, Y.; He, J.; Zhang, Y.; Xi, H.; Liu, M.; Ma, J.; Wu, L. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 2016, 7, 12626. [Google Scholar] [CrossRef]
- Sun, R.; Tian, X.; Li, Y.; Zhao, Y.; Wang, Z.; Hu, Y.; Zhang, L.; Wang, Y.; Gao, D.; Zheng, S.; et al. The m6A reader YTHDF3-mediated PRDX3 translation alleviates liver fibrosis. Redox Biol. 2022, 54, 102378. [Google Scholar] [CrossRef]
- Chang, G.; Shi, L.; Ye, Y.; Shi, H.; Zeng, L.; Tiwary, S.; Huse, J.T.; Huo, L.; Ma, L.; Ma, Y.; et al. YTHDF3 Induces the Translation of m(6)A-Enriched Gene Transcripts to Promote Breast Cancer Brain Metastasis. Cancer Cell 2020, 38, 857–871.e857. [Google Scholar] [CrossRef]
- Wang, J.; Sun, D.; Wang, M.; Cheng, A.; Zhu, Y.; Mao, S.; Ou, X.; Zhao, X.; Huang, J.; Gao, Q.; et al. Multiple functions of heterogeneous nuclear ribonucleoproteins in the positive single-stranded RNA virus life cycle. Front. Immunol. 2022, 13, 989298. [Google Scholar] [CrossRef] [PubMed]
- Geuens, T.; Bouhy, D.; Timmerman, V. The hnRNP family: Insights into their role in health and disease. Hum. Genet. 2016, 135, 851–867. [Google Scholar] [CrossRef] [PubMed]
- Schultz, A.S.; Preussner, M.; Bunse, M.; Karni, R.; Heyd, F. Activation-Dependent TRAF3 Exon 8 Alternative Splicing Is Controlled by CELF2 and hnRNP C Binding to an Upstream Intronic Element. Mol. Cell. Biol. 2017, 37, e00488-16. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Tang, X.; Tang, C.; Hua, Z.; Ke, M.; Wang, C.; Zhao, J.; Gao, S.; Jurczyszyn, A.; Janz, S.; et al. HNRNPA2B1 promotes multiple myeloma progression by increasing AKT3 expression via m6A-dependent stabilization of ILF3 mRNA. J. Hematol. Oncol. 2021, 14, 54. [Google Scholar] [CrossRef]
- Wang, H.; Han, L.; Zhao, G.; Shen, H.; Wang, P.; Sun, Z.; Xu, C.; Su, Y.; Li, G.; Tong, T.; et al. hnRNP A1 antagonizes cellular senescence and senescence-associated secretory phenotype via regulation of SIRT1 mRNA stability. Aging Cell 2016, 15, 1063–1073. [Google Scholar] [CrossRef]
- Alarcon, C.R.; Lee, H.; Goodarzi, H.; Halberg, N.; Tavazoie, S.F. N6-methyladenosine marks primary microRNAs for processing. Nature 2015, 519, 482–485. [Google Scholar] [CrossRef]
- Dai, W.; Sun, Y.; Jiang, Z.; Du, K.; Xia, N.; Zhong, G. Key genes associated with non-alcoholic fatty liver disease and acute myocardial infarction. Med. Sci. Monit. 2020, 26, e922492. [Google Scholar] [CrossRef]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.G.; et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.M.; Li, C.J.; Vagbo, C.B.; Shi, Y.; Wang, W.L.; Song, S.H.; et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef]
- Blakemore, A.I.F.; Froguel, P. Is Obesity Our Genetic Legacy? J. Clin. Endocrinol. Metab. 2008, 93, s51–s56. [Google Scholar] [CrossRef] [PubMed]
- Church, C.; Lee, S.; Bagg, E.A.L.; McTaggart, J.S.; Deacon, R.; Gerken, T.; Lee, A.; Moir, L.; Mecinović, J.; Quwailid, M.M.; et al. A Mouse Model for the Metabolic Effects of the Human Fat Mass and Obesity Associated FTO Gene. PLoS Genet. 2009, 5, e1000599. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Yang, Y.; Sun, B.F.; Shi, Y.; Yang, X.; Xiao, W.; Hao, Y.J.; Ping, X.L.; Chen, Y.S.; Wang, W.J.; et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014, 24, 1403–1419. [Google Scholar] [CrossRef] [PubMed]
- Fischer, J.; Koch, L.; Emmerling, C.; Vierkotten, J.; Peters, T.; Bruning, J.C.; Ruther, U. Inactivation of the Fto gene protects from obesity. Nature 2009, 458, 894–898. [Google Scholar] [CrossRef]
- Yang, S.; Wei, J.; Cui, Y.H.; Park, G.; Shah, P.; Deng, Y.; Aplin, A.E.; Lu, Z.; Hwang, S.; He, C.; et al. m(6)A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat. Commun. 2019, 10, 2782. [Google Scholar] [CrossRef] [PubMed]
- Nakano, M.; Ondo, K.; Takemoto, S.; Fukami, T.; Nakajima, M. Methylation of adenosine at the N6 position post-transcriptionally regulates hepatic P450s expression. Biochem. Pharmacol. 2020, 171, 113697. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Li, Y.; Yu, J.; Gan, Z.; Wei, W.; Wang, C.; Zhang, L.; Wang, T.; Zhong, X. Resveratrol Attenuates High-Fat Diet Induced Hepatic Lipid Homeostasis Disorder and Decreases m(6)A RNA Methylation. Front. Pharmacol. 2020, 11, 568006. [Google Scholar] [CrossRef]
- Ghazi, T.; Nagiah, S.; Chuturgoon, A.A. Fusaric acid induces hepatic global m6A RNA methylation and differential expression of m6A regulatory genes in vivo—A pilot study. Epigenetics 2022, 17, 695–703. [Google Scholar] [CrossRef]
- Mauer, J.; Luo, X.; Blanjoie, A.; Jiao, X.; Grozhik, A.V.; Patil, D.P.; Linder, B.; Pickering, B.F.; Vasseur, J.-J.; Chen, Q.; et al. Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature 2017, 541, 371–375. [Google Scholar] [CrossRef]
- Mauer, J.; Sindelar, M.; Despic, V.; Guez, T.; Hawley, B.R.; Vasseur, J.-J.; Rentmeister, A.; Gross, S.S.; Pellizzoni, L.; Debart, F.; et al. FTO controls reversible m6Am RNA methylation during snRNA biogenesis. Nat. Chem. Biol. 2019, 15, 340–347. [Google Scholar] [CrossRef]
- Mauer, J.; Jaffrey, S.R. FTO, m(6) A(m), and the hypothesis of reversible epitranscriptomic mRNA modifications. FEBS Lett. 2018, 592, 2012–2022. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Yan, J.; Li, Q.; Li, J.; Gong, S.; Zhou, H.; Gan, J.; Jiang, H.; Jia, G.F.; Luo, C.; et al. Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res. 2015, 43, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.J.; Wang, R.L.; Wang, D.; Liu, L.; Cheng, L. Visible-light-mediated oxidative demethylation of N(6)-methyl adenines. Chem. Commun. 2017, 53, 10734–10737. [Google Scholar] [CrossRef]
