Loss of a Premature Stop Codon in the Rice Wall-Associated Kinase 91 (WAK91) Gene Is a Candidate for Improving Leaf Sheath Blight Disease Resistance
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Inoculation
2.2. RNA Extraction and Sequencing
2.3. Sequence Quality Control and Read Alignments
2.4. Differential Gene Expression (DGE)
2.5. SNP Discovery and Variant Prediction
2.6. Population Study
2.7. Sampling DNA from Rice Varieties
2.8. Data Mining and Functional Annotation
3. Results
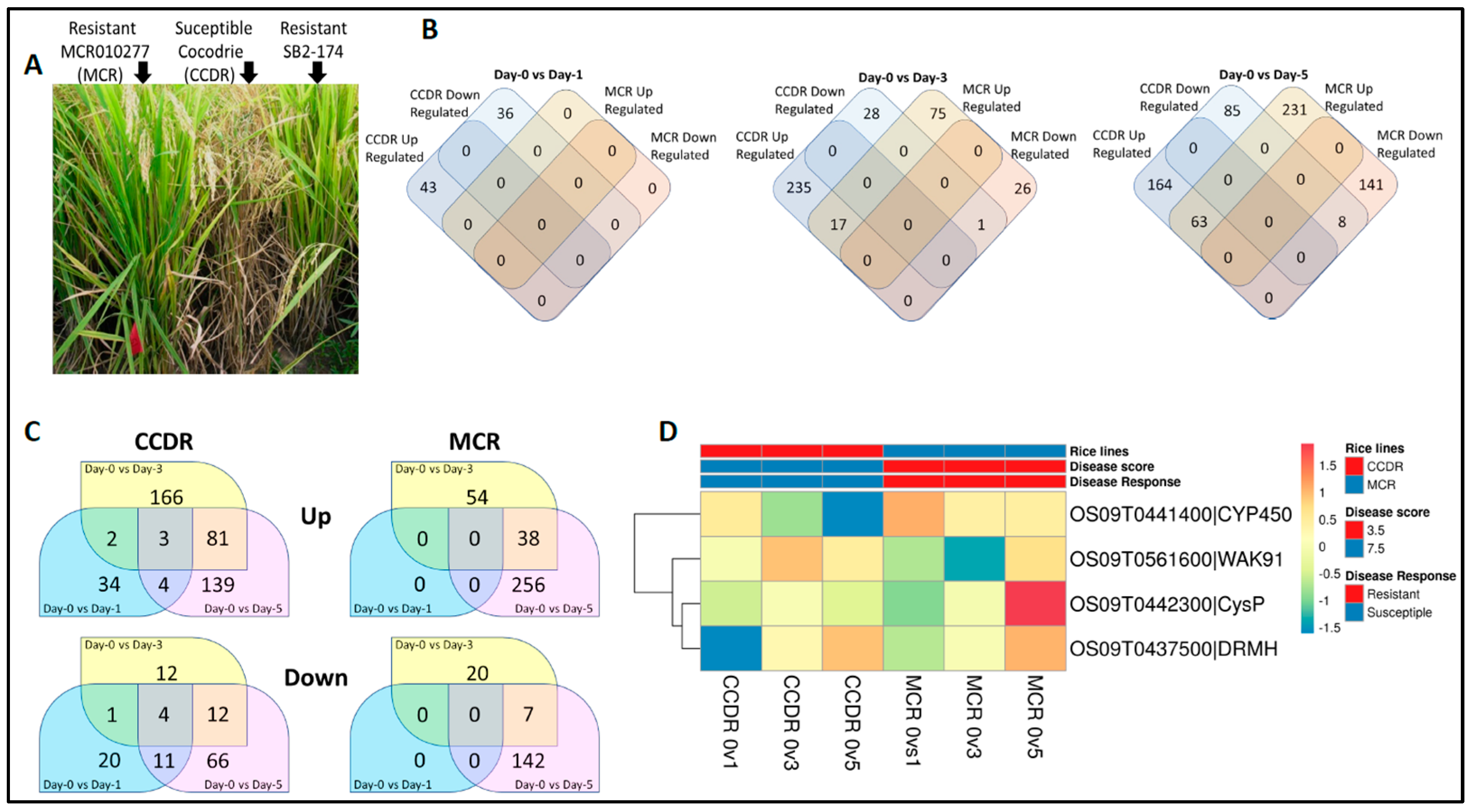
3.1. Transcriptome Analyses
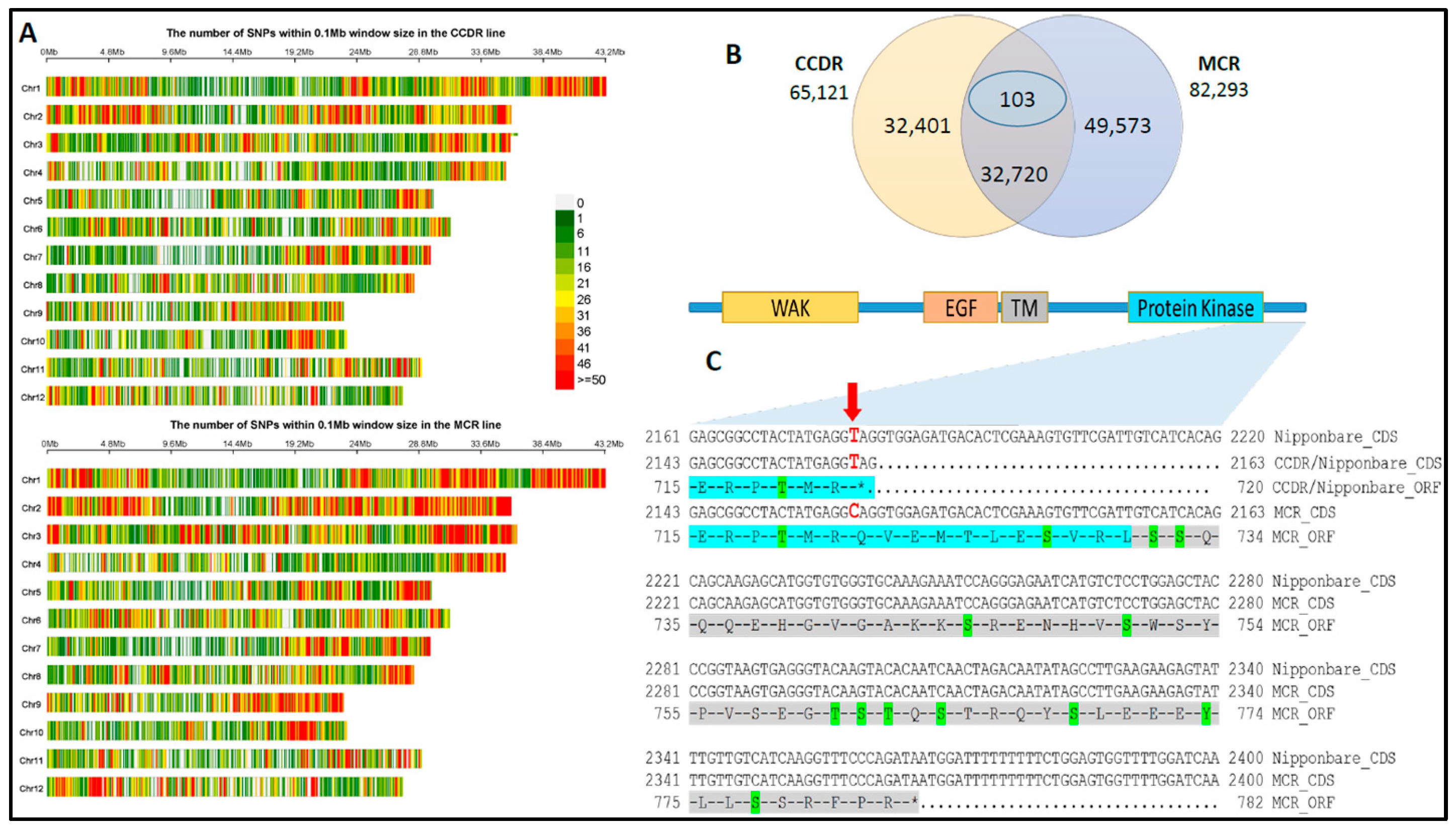
3.2. SNP Marker Discovery and Variant Cause Prediction
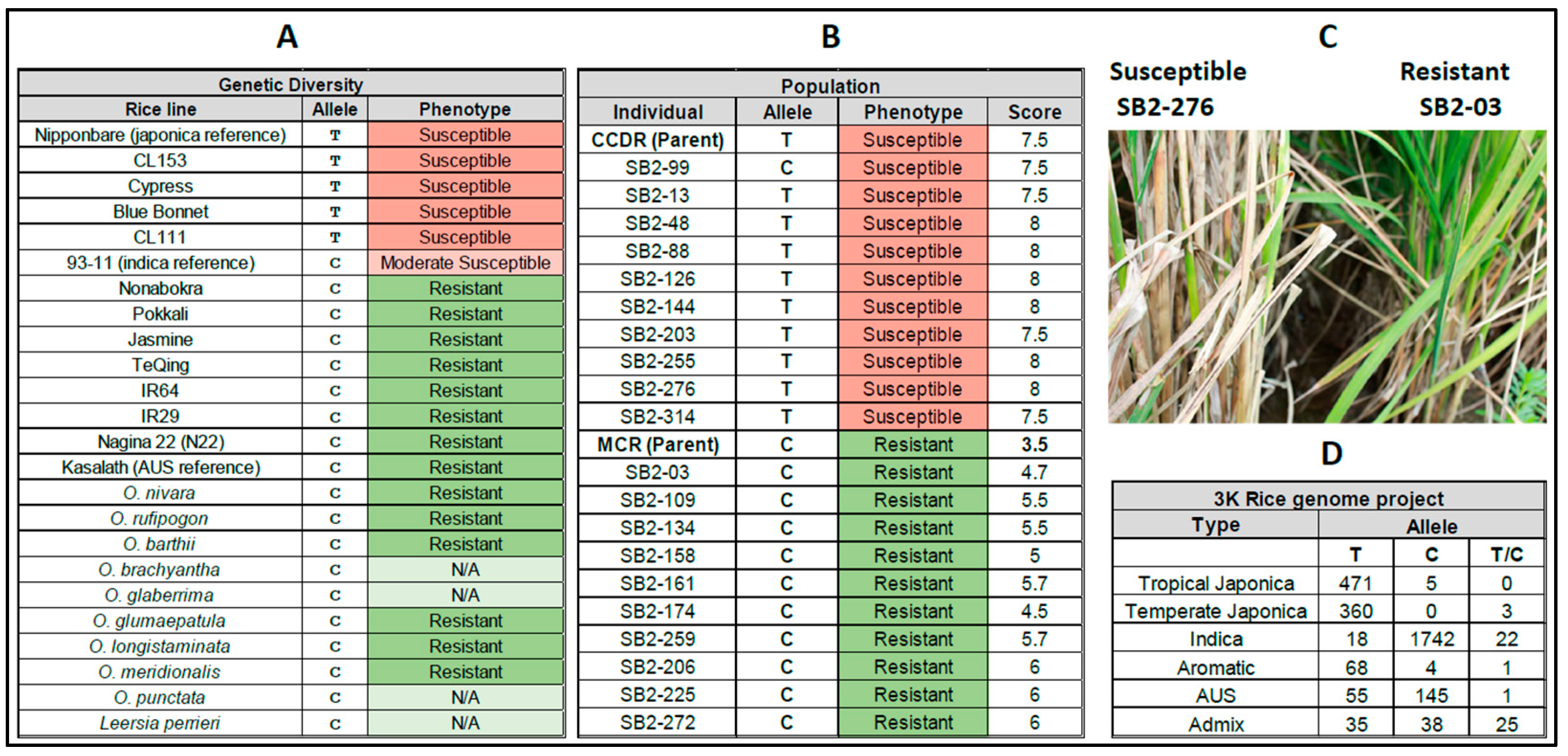
3.3. Double Haploid Population Study
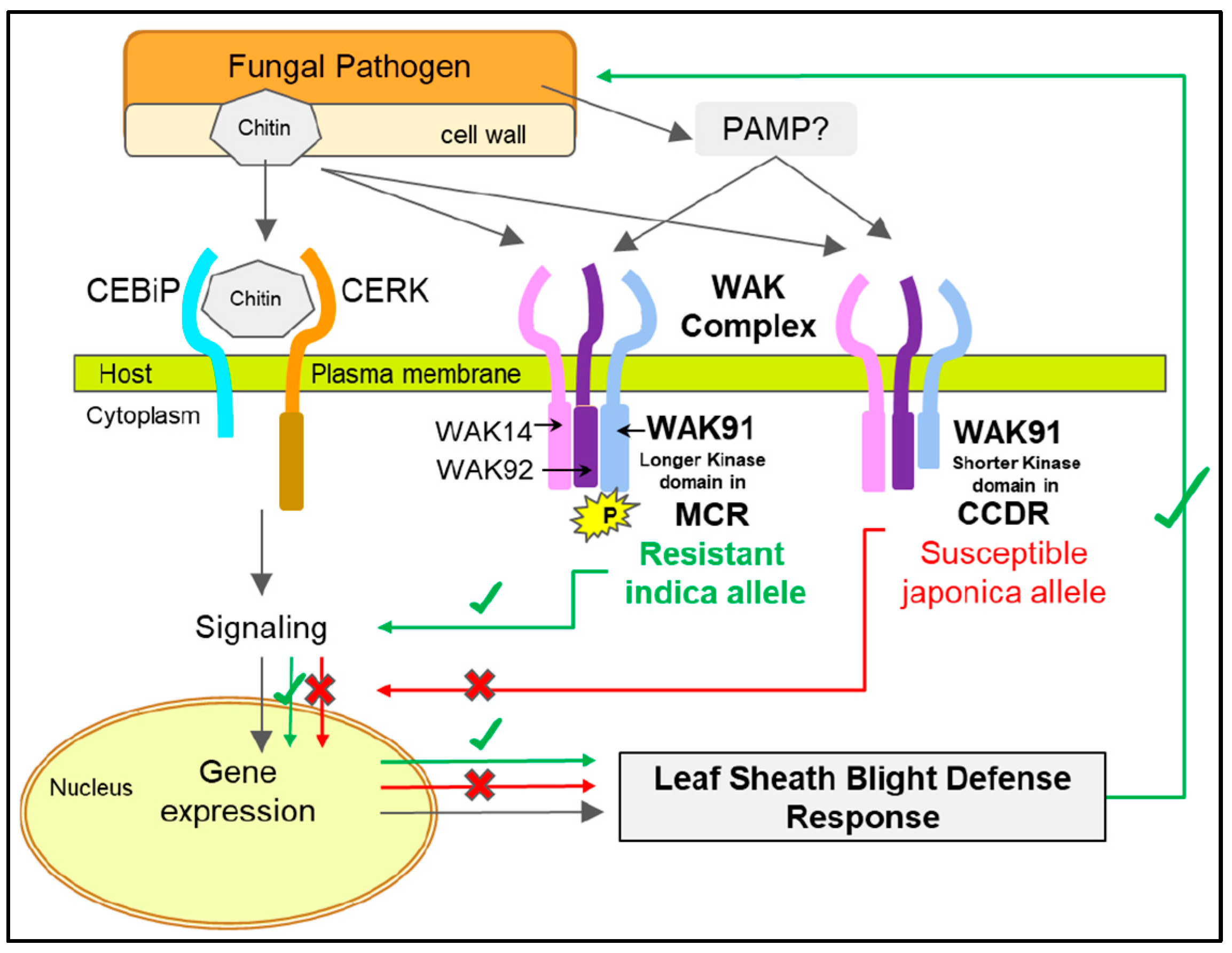
