Relating the Biogenesis and Function of P Bodies in Drosophila to Human Disease
Abstract
:1. Introduction
2. Drosophila P bodies Have Complex Compositions and Multiple Functions and Are Present in Diverse Tissues
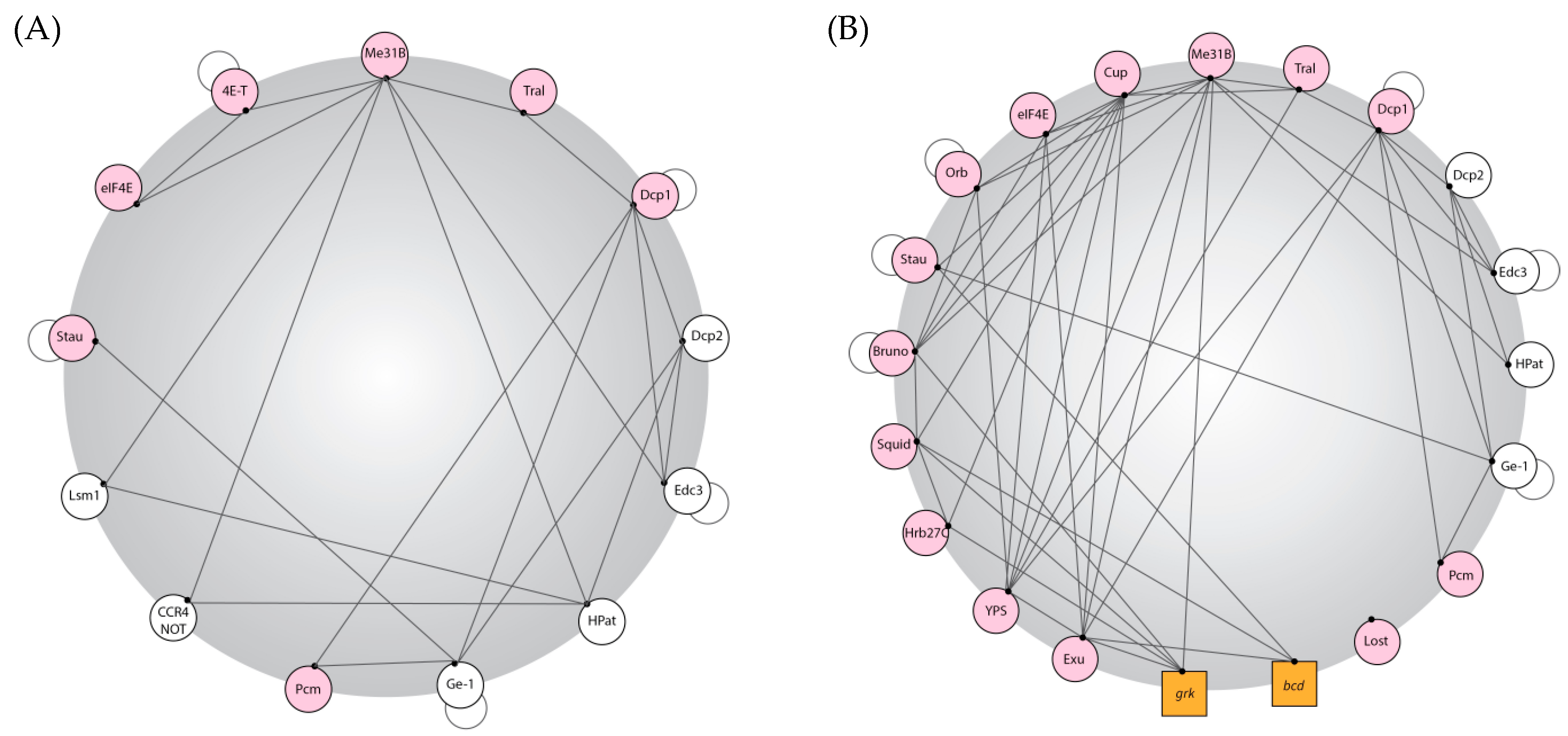
2.1. Focus on: P Body Protein Content in Drosophila
- Co-localisation studies in the female germline have shown unique RNA binding proteins present in these P bodies, such as Cup, Oo18 RNA binding protein (Orb), Squid (Sqd), Heterogeneous nuclear ribonucleoprotein at 27C (Hrb27C), Exuperantia (Exu), Ypsilon Schachtel (Yps), Lost, and Bruno 1 (Bru1) [35,40,42].