- Meng, Q.; Schatten, H.; Zhou, Q.; Chen, J. Crosstalk between m6A and coding/non-coding RNA in cancer and detection methods of m6A modification residues. Aging 2023, 15, 6577–6619. [Google Scholar] [CrossRef]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Salmon-Divon, M.; Amariglio, N.; Rechavi, G. Transcriptome-wide mapping of N(6)-methyladenosine by m(6)A-seq based on immunocapturing and massively parallel sequencing. Nat. Protoc. 2013, 8, 176–189. [Google Scholar] [CrossRef] [PubMed]
- Roberts, J.T.; Porman, A.M.; Johnson, A.M. Identification of m6A residues at single-nucleotide resolution using eCLIP and an accessible custom analysis pipeline. RNA 2020, 27, 527–541. [Google Scholar] [CrossRef]
- Thuring, K.; Schmid, K.; Keller, P.; Helm, M. Analysis of RNA modifications by liquid chromatography-tandem mass spectrometry. Methods 2016, 107, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.D. DART-seq: An antibody-free method for global m(6)A detection. Nat. Methods 2019, 16, 1275–1280. [Google Scholar] [CrossRef] [PubMed]
- Tegowski, M.; Zhu, H.; Meyer, K.D. Detecting m(6)A with In Vitro DART-Seq. Methods Mol. Biol. 2022, 2404, 363–374. [Google Scholar] [PubMed]
- Kono, N.; Arakawa, K. Nanopore sequencing: Review of potential applications in functional genomics. Dev. Growth Differ. 2019, 61, 316–326. [Google Scholar] [CrossRef]
- Parker, M.T.; Knop, K.; Sherwood, A.V.; Schurch, N.J.; Mackinnon, K.; Gould, P.D.; Hall, A.J.; Barton, G.J.; Simpson, G.G. Nanopore direct RNA sequencing maps the complexity of Arabidopsis mRNA processing and m(6)A modification. eLife 2020, 9, e49658. [Google Scholar] [CrossRef] [PubMed]
- Pratanwanich, P.N.; Yao, F.; Chen, Y.; Koh, C.W.Q.; Wan, Y.K.; Hendra, C.; Poon, P.; Goh, Y.T.; Yap, P.M.L.; Chooi, J.Y.; et al. Identification of differential RNA modifications from nanopore direct RNA sequencing with xPore. Nat. Biotechnol. 2021, 39, 1394–1402. [Google Scholar] [CrossRef]
- Hendra, C.; Pratanwanich, P.N.; Wan, Y.K.; Goh, W.S.S.; Thiery, A.; Goke, J. Detection of m6A from direct RNA sequencing using a multiple instance learning framework. Nat. Methods 2022, 19, 1590–1598. [Google Scholar] [CrossRef] [PubMed]
- Körtel, N.; Rücklé, C.; Zhou, Y.; Busch, A.; Hoch-Kraft, P.; Sutandy, F.X.R.; Haase, J.; Pradhan, M.; Musheev, M.; Ostareck, D.; et al. Deep and accurate detection of m6A RNA modifications using miCLIP2 and m6Aboost machine learning. Nucleic Acids Res. 2021, 49, e92. [Google Scholar] [CrossRef]
- Li, C.; Liu, W.; Liu, C.; Luo, Q.; Luo, K.; Wei, C.; Li, X.; Qin, J.; Zheng, C.; Lan, C.; et al. Integrating machine learning and bioinformatics analysis to m6A regulator-mediated methylation modification models for predicting glioblastoma patients’ prognosis and immunotherapy response. Aging 2023, 15, 4051–4070. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Hamada, M. DeepM6ASeq: Prediction and characterization of m6A-containing sequences using deep learning. BMC Bioinform. 2018, 19, 524. [Google Scholar] [CrossRef]
- Liu, H.; Jiang, Y.; Lu, J.; Peng, C.; Ling, Z.; Chen, Y.; Chen, D.; Tong, R.; Zheng, S.; Wu, J. m(6)A-modification regulated circ-CCT3 acts as the sponge of miR-378a-3p to promote hepatocellular carcinoma progression. Epigenetics 2023, 18, 2204772. [Google Scholar] [CrossRef] [PubMed]
- Hua, Y.; Li, X.; Yin, B.; Huang, J.; Lu, S.; Wang, C.; Ke, S.; Xu, Y.; Qian, B.; Feng, Z.; et al. Comprehensive analysis of transcriptome-wide M(6)A methylation for hepatic ischaemia reperfusion injury in mice. Epigenetics 2023, 18, 2201716. [Google Scholar] [CrossRef] [PubMed]
- Deblois, G.; St-Pierre, J.; Giguere, V. The PGC-1/ERR signaling axis in cancer. Oncogene 2013, 32, 3483–3490. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Xiong, L.; Wei, T.; Liu, Q.; Yan, L.; Chen, J.; Dai, L.; Shi, L.; Zhang, W.; Yang, J.; et al. Hypoxia-responsive PPARGC1A/BAMBI/ACSL5 axis promotes progression and resistance to lenvatinib in hepatocellular carcinoma. Oncogene 2023, 42, 1509–1523. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yu, H.; Dong, W.; Zhang, C.; Hu, M.; Ma, W.; Jiang, X.; Li, H.; Yang, P.; Xiang, D. N6-Methyladenosine–Mediated Up-Regulation of FZD10 Regulates Liver Cancer Stem Cells’ Properties and Lenvatinib Resistance Through WNT/β-Catenin and Hippo Signaling Pathways. Gastroenterology 2023, 164, 990–1005. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yang, Q.; Zhou, Q.; Fang, F.; Lei, K.; Liu, Z.; Zheng, G.; Zhu, L.; Huo, J.; Li, X.; et al. METTL3-m6A-EGFR-axis drives lenvatinib resistance in hepatocellular carcinoma. Cancer Lett. 2023, 559, 216122. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Tang, A.; Cui, Y.; Gong, H.; Li, H. LRPPRC facilitates tumor progression and immune evasion through upregulation of m(6)A modification of PD-L1 mRNA in hepatocellular carcinoma. Front. Immunol. 2023, 14, 1144774. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Wang, M.; Wang, G.; Mai, Z.; Zhou, B.; Han, Y.; Zhuang, J.; Xia, W. lncRNA miR4458HG modulates hepatocellular carcinoma progression by activating m6A-dependent glycolysis and promoting the polarization of tumor-associated macrophages. Cell. Mol. Life Sci. 2023, 80, 99. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Chen, G.; Zhao, Y.; Gao, H.; Li, L.; Yin, Y.; Jiang, J.; Wang, L.; Mang, Y.; Gao, Y.; et al. Exosome-derived circCCAR1 promotes CD8 + T-cell dysfunction and anti-PD1 resistance in hepatocellular carcinoma. Mol. Cancer 2023, 22, 55. [Google Scholar] [CrossRef]