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Elert, E. Rice by the numbers: A good grain. Nature 2014, 514, S50–S51. [Google Scholar] [CrossRef] [PubMed]
- Senapati, M.; Tiwari, A.; Sharma, N.; Chandra, P.; Bashyal, B.M.; Ellur, R.K.; Bhowmick, P.K.; Bollinedi, H.; Vinod, K.K.; Singh, A.K.; et al. Rhizoctonia solani Kuhn Pathophysiology: Status and Prospects of Sheath Blight Disease Management in Rice. Front. Plant Sci. 2022, 13, 881116. [Google Scholar] [PubMed]
- Molla, K.A.; Karmakar, S.; Molla, J.; Bajaj, P.; Varshney, R.K.; Datta, S.K.; Datta, K. Understanding sheath blight resistance in rice: The road behind and the road ahead. Plant Biotechnol. J. 2020, 18, 895–915. [Google Scholar] [CrossRef] [PubMed]
- Maeda, S.; Dubouzet, J.G.; Kondou, Y.; Jikumaru, Y.; Seo, S.; Oda, K.; Matsui, M.; Hirochika, H.; Mori, M. The rice CYP78A gene BSR2 confers resistance to Rhizoctonia solani and affects seed size and growth in Arabidopsis and rice. Sci. Rep. 2019, 9, 587. [Google Scholar] [CrossRef]
- Li, D.; Li, S.; Wei, S.; Sun, W. Strategies to Manage Rice Sheath Blight: Lessons from Interactions between Rice and Rhizoctonia solani. Rice 2021, 14, 21. [Google Scholar] [PubMed]
- Lee, F. Rice Sheath Blight: A Major Rice Disease. Plant Dis. 1983, 67. [Google Scholar] [CrossRef]
- International Network for Genetic Evaluation of Rice; International Rice Research Institute. Standard Evaluation System for Rice, 4th ed.; IRRI, International Rice Research Institute: Manila, Philippines, 1996. [Google Scholar]
- Groth, D.E. Foliar Fungicides for Use in the Management of Rice Diseases; LSU Agricultural Experiment Station Reports. 1993. Available online: http://digitalcommons.lsu.edu/agexp/318 (accessed on 24 December 2022).