2.2. In Vivo P Bodies Can Undergo Regulated Changes
3. Current Understanding of the Requirements for Drosophila P Body Formation
- Me31B can act as a scaffold for phase separation in vitro [38], and loss of Me31B results in the disassembly of P bodies in S2 cells [13]. However, in Drosophila nurse cells, when Me31B was mutated, such that it was not able to self-aggregate or be recruited to condensates, Cup and Tral still formed condensates [84]. This suggests that Me31B is not specifically required for condensate formation in this scenario [30,84].
4. In Vivo Exploration of P Body Biogenesis
4.1. Focus on: Techniques for the Study of P Bodies in Drosophila

| Technique | Information Gained | Live or Fixed | Single- Molecule Resolution Feasible | Multiplexing Possible (Currently) | Signal to Noise Ratio | Potential to Affect RNA Localisation | Super- Resolution Imaging Possible | Adapted to Drosophila |
| Stem loop and coat binding system | RNA localisation [126] | Live [126] | Yes [128] | Yes [128] | Low [126] | Yes [142] | Yes [128] | Yes [127,129,130] |
| smFISH | RNA localisation [131] | Fixed [131] | Yes [131] | Yes [131] | High [131] | No [131] | Yes [54,101,102,103,104,105,106] | Yes [54,101,102,103,104,105,106] |
| Multiplexed smFISH | mRNA decay [132] | Fixed [132] | Yes [132] | Yes [132] | High [132] | No [132] | Yes [132] | Yes [54] |
| TRICK | Nascent translation [133,134] | Live [133,134] | Yes [133,134] | No | Low [133,134] | Yes [133,134] | Yes [133,134] | Yes [133,134] |
| SunTag | Nascent translation [136,137,138,139] | Live [129,136,137,138,139] | Yes [136,137,138,139] | No | High [136,137,138,139] | Low [136,137,138,139] | Yes [136,137,138,139] | Yes [129,140,141] |
5. Using Drosophila as a Model to Understand the Role of P Bodies in Human Diseases
6. Concluding Remarks: Drosophila as a Model for the Future Study of P Bodies
- What regulates the assembly and disassembly of condensates in vivo?
- What effect do proteins, and their specific domains, have on condensate properties?
- What effect do post-translational modifications have on condensate integrity?
- How do RNAs, and certain motifs, contribute to the formation of RNP granules?
- Which RNA structures, sequences, and post-transcriptional modifications affect the ability of RNA to associate with RNP granules?
- Are the material properties of RNP granules intrinsic to their biological function?
- What are the biological functions of phase separation?
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Banani, S.F.; Lee, H.O.; Hyman, A.A.; Rosen, M.K. Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017, 18, 285–298. [Google Scholar] [CrossRef] [PubMed]
- Parker, M.W.; Bell, M.; Mir, M.; Kao, J.A.; Darzacq, X.; Botchan, M.R.; Berger, J.M. A new class of disordered elements controls DNA replication through initiator self-assembly. eLife 2019, 8, e48562. [Google Scholar] [CrossRef] [PubMed]
- Feric, M.; Vaidya, N.; Harmon, T.S.; Mitrea, D.M.; Zhu, L.; Richardson, T.M.; Kriwacki, R.W.; Pappu, R.V.; Brangwynne, C.P. Coexisting liquid phases underlie nucleolar sub-compartments. Cell 2016, 165, 1686–1697. [Google Scholar] [CrossRef] [PubMed]
- Brangwynne, C.P.; Mitchison, T.J.; Hyman, A.A. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl. Acad. Sci. USA 2011, 108, 4334–4339. [Google Scholar] [CrossRef]
- Horvathova, I.; Voigt, F.; Kotrys, A.V.; Zhan, Y.; Artus-Revel, C.G.; Eglinger, J.; Stadler, M.B.; Giorgetti, L.; Chao, J.A. The Dynamics of mRNA Turnover Revealed by Single-Molecule Imaging in Single Cells. Mol. Cell 2017, 68, 615–625.e9. [Google Scholar] [CrossRef]