- Kuang, Y.; Cheng, Y.; Wang, J.; Li, H.; Cao, X.; Wang, Y. KIAA1429 mediates epithelial mesenchymal transition in sorafenib-resistant hepatocellular carcinoma through m6A methylation modification. Cancer Med. 2023, 12, 7222–7233. [Google Scholar] [CrossRef]
- Lan, T.; Li, H.; Zhang, D.; Xu, L.; Liu, H.; Hao, X.; Yan, X.; Liao, H.; Chen, X.; Xie, K.; et al. KIAA1429 contributes to liver cancer progression through N6-methyladenosine-dependent post-transcriptional modification of GATA3. Mol. Cancer 2019, 18, 186. [Google Scholar] [CrossRef]
- Fang, J.; Wu, X.; He, J.; Zhang, H.; Chen, X.; Zhang, H.; Novakovic, B.; Qi, H.; Yu, X. RBM15 suppresses hepatic insulin sensitivity of offspring of gestational diabetes mellitus mice via m6A-mediated regulation of CLDN4. Mol. Med. 2023, 29, 23. [Google Scholar] [CrossRef]
- Yang, Y.; Yan, Y.; Yin, J.; Tang, N.; Wang, K.; Huang, L.; Hu, J.; Feng, Z.; Gao, Q.; Huang, A. O-GlcNAcylation of YTHDF2 promotes HBV-related hepatocellular carcinoma progression in an N(6)-methyladenosine-dependent manner. Signal Transduct. Target. Ther. 2023, 8, 63. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhou, P.; Deng, Y.; Cai, X.; Sun, M.; Sun, Y.; Wu, D. ALKBH5-mediated m(6) A demethylation of TIRAP mRNA promotes radiation-induced liver fibrosis and decreases radiosensitivity of hepatocellular carcinoma. Clin. Transl. Med. 2023, 13, e1198. [Google Scholar] [CrossRef]
- Lyu, Z.; Huang, B.; Zhang, J.; Qian, Q.; Pu, X.; Cui, N.; Ou, Y.; Li, B.; You, Z.; Lian, M.; et al. Suppression of YTHDF2 attenuates autoimmune hepatitis by expansion of myeloid-derived suppressor cells. J. Autoimmun. 2023, 135, 102993. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Cai, K.; Du, Y.; Liu, Z.; Gong, Y. HDAC1 overexpression promoted by METTL3-IGF2BP2 inhibits FGF21 expression in metabolic syndrome-related liver injury. Biochem. Cell Biol. 2023, 101, 52–63. [Google Scholar] [CrossRef]
- Zhou, S.; Yang, K.; Chen, S.; Lian, G.; Huang, Y.; Yao, H.; Zhao, Y.; Huang, K.; Yin, D.; Lin, H.; et al. CCL3 secreted by hepatocytes promotes the metastasis of intrahepatic cholangiocarcinoma by VIRMA-mediated N6-methyladenosine (m(6)A) modification. J. Transl. Med. 2023, 21, 43. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Hou, J.; Xu, H.; Lei, Z.; Li, Z.; Zhu, H.; Yu, X.; Yang, Z.; Jin, X.; Sun, J. The Prognostic Value of a lncRNA Risk Model Consists of 9 m6A Regulator-Related lncRNAs in Hepatocellular Carcinoma (HCC). Evol. Bioinform. 2023, 19, 11769343221142013. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Du, M.; Meng, L.; Yang, Y.; He, S.; Zhu, Y.; Ren, X.; Wei, M.; Dong, R.; Zheng, S.; et al. Integrative analysis implicates the significance of m6A in the liver fibrosis of biliary atresia by regulating THY1. Hepatol. Commun. 2023, 7, e0004. [Google Scholar] [CrossRef]
- Liu, Y.; Sun, L.; Guo, H.; Zhou, S.; Wang, C.; Ji, C.; Meng, F.; Liang, S.; Zhang, B.; Yuan, Y.; et al. Targeting SLP2-mediated lipid metabolism reprograming restricts proliferation and metastasis of hepatocellular carcinoma and promotes sensitivity to Lenvatinib. Oncogene 2023, 42, 374–388. [Google Scholar] [CrossRef]
- Chen, S.; Xia, H.; Sheng, L. WTAP-mediated m6A modification on circCMTM3 inhibits hepatocellular carcinoma ferroptosis by recruiting IGF2BP1 to increase PARK7 stability. Dig. Liver Dis. 2022, 55, 967–981. [Google Scholar] [CrossRef]
- Wang, L.; Yi, X.; Xiao, X.; Zheng, Q.; Ma, L.; Li, B. Exosomal miR-628-5p from M1 polarized macrophages hinders m6A modification of circFUT8 to suppress hepatocellular carcinoma progression. Cell. Mol. Biol. Lett. 2022, 27, 106. [Google Scholar] [CrossRef]
- Zhou, R.; Ni, W.; Qin, C.; Zhou, Y.; Li, Y.; Huo, J.; Bian, L.; Zhou, A.; Li, J. A functional loop between YTH domain family protein YTHDF3 mediated m(6)A modification and phosphofructokinase PFKL in glycolysis of hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2022, 41, 334. [Google Scholar] [CrossRef]
- Yang, I.; Oh, S.Y.; Jang, S.; Kim, I.Y.; Sung, Y.M.; Seong, J.K. Mettl14 mutation restrains liver regeneration by attenuating mitogens derived from non-parenchymal liver cells. BMB Rep. 2022, 55, 633–638. [Google Scholar] [CrossRef] [PubMed]
- Kong, F.; Wang, K.; Wang, L. Systematic analysis of the expression profile and prognostic significance of m6A regulators and PD-L1 in hepatocellular carcinoma. Discov. Oncol. 2022, 13, 131. [Google Scholar] [CrossRef]
- Zhang, X.; Yin, H.; Zhang, X.; Jiang, X.; Liu, Y.; Zhang, H.; Peng, Y.; Li, D.; Yu, Y.; Zhang, J.; et al. N6-methyladenosine modification governs liver glycogenesis by stabilizing the glycogen synthase 2 mRNA. Nat. Commun. 2022, 13, 7038. [Google Scholar] [CrossRef]
- Gao, H.; Wang, X.; Ma, H.; Lin, S.; Zhang, D.; Wu, W.; Liao, Z.; Chen, M.; Li, Q.; Lin, M.; et al. METTL16 regulates m(6)A methylation on chronic hepatitis B associated gene HLA-DPB1 involved in liver fibrosis. Front. Genet. 2022, 13, 996245. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, N.; Zeng, J.; Wang, T.; Shen, Y.; Ma, C.; Yang, M. N(6)-methyladenosine-modified lncRNA ARHGAP5-AS1 stabilises CSDE1 and coordinates oncogenic RNA regulons in hepatocellular carcinoma. Clin. Transl. Med. 2022, 12, e1107. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhou, X.; Wu, W.; Wang, X.; Wang, Y. FTO-dependent function of N6-methyladenosine is involved in the hepatoprotective effects of betaine on adolescent mice. J. Physiol. Biochem. 2015, 71, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Pascale, R.M.; Simile, M.M.; Calvisi, D.F.; Feo, C.F.; Feo, F. S-Adenosylmethionine: From the Discovery of Its Inhibition of Tumorigenesis to Its Use as a Therapeutic Agent. Cells 2022, 11, 409. [Google Scholar] [CrossRef]