- Prasad, B.; Eizenga, G.C. Rice Sheath Blight Disease Resistance Identified in Oryza spp. Accessions. Plant Dis. 2008, 92, 1503–1509. [Google Scholar] [CrossRef]
- Pan, X.B.; Rush, M.C.; Sha, X.Y.; Xie, Q.J.; Linscombe, S.D.; Stetina, S.R.; Oard, J.H. Major Gene, Nonallelic Sheath Blight Resistance from the Rice Cultivars Jasmine 85 and Teqing. Crop Sci. 1999, 39, 338–346. [Google Scholar] [CrossRef]
- Singh, R.; Sunder, S.; Kumar, P. Sheath blight of rice: Current status and perspectives. Indian Phytopathol. 2016, 69, 340–351. [Google Scholar]
- USDA NASS. 2021 agricultural chemical use survey: Rice. In NASS Highlights; USDA NASS: Highlights, 2022. Available online: https://www.nass.usda.gov/Surveys/Guide_to_NASS_Surveys/Chemical_Use/2021_Field_Crops/chemhighlights-rice.pdf (accessed on 15 January 2023).
- Nalley, L.; Tsiboe, F.; Durand-Morat, A.; Shew, A.; Thoma, G. Economic and Environmental Impact of Rice Blast Pathogen (Magnaporthe oryzae) Alleviation in the United States. PLoS ONE 2016, 11, e0167295. [Google Scholar] [CrossRef]
- Sharma, A.; Kumar, V.; Shahzad, B.; Tanveer, M.; Sidhu, G.P.S.; Handa, N.; Kohli, S.; Yadav, P.; Bali, A.; Parihar, R.; et al. Worldwide pesticide usage and its impacts on ecosystem. SN Appl. Sci. 2019, 1, 1446. [Google Scholar] [CrossRef]
- Zheng, A.; Lin, R.; Zhang, D.; Qin, P.; Xu, L.; Ai, P.; Ding, L.; Wang, Y.; Chen, Y.; Liu, Y.; et al. The evolution and pathogenic mechanisms of the rice sheath blight pathogen. Nat. Commun. 2013, 4, 1424. [Google Scholar] [CrossRef]
- Chuanqing, Z.; Liu, Y.; Ma, X.; Feng, Z.; Ma, Z.H. Characterization of sensitivity of Rhizoctonia solani, causing rice sheath blight, to mepronil and boscalid. Crop Prot. 2009, 28, 381–386. [Google Scholar]
- Webster, R.K.; Gunnell, P.S. Compendium of Rice Diseases; The Disease Compendium Series of the American Phytopathological Society; APS Press, The American Phytopathological Society: St. Paul, MN, USA, 1992. [Google Scholar]
- Ajayi-Oyetunde, O.O.; Bradley, C.A. Rhizoctonia solani: Taxonomy, population biology and management of rhizoctonia seedling disease of soybean. Plant Pathol. 2018, 67, 3–17. [Google Scholar] [CrossRef]
- Olaya, G. Detection of resistance to QoI fungicides in Rhizoctonia solani isolates from rice. Phytopathology 2012, 102, S4-88. [Google Scholar]
- Linscombe, S.; Jodari, F.; Bollich, P.; Groth, D.; White, L.M.; Chu, Q.R.; Dunand, R.T.; Sanders, D.E. Registration of ‘Cocodrie’ Rice. Crop Sci. 2000, 40, 294. [Google Scholar] [CrossRef]
- Silva, J.; Scheffler, B.; Sanabria, Y.; De Guzman, C.; Galam, D.; Farmer, A.; Woodward, J.; May, G.; Oard, J. Identification of candidate genes in rice for resistance to sheath blight disease by whole genome sequencing. Theor. Appl. Genet. 2012, 124, 63–74. [Google Scholar] [CrossRef]
- Li, Z.; Pinson, S.R.; Marchetti, M.A.; Stansel, J.W.; Park, W.D. Characterization of quantitative trait loci (QTLs) in cultivated rice contributing to field resistance to sheath blight (Rhizoctonia solani). Theor. Appl. Genet. 1995, 91, 382–388. [Google Scholar] [CrossRef]
- Galam, D. Development of Sheath Blight-Resistant Breeding Lines for Southern U.S. Environments and Morphological and Genetic Survey of Giant Salvinia Populations in Louisiana and Texas. Ph.D. Thesis, Louisiana State University, Baton Rouge, LA, USA, 2019. [Google Scholar]
- Ohyanagi, H.; Tanaka, T.; Sakai, H.; Shigemoto, Y.; Yamaguchi, K.; Habara, T.; Fujii, Y.; Antonio, B.A.; Nagamura, Y.; Imanishi, T.; et al. The Rice Annotation Project Database (RAP-DB): Hub for Oryza sativa ssp. japonica genome information. Nucleic Acids Res. 2006, 34, D741–D744. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Koboldt, D.C.; Zhang, Q.; Larson, D.E.; Shen, D.; McLellan, M.D.; Lin, L.; Miller, C.A.; Mardis, E.R.; Ding, L.; Wilson, R.K. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012, 22, 568–576. [Google Scholar] [CrossRef] [PubMed]
- Naithani, S.; Geniza, M.; Jaiswal, P. Variant Effect Prediction Analysis Using Resources Available at Gramene Database. Methods Mol. Biol. 2017, 1533, 279–297. [Google Scholar]