- Mato, J.M.; Martínez-Chantar, M.L.; Lu, S.C. S-adenosylmethionine metabolism and liver disease. Ann. Hepatol. 2013, 12, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Ramos, D.; Lopitz-Otsoa, F.; Millet, O.; Alonso, C.; Lu, S.C.; Mato, J.M. One Carbon Metabolism and S-Adenosylmethionine in Non-Alcoholic Fatty Liver Disease Pathogenesis and Subtypes. Livers 2022, 2, 243–257. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Feng, Y.; Zhang, L.; Jia, Y.; Cai, D.; Qian, S.B.; Du, M.; Zhao, R. GR-mediated FTO transactivation induces lipid accumulation in hepatocytes via demethylation of m(6)A on lipogenic mRNAs. RNA Biol. 2020, 17, 930–942. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Ling, Y.; Jiang, J.; Wang, D.; Wang, J.; Li, J.; Wang, X.; Wang, H. Differential mechanisms regarding triclosan vs. bisphenol A and fluorene-9-bisphenol induced zebrafish lipid-metabolism disorders by RNA-Seq. Chemosphere 2020, 251, 126318. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Zhang, Z.; Tai, L.; Zhang, L.; Sun, Z.; Zhou, L. Comprehensive analysis of differences of N(6)-methyladenosine RNA methylomes between high-fat-fed and normal mouse livers. Epigenomics 2019, 11, 1267–1282. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Liu, C.; Xu, L.; Yuan, Y.; Zhao, J.; Zhao, W.; Chen, Y.; Qiu, J.; Meng, M.; Zheng, Y.; et al. N6-Methyladenosine Reader Protein YT521-B Homology Domain-Containing 2 Suppresses Liver Steatosis by Regulation of mRNA Stability of Lipogenic Genes. Hepatology 2021, 73, 91–103. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Li, B.; Arumugam, S.; Lu, Q.; Mankash, S.M.; Li, J.; Sun, B.; Li, J.; Flavell, R.A.; Li, H.B.; et al. m(6)A mRNA methylation-directed myeloid cell activation controls progression of NAFLD and obesity. Cell Rep. 2021, 37, 109968. [Google Scholar] [CrossRef]
- Luo, Y.; Zhang, Z.; Xiang, L.; Zhou, B.; Wang, X.; Lin, Y.; Ding, X.; Liu, F.; Lu, Y.; Peng, Y. Analysis of N6-Methyladenosine Methylation Modification in Fructose-Induced Non-Alcoholic Fatty Liver Disease. Front. Endocrinol. 2021, 12, 780617. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Gong, Y.; Wang, X.; He, W.; Wu, L.; Zhang, L.; Xiong, L.; Huang, Y.; Su, L.; Shi, P.; et al. METTL3-m(6)A-Rubicon axis inhibits autophagy in nonalcoholic fatty liver disease. Mol. Ther. 2022, 30, 932–946. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Sun, C.; Yan, Y.; Niu, Z.; Li, Y.; Xu, X.; Zhang, J.; Wu, Y.; Li, Y.; Wang, L.; et al. Aberrant elevation of FTO levels promotes liver steatosis by decreasing the m6A methylation and increasing the stability of SREBF1 and ChREBP mRNAs. J. Mol. Cell Biol. 2023, 14, mjac061. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yang, Y.; Sun, F.; Luo, Y.; Yang, Y.; Li, J.; Hu, W.; Tao, H.; Lu, C.; Yang, J.-J. ALKBH5 attenuates mitochondrial fission and ameliorates liver fibrosis by reducing Drp1 methylation. Pharmacol. Res. 2023, 187, 106608. [Google Scholar] [CrossRef]
- Wang, S.; Friedman, S.L. Hepatic fibrosis: A convergent response to liver injury that is reversible. J. Hepatol. 2020, 73, 210–211. [Google Scholar] [CrossRef]
- Li, Y.; Kang, X.; Zhou, Z.; Pan, L.; Chen, H.; Liang, X.; Chu, J.; Dong, S.; Liu, C.; Yu, S.; et al. The m6A methyltransferase Mettl3 deficiency attenuates hepatic stellate cell activation and liver fibrosis. Mol. Ther. 2022, 30, 3714–3728. [Google Scholar] [CrossRef]
- Cheng, W.; Li, M.; Zhang, L.; Zhou, C.; Yu, S.; Peng, X.; Zhang, W.; Zhang, W. New roles of N6-methyladenosine methylation system regulating the occurrence of non-alcoholic fatty liver disease with N6-methyladenosine-modified MYC. Front. Pharmacol. 2022, 13, 973116. [Google Scholar] [CrossRef]
- Liu, S.; He, L.; Bannister, O.B.; Li, J.; Schnegelberger, R.D.; Vanderpuye, C.-M.; Althouse, A.D.; Schopfer, F.J.; Wahlang, B.; Cave, M.C.; et al. Western diet unmasks transient low-level vinyl chloride-induced tumorigenesis; potential role of the (epi-)transcriptome. Toxicol. Appl. Pharmacol. 2023, 468, 116514. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Vatsalya, V.; Ma, X.; Klinge, C.M.; Cave, M.C.; Feng, W.; McClain, C.J.; Zhang, X. Metabolic Analysis of Nucleosides/Bases in the Urine and Serum of Patients with Alcohol-Associated Liver Disease. Metabolites 2022, 12, 1187. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zeng, F.; Zeng, M.; Han, X.; Cai, L.; Zhang, J.; Weng, J.; Gao, Y. Identification and Characterization of Alcohol-related Hepatocellular Carcinoma Prognostic Subtypes based on an Integrative N6-methyladenosine methylation Model. Int. J. Biol. Sci. 2021, 17, 3554–3572. [Google Scholar] [CrossRef] [PubMed]
- Rinella, M.E. Nonalcoholic fatty liver disease: A systematic review. JAMA 2015, 313, 2263–2273. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.T.; Kim, D.-Y.; Lee, Y.-G.; Lee, Y.-S.; Truong, X.T.; Lee, J.-H.; Song, D.-K.; Kwon, T.K.; Park, S.-H.; Jung, C.H. SREBP-1c impairs ULK1 sulfhydration-mediated autophagic flux to promote hepatic steatosis in high-fat-diet-fed mice. Mol. Cell 2021, 81, 3820–3832.e27. [Google Scholar] [CrossRef]
- Jeon, T.-I.; Osborne, T.F. SREBPs: Metabolic integrators in physiology and metabolism. Trends Endocrinol. Metab. 2012, 23, 65–72. [Google Scholar] [CrossRef]
- Yadav, P.K.; Rajvanshi, P.K.; Rajasekharan, R. The role of yeast m6A methyltransferase in peroxisomal fatty acid oxidation. Curr. Genet. 2018, 64, 417–422. [Google Scholar] [CrossRef]
- Yang, Y.; Cai, J.; Yang, X.; Wang, K.; Sun, K.; Yang, Z.; Zhang, L.; Yang, L.; Gu, C.; Huang, X.; et al. Dysregulated m6A modification promotes lipogenesis and development of non-alcoholic fatty liver disease and hepatocellular carcinoma. Mol. Ther. 2022, 30, 2342–2353. [Google Scholar] [CrossRef]
- Xu, Z.; Qin, Y.; Lv, B.; Tian, Z.; Zhang, B. Intermittent Fasting Improves High-Fat Diet-Induced Obesity Cardiomyopathy via Alleviating Lipid Deposition and Apoptosis and Decreasing m6A Methylation in the Heart. Nutrients 2022, 14, 251. [Google Scholar] [CrossRef]
- Petri, B.J.; Piell, K.M.; Wahlang, B.; Head, K.Z.; Andreeva, K.; Rouchka, E.C.; Cave, M.C.; Klinge, C.M. Polychlorinated biphenyls alter hepatic m6A mRNA methylation in a mouse model of environmental liver disease. Environ. Res. 2022, 216, 114686. [Google Scholar] [CrossRef]
- He, S.; Wang, H.; Liu, R.; He, M.; Che, T.; Jin, L.; Deng, L.; Tian, S.; Li, Y.; Lu, H. mRNA N6-methyladenosine methylation of postnatal liver development in pig. PLoS ONE 2017, 12, e0173421. [Google Scholar] [CrossRef]
- Xie, W.; Ma, L.L.; Xu, Y.Q.; Wang, B.H.; Li, S.M. METTL3 inhibits hepatic insulin sensitivity via N6-methyladenosine modification of Fasn mRNA and promoting fatty acid metabolism. Biochem. Biophys. Res. Commun. 2019, 518, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Yu, J.; Frazier, K.; Weng, X.; Li, Y.; Cham, C.M.; Dolan, K.; Zhu, X.; Hubert, N.; Tao, Y. Circadian clock regulation of hepatic lipid metabolism by modulation of m6A mRNA methylation. Cell Rep. 2018, 25, 1816–1828.e1814. [Google Scholar] [CrossRef] [PubMed]
- Adlanmerini, M.; Lazar, M.A. The REV-ERB Nuclear Receptors: Timekeepers for the Core Clock Period and Metabolism. Endocrinology 2023, 164, bqad069. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Ren, W.; Li, A.; Ding, Y.; Guo, W.; Su, D.; Hu, C.; Xu, K.; Chen, H.; Xu, X. Fat mass and obesity-associated gene enhances oxidative stress and lipogenesis in nonalcoholic fatty liver disease. Dig. Dis. Sci. 2013, 58, 1004–1009. [Google Scholar] [CrossRef] [PubMed]
- Lim, A.; Zhou, J.; Sinha, R.A.; Singh, B.K.; Ghosh, S.; Lim, K.-H.; Chow, P.K.-H.; Woon, E.C.; Yen, P.M. Hepatic FTO expression is increased in NASH and its silencing attenuates palmitic acid-induced lipotoxicity. Biochem. Biophys. Res. Commun. 2016, 479, 476–481. [Google Scholar] [CrossRef]
- Yang, Y.; Shen, F.; Huang, W.; Qin, S.; Huang, J.-T.; Sergi, C.; Yuan, B.-F.; Liu, S.-M. Glucose Is Involved in the Dynamic Regulation of m6A in Patients with Type 2 Diabetes. J. Clin. Endocrinol. Metab. 2018, 104, 665–673. [Google Scholar] [CrossRef]
- Church, C.; Moir, L.; McMurray, F.; Girard, C.; Banks, G.T.; Teboul, L.; Wells, S.; Brüning, J.C.; Nolan, P.M.; Ashcroft, F.M. Overexpression of Fto leads to increased food intake and results in obesity. Nat. Genet. 2010, 42, 1086–1092. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Zhang, Z.; Yu, L.; Li, Y.; Liang, M.; Zhou, L. FTO reduces mitochondria and promotes hepatic fat accumulation through RNA demethylation. J. Cell. Biochem. 2018, 119, 5676–5685. [Google Scholar] [CrossRef]
- Chen, A.; Chen, X.; Cheng, S.; Shu, L.; Yan, M.; Yao, L.; Wang, B.; Huang, S.; Zhou, L.; Yang, Z. FTO promotes SREBP1c maturation and enhances CIDEC transcription during lipid accumulation in HepG2 cells. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2018, 1863, 538–548. [Google Scholar] [CrossRef]
- Xu, Y.; Zhou, Z.; Kang, X.; Pan, L.; Liu, C.; Liang, X.; Chu, J.; Dong, S.; Li, Y.; Liu, Q.; et al. Mettl3-mediated mRNA m(6)A modification controls postnatal liver development by modulating the transcription factor Hnf4a. Nat. Commun. 2022, 13, 4555. [Google Scholar] [CrossRef] [PubMed]
- Bechmann, L.P.; Hannivoort, R.A.; Gerken, G.; Hotamisligil, G.S.; Trauner, M.; Canbay, A. The interaction of hepatic lipid and glucose metabolism in liver diseases. J. Hepatol. 2012, 56, 952–964. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Li, P.; Fu, S.; Calay, E.S.; Hotamisligil, G.S. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010, 11, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Giugliano, D.; Ceriello, A.; Esposito, K. Glucose metabolism and hyperglycemia. Am. J. Clin. Nutr. 2008, 87, 217S–222S. [Google Scholar] [CrossRef] [PubMed]
- Alves-Bezerra, M.; Cohen, D.E. Triglyceride Metabolism in the Liver. Compr. Physiol. 2017, 8, 1–8. [Google Scholar]
- Li, Y.; Zhang, Q.; Cui, G.; Zhao, F.; Tian, X.; Sun, B.F.; Yang, Y.; Li, W. m(6)A Regulates Liver Metabolic Disorders and Hepatogenous Diabetes. Genom. Proteom. Bioinform. 2020, 18, 371–383. [Google Scholar] [CrossRef]
- Onalan, E.; Yakar, B.; Onalan, E.E.; Karakulak, K.; Kaymaz, T.; Donder, E. m(6)A RNA, FTO, ALKBH5 Expression in Type 2 Diabetic and Obesity Patients. J. Coll. Physicians Surg. Pak. 2022, 32, 1143–1148. [Google Scholar]
- Yang, K.; Sun, J.; Zhang, Z.; Xiao, M.; Ren, D.; Liu, S.-M. Reduction of mRNA m6A associates with glucose metabolism via YTHDC1 in human and mice. Diabetes Res. Clin. Pract. 2023, 198, 110607. [Google Scholar] [CrossRef]
- Ng, M.C.; Shriner, D.; Chen, B.H.; Li, J.; Chen, W.M.; Guo, X.; Liu, J.; Bielinski, S.J.; Yanek, L.R.; Nalls, M.A.; et al. Meta-analysis of genome-wide association studies in African Americans provides insights into the genetic architecture of type 2 diabetes. PLoS Genet. 2014, 10, e1004517. [Google Scholar] [CrossRef]
- Fuchsberger, C.; Flannick, J.; Teslovich, T.M.; Mahajan, A.; Agarwala, V.; Gaulton, K.J.; Ma, C.; Fontanillas, P.; Moutsianas, L.; McCarthy, D.J.; et al. The genetic architecture of type 2 diabetes. Nature 2016, 536, 41–47. [Google Scholar] [CrossRef]