- Mansueto, L.; Fuentes, R.R.; Borja, F.N.; Detras, J.; Abriol-Santos, J.M.; Chebotarov, D.; Sanciangco, M.; Palis, K.; Copetti, D.; Poliakov, A.; et al. Rice SNP-seek database update: New SNPs, indels, and queries. Nucleic Acids Res. 2016, 45, D1075–D1081. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Mauleon, R.; Hu, Z.; Chebotarov, D.; Tai, S.; Wu, Z.; Li, M.; Zheng, T.; Fuentes, R.R.; Zhang, F.; et al. Genomic variation in 3010 diverse accessions of Asian cultivated rice. Nature 2018, 557, 43–49. [Google Scholar] [CrossRef]
- Lu, J.; Wang, C.; Zeng, D.; Li, J.; Shi, X.; Shi, Y.; Zhou, Y. Genome-Wide Association Study Dissects Resistance Loci against Bacterial Blight in a Diverse Rice Panel from the 3000 Rice Genomes Project. Rice 2021, 14, 22. [Google Scholar] [CrossRef]
- Tello-Ruiz, M.K.; Naithani, S.; Gupta, P.; Olson, A.; Wei, S.; Preece, J.; Jiao, Y.; Wang, B.; Chougule, K.; Garg, P.; et al. Gramene 2021: Harnessing the power of comparative genomics and pathways for plant research. Nucleic Acids Res. 2021, 49, D1452–D1463. [Google Scholar]
- Gupta, P.; Naithani, S.; Tello-Ruiz, M.K.; Chougule, K.; D’Eustachio, P.; Fabregat, A.; Jiao, Y.; Keays, M.; Lee, Y.K.; Kumari, S.; et al. Gramene Database: Navigating Plant Comparative Genomics Resources. Curr. Plant Biol. 2016, 7–8, 10–15. [Google Scholar] [CrossRef]
- Chu, Q.R.; Linscombe, S.; Rush, M.; Groth, D.; Oard, J.; Sha, X.; Utomo, H. Registration of a C/M Doubled Haploid Mapping Population of Rice. Crop Sci. 2006, 46, 1417–1418. [Google Scholar] [CrossRef][Green Version]
- Nelson, J.; Oard, J.; Groth, D.; Utomo, H.; Jia, Y.; Liu, G.; Moldenhauer, K.; Correa-Victoria, F.J.; Fjellstrom, R.; Scheffler, B. Sheath-blight resistance QTLS in japonica rice germplasm. Euphytica 2012, 184, 23–34. [Google Scholar] [CrossRef]
- Tello-Ruiz, M.K.; Jaiswal, P.; Ware, D. Gramene: A Resource for Comparative Analysis of Plants Genomes and Pathways. Methods Mol. Biol. 2022, 2443, 101–131. [Google Scholar] [PubMed]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef] [PubMed]
- Magbanua, Z.V.; Arick, M., 2nd; Buza, T.; Hsu, C.Y.; Showmaker, K.C.; Chouvarine, P.; Deng, P.; Peterson, D.G.; Lu, S. Transcriptomic dissection of the rice-Burkholderia glumae interaction. BMC Genom. 2014, 15, 755. [Google Scholar] [CrossRef]
- Wilkins, K.E.; Booher, N.J.; Wang, L.; Bogdanove, A.J. TAL effectors and activation of predicted host targets distinguish Asian from African strains of the rice pathogen Xanthomonas oryzae pv. oryzicola while strict conservation suggests universal importance of five TAL effectors. Front. Plant Sci. 2015, 6, 536. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; McDowell, I.C.; Nodzenski, M.; Scholtens, D.M.; Allen, A.S.; Lowe, W.L.; Reddy, T.E. Transversions have larger regulatory effects than transitions. BMC Genom. 2017, 18, 394. [Google Scholar] [CrossRef]
- Li, G.; Chern, M.; Jain, R.; Martin, J.A.; Schackwitz, W.S.; Jiang, L.; Vega-Sánchez, M.E.; Lipzen, A.M.; Barry, K.W.; Schmutz, J.; et al. Genome-Wide Sequencing of 41 Rice (Oryza sativa L.) Mutated Lines Reveals Diverse Mutations Induced by Fast-Neutron Irradiation. Mol. Plant 2016, 9, 1078–1081. [Google Scholar] [CrossRef]
- Sato, H.; Ideta, O.; Ando, I.; Kunihiro, Y.; Hirabayashi, H.; Iwano, M.; Miyasaka, A.; Nemoto, H.; Imbe, T. Mapping QTLs for sheath blight resistance in the rice line WSS2. Breed. Sci. 2004, 54, 265–271. [Google Scholar] [CrossRef]
- Pinson, S.R.M.; Capdevielle, F.M.; Oard, J.H. Confirming QTLs and Finding Additional Loci Conditioning Sheath Blight Resistance in Rice Using Recombinant Inbred Lines. Crop Sci. 2005, 45, 503–510. [Google Scholar] [CrossRef]
- Chen, J.; Xuan, Y.; Yi, J.; Xiao, G.; Yuan, P.; Li, D. Progress in rice sheath blight resistance research. Front. Plant Sci. 2023, 14, 1141697. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.-X.; Xia, L.-Z.; Wen, Z.-H.; Ji, Z.-J.; Zeng, D.-L.; Qian, Q.; Yang, C.-D. Mapping resistant QTLs for rice sheath blight disease with a doubled haploid population. J. Integr. Agric. 2015, 14, 801–810. [Google Scholar] [CrossRef]
- Jia, Y.; Liu, G.; Correa-Victoria, F.J.; McClung, A.M.; Oard, J.H.; Bryant, R.J.; Jia, M.H.; Correll, J.C. Registration of Four Rice Germplasm Lines with Improved Resistance to Sheath Blight and Blast Diseases. J. Plant Regist. 2012, 6, 95–100. [Google Scholar] [CrossRef]
- Taguchi-Shiobara, F.; Ozaki, H.; Sato, H.; Maeda, H.; Kojima, Y.; Ebitani, T.; Yano, M. Mapping and validation of QTLs for rice sheath blight resistance. Breed. Sci. 2013, 63, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Costanzo, S.; Jackson, A.K.; Brooks, S.A. High-resolution mapping of Rsn1, a locus controlling sensitivity of rice to a necrosis-inducing phytotoxin from Rhizoctonia solani AG1-IA. Theor. Appl. Genet. 2011, 123, 33–41. [Google Scholar] [CrossRef]