- García-Chapa, E.G.; Leal-Ugarte, E.; Peralta-Leal, V.; Durán-González, J.; Meza-Espinoza, J.P. Genetic Epidemiology of Type 2 Diabetes in Mexican Mestizos. Biomed. Res. Int. 2017, 2017, 3937893. [Google Scholar] [CrossRef] [PubMed]
- Hafner, M.; Landthaler, M.; Burger, L.; Khorshid, M.; Hausser, J.; Berninger, P.; Rothballer, A.; Ascano, M.; Jungkamp, A.-C.; Munschauer, M. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 2010, 141, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Conway, A.E.; Van Nostrand, E.L.; Pratt, G.A.; Aigner, S.; Wilbert, M.L.; Sundararaman, B.; Freese, P.; Lambert, N.J.; Sathe, S.; Liang, T.Y. Enhanced CLIP uncovers IMP protein-RNA targets in human pluripotent stem cells important for cell adhesion and survival. Cell Rep. 2016, 15, 666–679. [Google Scholar] [CrossRef] [PubMed]
- Senoo, H. Structure and function of hepatic stellate cells. Med. Electron Microsc. 2004, 37, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Xie, L.; Yang, D.; Luo, K.; Li, X. Small-molecule natural plants for reversing liver fibrosis based on modulation of hepatic stellate cells activation: An update. Phytomedicine 2023, 113, 154721. [Google Scholar] [CrossRef]
- Puche, J.E.; Saiman, Y.; Friedman, S.L. Hepatic stellate cells and liver fibrosis. Compr. Physiol. 2013, 3, 1473–1492. [Google Scholar]
- Tsuchida, T.; Friedman, S.L. Mechanisms of hepatic stellate cell activation. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 397–411. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.J.; Wang, J.; Yang, Y.; Yang, Y.; Li, J.; Lu, D.; Lu, C. ALKBH5 ameliorated liver fibrosis and suppressed HSCs activation via triggering PTCH1 activation in an m(6)A dependent manner. Eur. J. Pharmacol. 2022, 922, 174900. [Google Scholar] [CrossRef]
- Shen, M.; Li, Y.; Wang, Y.; Shao, J.; Zhang, F.; Yin, G.; Chen, A.; Zhang, Z.; Zheng, S. N(6)-methyladenosine modification regulates ferroptosis through autophagy signaling pathway in hepatic stellate cells. Redox Biol. 2021, 47, 102151. [Google Scholar] [CrossRef]
- Shen, M.; Guo, M.; Li, Y.; Wang, Y.; Qiu, Y.; Shao, J.; Zhang, F.; Xu, X.; Yin, G.; Wang, S.; et al. m(6)A methylation is required for dihydroartemisinin to alleviate liver fibrosis by inducing ferroptosis in hepatic stellate cells. Free. Radic. Biol. Med. 2022, 182, 246–259. [Google Scholar] [CrossRef]
- Dewidar, B.; Meyer, C.; Dooley, S.; Meindl-Beinker, N. TGF-β in hepatic stellate cell activation and liver fibrogenesis—Updated 2019. Cells 2019, 8, 1419. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, J.; Tsui, Y.M.; Shi, C.; Wang, Y.; Zhang, X.; Yan, Q.; Chen, M.; Jiang, C.; Yuan, Y.F.; et al. RALYL increases hepatocellular carcinoma stemness by sustaining the mRNA stability of TGF-β2. Nat. Commun. 2021, 12, 1518. [Google Scholar] [CrossRef] [PubMed]
- Neuschwander-Tetri, B.A. Hepatic lipotoxicity and the pathogenesis of nonalcoholic steatohepatitis: The central role of nontriglyceride fatty acid metabolites. Hepatology 2010, 52, 774–788. [Google Scholar] [CrossRef] [PubMed]
- Guilliams, M.; Scott, C.L. Liver macrophages in health and disease. Immunity 2022, 55, 1515–1529. [Google Scholar] [CrossRef]
- Zwicker, C.; Bujko, A.; Scott, C.L. Hepatic macrophage responses in inflammation, a function of plasticity, heterogeneity or both? Front. Immunol. 2021, 12, 690813. [Google Scholar] [CrossRef]
- Ramadori, G.; Armbrust, T. Cytokines in the liver. Eur. J. Gastroenterol. Hepatol. 2001, 13, 777–784. [Google Scholar] [CrossRef]
- Rivera, C.A.; Adegboyega, P.; van Rooijen, N.; Tagalicud, A.; Allman, M.; Wallace, M. Toll-like receptor-4 signaling and Kupffer cells play pivotal roles in the pathogenesis of non-alcoholic steatohepatitis. J. Hepatol. 2007, 47, 571–579. [Google Scholar] [CrossRef]
- Schwabe, R.F.; Seki, E.; Brenner, D.A. Toll-like receptor signaling in the liver. Gastroenterology 2006, 130, 1886–1900. [Google Scholar] [CrossRef]
- Feng, Y.; Dong, H.; Sun, B.; Hu, Y.; Yang, Y.; Jia, Y.; Jia, L.; Zhong, X.; Zhao, R. METTL3/METTL14 Transactivation and m6A-Dependent TGF-β1 Translation in Activated Kupffer Cells. Cell. Mol. Gastroenterol. Hepatol. 2021, 12, 839–856. [Google Scholar] [CrossRef]
- Chen, G.; Zhao, Q.; Yuan, B.; Wang, B.; Zhang, Y.; Li, Z.; Du, S.; Zeng, Z. ALKBH5-Modified HMGB1-STING Activation Contributes to Radiation Induced Liver Disease via Innate Immune Response. Int. J. Radiat. Oncol. Biol. Phys. 2021, 111, 491–501. [Google Scholar] [CrossRef]
- Xu, M.; Zhuo, R.; Tao, S.; Liang, Y.; Liu, C.; Liu, Q.; Wang, T.; Zhong, X. M(6)A RNA Methylation Mediates NOD1/NF-kB Signaling Activation in the Liver of Piglets Challenged with Lipopolysaccharide. Antioxidants 2022, 11, 1954. [Google Scholar] [CrossRef] [PubMed]
- Martinez, N.M.; Su, A.; Burns, M.C.; Nussbacher, J.K.; Schaening, C.; Sathe, S.; Yeo, G.W.; Gilbert, W.V. Pseudouridine synthases modify human pre-mRNA co-transcriptionally and affect pre-mRNA processing. Mol. Cell 2022, 82, 645–659.e9. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, J.; Li, X.; Xiong, X.; Wang, J.; Zhou, Z.; Zhu, X.; Gu, Y.; Dominissini, D.; He, L.; et al. N(1)-methyladenosine methylation in tRNA drives liver tumourigenesis by regulating cholesterol metabolism. Nat. Commun. 2021, 12, 6314. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Liu, Y.; Jiang, X.-W.; Li, W.-F.; Guo, G.; Gong, J.-P.; Ding, X. Clinicopathological and prognostic significance of Yes-associated protein expression in hepatocellular carcinoma and hepatic cholangiocarcinoma. Tumor Biol. 2016, 37, 13499–13508. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Zhu, M.; Li, W.; Zhou, Z.; Wan, X. m5C and m6A modification of long noncoding NKILA accelerates cholangiocarcinoma progression via the miR-582-3p-YAP1 axis. Liver Int. 2022, 42, 1144–1157. [Google Scholar] [CrossRef] [PubMed]