- Ontoy, J.C.; Shrestha, B.; Karki, H.S.; Barphagha, I.; Angira, B.; Famoso, A.; Ham, J.H. Genetic Characterization of the Partial Disease Resistance of Rice to Bacterial Panicle Blight and Sheath Blight by Combined QTL Linkage and QTL-seq Analyses. Plants 2023, 12, 559. [Google Scholar] [CrossRef]
- Goad, D.M.; Jia, Y.; Gibbons, A.; Liu, Y.; Gealy, D.; Caicedo, A.L.; Olsen, K.M. Identification of Novel QTL Conferring Sheath Blight Resistance in Two Weedy Rice Mapping Populations. Rice 2020, 13, 21. [Google Scholar] [CrossRef]
- Zuo, S.; Zhang, Y.; Yin, Y.; Li, G.; Zhang, G.; Wang, H.; Chen, Z.; Pan, X. Fine-mapping of qSB-9 TQ, a gene conferring major quantitative resistance to rice sheath blight. Mol. Breed. 2014, 34, 2191–2203. [Google Scholar] [CrossRef]
- Zuo, S.; Yin, Y.; Pan, C.; Chen, Z.; Zhang, Y.; Gu, S.; Zhu, L.; Pan, X. Fine mapping of qSB-11(LE), the QTL that confers partial resistance to rice sheath blight. Theor. Appl. Genet. 2013, 126, 1257–1272. [Google Scholar] [CrossRef]
- Masajo, T. Varietal Resistance in Rice to Sheath Blight Disease Caused by Thanatephorus cucumeris (Frank) Donk (Rhizoctonia solani Kuhn). Ph.D. Thesis, Louisiana State University, Baton Rouge, LA, USA, 1976. [Google Scholar]
- Wamishe, Y.; Cartwright, R.; Lee, F. Management of Rice Diseases. In Compendium of Rice Diseases and Pests; Hardke, J.T., Ed.; University of Arkansas Cooperative Extension Service: Little Rock, AR, USA, 2013; pp. 123–223. [Google Scholar]
- Eizenga, G.C.; Lee, F.N.; Rutger, J.N. Screening Oryza Species Plants for Rice Sheath Blight Resistance. Plant Dis. 2002, 86, 808–812. [Google Scholar] [CrossRef]
- Groth, D.E.; Bond, J.A. Effects of Cultivars and Fungicides on Rice Sheath Blight, Yield, and Quality. Plant Dis. 2007, 91, 1647–1650. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Correa-Victoria, F.; McClung, A.; Zhu, L.; Liu, G.; Wamishe, Y.; Xie, J.; Marchetti, M.A.; Pinson, S.R.M.; Rutger, J.N.; et al. Rapid Determination of Rice Cultivar Responses to the Sheath Blight Pathogen Rhizoctonia solani Using a Micro-Chamber Screening Method. Plant Dis. 2007, 91, 485–489. [Google Scholar] [CrossRef] [PubMed]
- Srinivasachary; Willocquet, L.; Savary, S. Resistance to rice sheath blight (Rhizoctonia solani Kühn) [(teleomorph: Thanatephorus cucumeris (A.B. Frank) Donk.] disease: Current status and perspectives. Euphytica 2011, 178, 1–22. [Google Scholar] [CrossRef]
- XiaoXing, L.; Ping, L.; AiPing, Z. In vitro selection of sheath blight resistance germplams in rice. Afr. J. Microbiol. Res. 2013, 7, 4422–4429. [Google Scholar]
- IRRI-CGIAR. The International Rice Genebank Collection Information System; IRRI-CGIAR: Manila, Philippines, 2019. [Google Scholar]
- Hossain, M.K.; Tze, O.S.; Nadarajah, K.; Jena, K.; Rahman Bhuiyan, M.A.; Ratnam, W. Identification and validation of sheath blight resistance in rice (Oryza sativa L.) cultivars against Rhizoctonia solani. Can. J. Plant Pathol. 2014, 36, 482–490. [Google Scholar] [CrossRef]
- Bedanand, C. Evaluating Sheath Blight Resistance in Rice Using Detached Tiller and Field Screening Method. J. Nepal Agric. Res. Counc. 2015, 1, 1–8. [Google Scholar]
- Yu, J.; Hu, S.; Wang, J.; Wong, G.K.; Li, S.; Liu, B.; Deng, Y.; Dai, L.; Zhou, Y.; Zhang, X.; et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 2002, 296, 79–92. [Google Scholar] [CrossRef]
- Xia, Y.; Fei, B.; He, J.; Zhou, M.; Zhang, D.; Pan, L.; Li, S.; Liang, Y.; Wang, L.; Zhu, J.; et al. Transcriptome analysis reveals the host selection fitness mechanisms of the Rhizoctonia solani AG1IA pathogen. Sci. Rep. 2017, 7, 10120. [Google Scholar] [CrossRef]
- Lin, W.; Wang, Y.; Liu, X.; Shang, J.X.; Zhao, L. OsWAK112, A Wall-Associated Kinase, Negatively Regulates Salt Stress Responses by Inhibiting Ethylene Production. Front. Plant Sci. 2021, 12, 751965. [Google Scholar] [CrossRef]
- Ram, T.; Majumder, N.D.; Laha, G.S.; Ansari, M.M.; Kar, C.S.; Mishra, B. Identification of donors for sheath blight resistance in wild species of rice. Indian J. Genet. Plant Breed. 2008, 68, 317–319. [Google Scholar]