- Tóth, M.; Wehling, L.; Thiess, L.; Rose, F.; Schmitt, J.; Weiler, S.M.E.; Sticht, C.; De La Torre, C.; Rausch, M.; Albrecht, T.; et al. Co-expression of YAP and TAZ associates with chromosomal instability in human cholangiocarcinoma. BMC Cancer 2021, 21, 1079. [Google Scholar] [CrossRef]
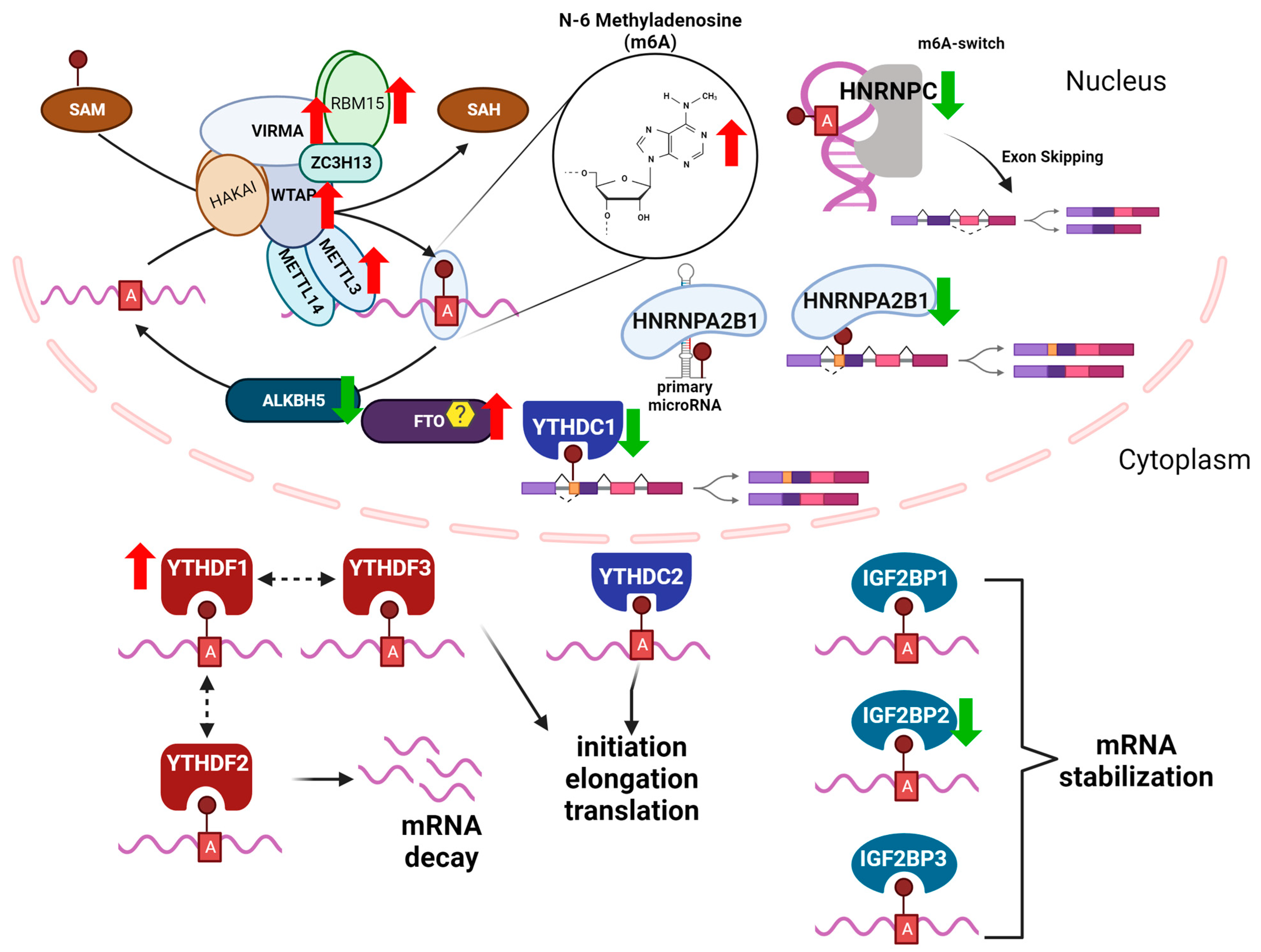

| Function | Description | Examples |
|---|---|---|
| mRNA stability | m6A can influence RNA stability by either increasing or decreasing the half-life of the transcript. | m6A increased the stability of the circadian clock gene transcript NR1D1, activating hepatic stellate cells (HSC) in a liver fibrosis cell model [49]. m6A decreased the stability of IFNB in human foreskin fibroblasts and m6A depletion enhanced IFNA and IFNB mRNA in response to viral infection [50]. |
| mRNA processing | m6A affects mRNA polyadenylation and other processing events. The presence of m6A near alternative splicing sites can enhance or inhibit the recognition of the spliceosome, leading to changes in the splicing pattern of the pre-mRNA. | In a cell model of pancreatic ductal adenocarcinoma, HNRNPC interacted with m6A sites in TAF8 to increase exon skipping and increased the transcript abundance of the pro-metastatic isoform TAF8S [51]. HCV infection induces the loss of an m6A peak on CIRBP, a gene that encodes for a stress-induced binding protein, in HepG2 cells. The decreased m6A levels resulted differential CIRBP isoform, suggesting a role for HCV-induced m6A changes on alternative splicing [52]. |
| mRNA localization | m6A can affect the localization of mRNA transcripts within cells | In mouse hippocampal neurons, Mettl3 knockout reduced m6A in the 3′UTR of Camk2a and Map2, inhibiting mRNA localization and reducing transcript abundance in neurites [53]. Knockdown of the m6A reader YTHDC1 in HeLa cells reduced cytoplasmic abundance of mRNA [54]. |
| mRNA translation | m6A affects the translation efficiency of mRNA transcripts. | Knockdown of the m6A eraser FTO increased m6A in axonal GAP-43 mRNA and decreased GAP-43 translation in cultured dorsal root ganglia neurons [55]. In HEK-293 cells, METTL3 knockdown decreased m6A in coding regions of transcripts, lead to ribosomal pausing. and decreased translation efficiency [56]. In human gastric cancer cells, translation efficiency of USP14 was increased by YTHDF1 and dependent on m6A methylation in the 3′UTR of the USP14 transcript [57]. |
| m6A Function in Liver Disease | M6A Modifiers |
|---|---|
| m6A levels in Circ-CCT3, a circular RNA upregulated in human HCC tissue that promoted growth and migration of HCC cells by sponging miR-378a-3p, were decreased by METTL3 knockdown and increased by ALKBH5 knockdown. Additionally, knockdown of METTL3 increased Circ-CCT3 expression in HCC cells [128]. | ALKBH5 (eraser), METTL3 (writer) |
| m6A transcriptome-wide profiling in livers of C57BL/6J mice identified 256 differentially methylated peaks in mRNA transcripts in pathways associated with hepatic ischemia reperfusion [129]. | |
| PPARGC1A plays an anti-oncogenic role in human and rodent models [130]. Knockdown of METTL3 increased PPARGC1A mRNA by decreasing four m6A sites near the stop codon of the PPARGC1A 3′UTR in Huh7 cells and YTHDF2 bound directly to PPARGC1A mRNA and promoted degradation of the transcript [131]. | METTL3 (writer), YTHDF2 (reader) |
| FZD10 induced liver cancer stem cell (CSC) expansion. Knockdown of METTL3 or YTHDF2 reduced FZD10 mRNA in HCC cells [132]. | METTL3 (writer), YTHDF2 (reader) |
| Mettl3 and m6A levels were upregulated in lenvatinib-resistant Huh7 cells compared to parental cells and Mettl3 promoted EGFR translation and lenvatinib resistance [133]. | METTL3 (writer) |
| LRPPRC upregulated PD-L1 by increasing m6A modifications in HepG2 and Hep3B cells. Knockdown of LRPPRC increased apoptosis and reduced migratory ability of HCC cells [134]. | LRPPRC (reader) |
| lncRNA miR4458HG was upregulated in human HCC tissue samples and activated the glycolysis pathway in HCC cells. miR4458HG interacted with IGF2BP2 to stabilize SLC2A1 and HK mRNA in BEL-7404 cells [135]. | IGF2BP2 (reader) |
| CircCCAR1 was increased in human HCC tissues and promoted tumor growth in a xenograft mouse model. m6A levels in CircCCAR1 were increased by WTAP and the interaction between WTAP and IGF2BP3 stabilized CircCCAR1 [136]. | WTAP (writer), IGF2BP3 (reader) |
| NRD1 deficiency promoted liver fibrosis in a CCl4-induced mouse liver fibrosis model. Additionally, ZCH3H13 was decreased and FTO was increased. Knockdown of ZCH3H13 and overexpression of FTO in HSC-LX2 cells decreased m6A methylation of NR1D1 mRNA, preventing YTHDC1 binding and stabilization of the NR1D1 transcript [49]. | ZCH3H13 (writer), FTO (eraser) |
| KIAA1429 was upregulated in sorafenib-resistant HCC cells and KIAA1429 depletion inhibited cell invasion and migratory ability [137]. KIAA1429 is elevated in HCC tissues and is associated with reduced overall survival [138]. KIAA1429 promotes m6A methylation of the 3′UTR of GATA3 pre-mRNA in the nucleus of SK-Hep1 and HCCLM3 cells resulting in transcript degradation and reduced GATA3 protein [138]. | KIAA1429 (writer) |
| RBM15 expression was upregulated and global m6A was increased in mouse fetal liver tissue of Gestational diabetes mellitus offspring and overexpression of RBM15 increased insulin resistance in primary mouse hepatocytes [139]. | RBM15 (writer) |