- Djedatin, G.; Ndjiondjop, M.N.; Mathieu, T.; Cruz, C.M.V.; Sanni, A.; Ghesquière, A.; Verdier, V. Evaluation of African Cultivated Rice Oryza glaberrima for Resistance to Bacterial Blight. Plant Dis. 2011, 95, 441–447. [Google Scholar] [CrossRef]
- Kohorn, B.D.; Kohorn, S.L. The cell wall-associated kinases, WAKs, as pectin receptors. Front. Plant Sci. 2012, 3, 88. [Google Scholar] [CrossRef] [PubMed]
- Anderson, C.M.; Wagner, T.A.; Perret, M.; He, Z.H.; He, D.; Kohorn, B.D. WAKs: Cell wall-associated kinases linking the cytoplasm to the extracellular matrix. Plant Mol. Biol. 2001, 47, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Wagner, T.A.; Kohorn, B.D. Wall-associated kinases are expressed throughout plant development and are required for cell expansion. Plant Cell 2001, 13, 303–318. [Google Scholar] [CrossRef] [PubMed]
- He, Z.H.; Cheeseman, I.; He, D.; Kohorn, B.D. A cluster of five cell wall-associated receptor kinase genes, Wak1-5, are expressed in specific organs of Arabidopsis. Plant Mol. Biol. 1999, 39, 1189–1196. [Google Scholar] [CrossRef]
- He, Z.H.; He, D.; Kohorn, B.D. Requirement for the induced expression of a cell wall associated receptor kinase for survival during the pathogen response. Plant J. 1998, 14, 55–63. [Google Scholar] [CrossRef]
- Hurni, S.; Scheuermann, D.; Krattinger, S.G.; Kessel, B.; Wicker, T.; Herren, G.; Fitze, M.N.; Breen, J.; Presterl, T.; Ouzunova, M.; et al. The maize disease resistance gene Htn1 against northern corn leaf blight encodes a wall-associated receptor-like kinase. Proc. Natl. Acad. Sci. USA 2015, 112, 8780–8785. [Google Scholar] [CrossRef]
- Yang, P.; Praz, C.; Li, B.; Singla, J.; Robert, C.A.M.; Kessel, B.; Scheuermann, D.; Lüthi, L.; Ouzunova, M.; Erb, M.; et al. Fungal resistance mediated by maize wall-associated kinase ZmWAK-RLK1 correlates with reduced benzoxazinoid content. New Phytol. 2019, 221, 976–987. [Google Scholar] [CrossRef] [PubMed]
- Saintenac, C.; Lee, W.S.; Cambon, F.; Rudd, J.J.; King, R.C.; Marande, W.; Powers, S.J.; Bergès, H.; Phillips, A.L.; Uauy, C.; et al. Wheat receptor-kinase-like protein Stb6 controls gene-for-gene resistance to fungal pathogen Zymoseptoria tritici. Nat. Genet. 2018, 50, 368–374. [Google Scholar] [CrossRef] [PubMed]
- Kaur, R.; Singh, K.; Singh, J. A root-specific wall-associated kinase gene, HvWAK1, regulates root growth and is highly divergent in barley and other cereals. Funct. Integr. Genom. 2013, 13, 167–177. [Google Scholar] [CrossRef]
- Kanneganti, V.; Gupta, A.K. Wall associated kinases from plants—An overview. Physiol. Mol. Biol. Plants 2008, 14, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Lv, Y.; Lei, W.; Li, X.; Chen, Y.; Zheng, L.; Xia, Y.; Shen, Z.G. Cloning and characterization of the Oryza sativa wall-associated kinase gene OsWAK11 and its transcriptional response to abiotic stresses. Plant Soil 2014, 384, 335–346. [Google Scholar] [CrossRef]
- Xia, Y.; Yin, S.; Zhang, K.; Shi, X.; Lian, C.; Zhang, H.; Hu, Z.; Shen, Z. OsWAK11, a rice wall-associated kinase, regulates Cu detoxification by alteration the immobilization of Cu in cell walls. Environ. Exp. Bot. 2018, 150, 99–105. [Google Scholar] [CrossRef]
- Delteil, A.; Gobbato, E.; Cayrol, B.; Estevan, J.; Michel-Romiti, C.; Dievart, A.; Kroj, T.; Morel, J.B. Several wall-associated kinases participate positively and negatively in basal defense against rice blast fungus. BMC Plant Biol. 2016, 16, 17. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhou, S.Y.; Zhao, W.S.; Su, S.C.; Peng, Y.L. A novel wall-associated receptor-like protein kinase gene, OsWAK1, plays important roles in rice blast disease resistance. Plant Mol. Biol. 2009, 69, 337–346. [Google Scholar] [PubMed]
- Wang, N.; Huang, H.J.; Ren, S.T.; Li, J.J.; Sun, Y.; Sun, D.Y.; Zhang, S.Q. The rice wall-associated receptor-like kinase gene OsDEES1 plays a role in female gametophyte development. Plant Physiol. 2012, 160, 696–707. [Google Scholar] [CrossRef]
- Hu, X.; Yu, P.; Zhang, Y.; Gao, Z.; Sun, B.; Wu, W.; Deng, C.; Abbas, A.; Hong, Y.; Sun, L.; et al. Mutation of DEFECTIVE EMBRYO SAC1 results in a low seed-setting rate in rice by regulating embryo sac development. J. Exp. Bot. 2023, 74, 1501–1516. [Google Scholar] [CrossRef]
- Harkenrider, M.; Sharma, R.; De Vleesschauwer, D.; Tsao, L.; Zhang, X.; Chern, M.; Canlas, P.; Zuo, S.; Ronald, P.C. Overexpression of Rice Wall-Associated Kinase 25 (OsWAK25) Alters Resistance to Bacterial and Fungal Pathogens. PLoS ONE 2016, 11, e0147310. [Google Scholar] [CrossRef]