| Post-translational O-GlcNAcylation of YTHDF2 at Ser263 contributed to HBV-related hepatocarcinogenesis by promoting stabilization of MCM2 and MCM5 mRNA in HepG2-HBV1.3 cells [140]. | YTHDF2 (reader) |
| MeRIP-seq of LX2 human HSCs with ALKBH5 knockdown revealed fewer m6A peaks and fewer m6A peak-containing genes that the LX2 control. Silencing of ALKBH5 also downregulated CCL5, however silencing YTHDF2 restored CCL5 expression and promoted monocyte recruitment and polarization of irradiated LX2 cells [141]. | ALKBH5 (eraser), YTHDF2 (reader) |
| YTHDF2 regulated myeloid cell homeostasis in immune hepatitis through targeted degradation of Rxra in mouse myeloid-derived suppressor cells (MDSCs). YTHDF2 depletion increased MDSC expansion and decreased apoptosis [142]. | YTHDF2 (reader) |
| m6A modifications in HDAC1 mRNA was increased in the livers of rats with diet-induced metabolic syndrome. Additionally, METTL3 was upregulated and RIP-assays revealed that IGF2BP2 bound to HDAC1 mRNA and increased HDAC1 expression [143]. | METTL3 (writer), IGF2BP3 (reader) |
| Upregulation of VIRMA in intrahepatic cholangiocarcinoma (ICC) cells increased cell proliferation and invasion. CCL3, a cytokine secreted by hepatocytes, interacted with VIRMA to alter m6A modifications in ICC cells, specifically upregulating SIRT1, a downstream target of VIRMA-mediated m6A modification [144]. | VIRMA (writer) |
| A genome-wide analysis of tumors from HCC patients demonstrated an association between specific oncogenic lncRNAs and m6A modification. The correlations between lncRNA and m6A were identified as prognostic markers that can be used in HCC risk-assessment [145]. | |
| m6A-RIP-seq on human liver samples revealed higher hepatic m6A and increased expression of m6A writers, including METTL3, METTL14, and WTAP in biliary astresia (BA) patients compared to normal human liver controls. Additionally, m6A levels were increased in BA patients with advanced stage fibrosis, compared to early stage BA patients [146]. | METTL3(writer), METTL14(writer), WTAP (writer) |
| SLP2, a prognostic marker for HCC, was decreased in HCC cells with the inhibition of METTL3 and METTL3 is positively correlated with SLP2 in HCC patients [147]. | METTL3 (writer) |
| PARK7 mRNA encodes the Ras-dependent oncoprotein DJ-1 which is significantly upregulated in HCC patients. Overexpression of WTAP in HCC cells increased m6A modifications in PARK7 and IGF2BP1 bound PARK7 at an m6A site, stabilizing PARK7 mRNA [148]. | WTAP (writer), IGF2BP1 (reader) |
| The oncogenic circRNA, circFUT8 was upregulated in HCC and METTL14 promoted the m6A modification of circFUT8 in HCC cells. Additionally, M1 macrophage-derived exosomal miR-628-5p reduced m6A modification of circFUT8 be reducing METTL14 expression in HCC cells [149]. | METT14 (writer) |
| YTHDF3 facilitated HCC progression by promoting glycolysis in HCC cells and preventing the degradation of phosphofructokinase (PFKL) mRNA, via binding to the m6A modification in the PFKL transcript [150] | YTHDF3 (reader) |
| After partial hepatectomy in C57BL/6 mice, global liver m6A levels were increased and were correlated to an increase in METTL14 and hepatocyte growth factor (HGF) expression. The hepatic regenerative ability of hepatocyte-specific Mettl14 knockout mice was significantly reduced compared to wild-type [151]. | METTL14 (writer) |
| m6A regulators, i.e., YTHDC1, RBM15, and METTL3 were associated with HCC stage in human HCC tissue. While they were found to be upregulated in early stages of HCC, the expression of these m6A regulators was decreased in stage 4 HCC, suggesting that m6A modifications are altered in an HCC stage-specific manner [152]. | YTHDC1(reader), RBM15(reader), METTL3 (writer) |
| Global inhibition of METT3 expression in livers from Mettl3 knockout (ko) mice showed reduced expression of AQP8, a channel protein associated with glycogen accumulation. STM2457-inhibition of METTL3 activity decreased m6A levels and decreased glycogen storage capacity in mouse hepatocytes [153]. | METTL3 (writer) |
| The expression of METTL16 was significantly higher in livers from chronic hepatitis B patients with severe fibrosis compared to those with only mild fibrosis and METTL16 was positively correlated the expression of genes associated with chronic hepatitis B (CHB), including HLA-DPB1 and HLA-DPA1 [154]. | METTL16 (writer) |
| Knockdown of METT14 in HepG2 cells resulted in the differential m6A modification of 8 lncRNAs associated with HCC in liver patients. ARHGAP5-AS1 had the most m6A changes. RIP-qPCR assays in HCC cells demonstrated that IGF2BP2 had the highest binding affinity with ARHGAP5-AS1 and IGF2BP2 deletion decreased ARHGAP5-AS1 expression [155]. | METTL14 (writer), IGF2BP2 (reader) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petri, B.J.; Cave, M.C.; Klinge, C.M. Changes in m6A in Steatotic Liver Disease. Genes 2023, 14, 1653. https://doi.org/10.3390/genes14081653
Petri BJ, Cave MC, Klinge CM. Changes in m6A in Steatotic Liver Disease. Genes. 2023; 14(8):1653. https://doi.org/10.3390/genes14081653
Chicago/Turabian StylePetri, Belinda J., Matthew C. Cave, and Carolyn M. Klinge. 2023. "Changes in m6A in Steatotic Liver Disease" Genes 14, no. 8: 1653. https://doi.org/10.3390/genes14081653
APA StylePetri, B. J., Cave, M. C., & Klinge, C. M. (2023). Changes in m6A in Steatotic Liver Disease. Genes, 14(8), 1653. https://doi.org/10.3390/genes14081653