- Seo, Y.S.; Chern, M.; Bartley, L.E.; Han, M.; Jung, K.H.; Lee, I.; Walia, H.; Richter, T.; Xu, X.; Cao, P.; et al. Towards establishment of a rice stress response interactome. PLoS Genet. 2011, 7, e1002020. [Google Scholar] [CrossRef]
- Ke, Y.; Deng, H.; Wang, S. Advances in understanding broad-spectrum resistance to pathogens in rice. Plant J. 2017, 90, 738–748. [Google Scholar] [CrossRef]
- Li, L.; Yu, Y.; Zhou, Z.; Zhou, J.M. Plant pattern-recognition receptors controlling innate immunity. Sci. China Life Sci. 2016, 59, 878–888. [Google Scholar] [PubMed]
- Cayrol, B.; Delteil, A.; Gobbato, E.; Kroj, T.; Morel, J.B. Three wall-associated kinases required for rice basal immunity form protein complexes in the plasma membrane. Plant Signal. Behav. 2016, 11, e1149676. [Google Scholar] [CrossRef] [PubMed]
- Boller, T.; Felix, G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 2009, 60, 379–406. [Google Scholar] [CrossRef]
- Macho, A.P.; Zipfel, C. Plant PRRs and the activation of innate immune signaling. Mol. Cell 2014, 54, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Hayafune, M.; Berisio, R.; Marchetti, R.; Silipo, A.; Kayama, M.; Desaki, Y.; Arima, S.; Squeglia, F.; Ruggiero, A.; Tokuyasu, K.; et al. Chitin-induced activation of immune signaling by the rice receptor CEBiP relies on a unique sandwich-type dimerization. Proc. Natl. Acad. Sci. USA 2014, 111, E404–E413. [Google Scholar] [CrossRef] [PubMed]
- Duitama, J.; Silva, A.; Sanabria, Y.; Cruz, D.F.; Quintero, C.; Ballen, C.; Lorieux, M.; Scheffler, B.; Farmer, A.; Torres, E.; et al. Whole genome sequencing of elite rice cultivars as a comprehensive information resource for marker assisted selection. PLoS ONE 2015, 10, e0124617. [Google Scholar]
- Monaco, M.K.; Stein, J.; Naithani, S.; Wei, S.; Dharmawardhana, P.; Kumari, S.; Amarasinghe, V.; Youens-Clark, K.; Thomason, J.; Preece, J.; et al. Gramene 2013: Comparative plant genomics resources. Nucleic Acids Res. 2014, 42, D1193–D1199. [Google Scholar] [CrossRef]
- Hossain, M.K.; Islam, M.R.; Sundaram, R.M.; Bhuiyan, M.A.R.; Wickneswari, R. Introgression of the QTL qSB11-1(TT) conferring sheath blight resistance in rice (Oryza sativa) into an elite variety, UKMRC 2, and evaluation of its backcross-derived plants. Front. Plant Sci. 2022, 13, 981345. [Google Scholar] [CrossRef]

| SNP | Substitution Type | CCDR | MCR |
|---|---|---|---|
| A→G | Transition | 8703 | 11,102 |
| T→C | Transition | 8896 | 11,228 |
| G→A | Transition | 8735 | 11,420 |
| C→T | Transition | 8878 | 11,622 |
| G→C | Transversion | 2817 | 3523 |
| C→G | Transversion | 2730 | 3559 |
| T→G | Transversion | 3274 | 4108 |
| G→T | Transversion | 3367 | 4158 |
| A→C | Transversion | 3316 | 3964 |
| C→A | Transversion | 3339 | 4175 |
| T→A | Transversion | 3734 | 4428 |
| A→T | Transversion | 3685 | 4397 |
| Total SNPs | 61,474 | 77,684 | |
| Total Indels | Insertions and deletions | 3647 | 4609 |
| Total variants | 65,121 | 82,293 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Bader, N.; Meier, A.; Geniza, M.; Gongora, Y.S.; Oard, J.; Jaiswal, P. Loss of a Premature Stop Codon in the Rice Wall-Associated Kinase 91 (WAK91) Gene Is a Candidate for Improving Leaf Sheath Blight Disease Resistance. Genes 2023, 14, 1673. https://doi.org/10.3390/genes14091673
Al-Bader N, Meier A, Geniza M, Gongora YS, Oard J, Jaiswal P. Loss of a Premature Stop Codon in the Rice Wall-Associated Kinase 91 (WAK91) Gene Is a Candidate for Improving Leaf Sheath Blight Disease Resistance. Genes. 2023; 14(9):1673. https://doi.org/10.3390/genes14091673
Chicago/Turabian StyleAl-Bader, Noor, Austin Meier, Matthew Geniza, Yamid Sanabria Gongora, James Oard, and Pankaj Jaiswal. 2023. "Loss of a Premature Stop Codon in the Rice Wall-Associated Kinase 91 (WAK91) Gene Is a Candidate for Improving Leaf Sheath Blight Disease Resistance" Genes 14, no. 9: 1673. https://doi.org/10.3390/genes14091673
APA StyleAl-Bader, N., Meier, A., Geniza, M., Gongora, Y. S., Oard, J., & Jaiswal, P. (2023). Loss of a Premature Stop Codon in the Rice Wall-Associated Kinase 91 (WAK91) Gene Is a Candidate for Improving Leaf Sheath Blight Disease Resistance. Genes, 14(9), 1673. https://doi.org/10.3390/genes14091673