- Hubstenberger, A.; Courel, M.; Bénard, M.; Souquere, S.; Ernoult-Lange, M.; Chouaib, R.; Yi, Z.; Morlot, J.-B.; Munier, A.; Fradet, M.; et al. P-Body Purification Reveals the Condensation of Repressed mRNA Regulons. Mol. Cell 2017, 68, 144–157.e5. [Google Scholar] [CrossRef]
- Sheth, U.; Parker, R. Decapping and Decay of Messenger RNA Occur in Cytoplasmic Processing Bodies. Science 2003, 300, 805–808. [Google Scholar] [CrossRef]
- Xu, J.; Chua, N.-H. Arabidopsis decapping 5 is required for mRNA decapping, P-body formation, and translational repression during postembryonic development. Plant Cell 2009, 21, 3270–3279. [Google Scholar] [CrossRef]
- Xu, J.; Yang, J.-Y.; Niu, Q.-W.; Chua, N.-H. Arabidopsis DCP2, DCP1, and VARICOSE Form a Decapping Complex Required for Postembryonic Development. Plant Cell 2006, 18, 3386–3398. [Google Scholar] [CrossRef]
- Gallo, C.M.; Munro, E.; Rasoloson, D.; Merritt, C.; Seydoux, G. Processing bodies and germ granules are distinct RNA granules that interact in C. elegans embryos. Dev. Biol. 2008, 323, 76–87. [Google Scholar] [CrossRef]
- Ding, L.; Spencer, A.; Morita, K.; Han, M. The developmental timing regulator AIN-1 interacts with miRISCs and may target the argonaute protein ALG-1 to cytoplasmic P bodies in C. elegans. Mol. Cell 2005, 19, 437–447. [Google Scholar] [CrossRef]
- Zhang, L.; Ding, L.; Cheung, T.H.; Dong, M.-Q.; Chen, J.; Sewell, A.K.; Liu, X.; Yates, J.R.; Han, M. Systematic identification of miRISC proteins, miRNAs, and their mRNA targets in C. elegans by their interactions with GW182 family proteins AIN-1 and AIN-2. Mol. Cell 2007, 28, 598–613. [Google Scholar] [CrossRef] [PubMed]
- Eulalio, A.; Behm-Ansmant, I.; Schweizer, D.; Izaurralde, E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell Biol. 2007, 27, 3970–3981. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.-D.; Jiao, X.; Grima, D.; Newbury, S.F.; Kiledjian, M.; Chou, T.-B. Drosophila processing bodies in oogenesis. Dev. Biol. 2008, 322, 276–288. [Google Scholar] [CrossRef]
- Flemr, M.; Ma, J.; Schultz, R.M.; Svoboda, P. P-Body Loss Is Concomitant with Formation of a Messenger RNA Storage Domain in Mouse Oocytes. Biol. Reprod. 2010, 82, 1008–1017. [Google Scholar] [CrossRef] [PubMed]
- Cougot, N.; Babajko, S.; Séraphin, B. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 2004, 165, 31–40. [Google Scholar] [CrossRef]
- Andrei, M.A.; Ingelfinger, D.; Heintzmann, R.; Achsel, T.; Rivera-Pomar, R.; Lührmann, R. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA 2005, 11, 717–727. [Google Scholar] [CrossRef]
- Xing, W.; Muhlrad, D.; Parker, R.; Rosen, M.K. A quantitative inventory of yeast P body proteins reveals principles of composition and specificity. eLife 2020, 9, e56525. [Google Scholar] [CrossRef]
- Standart, N.; Weil, D. P-Bodies: Cytosolic Droplets for Coordinated mRNA Storage. Trends Genet. 2018, 34, 612–626. [Google Scholar] [CrossRef]
- Luo, Y.; Na, Z.; Slavoff, S.A. P-Bodies: Composition, Properties, and Functions. Biochemistry 2018, 57, 2424–2431. [Google Scholar] [CrossRef]
- Wang, C.; Schmich, F.; Srivatsa, S.; Weidner, J.; Beerenwinkel, N.; Spang, A. Context-dependent deposition and regulation of mRNAs in P-bodies. eLife 2018, 7, e29815. [Google Scholar] [CrossRef] [PubMed]
- Riggs, C.L.; Kedersha, N.; Ivanov, P.; Anderson, P. Mammalian stress granules and P bodies at a glance. J. Cell Sci. 2020, 133, jcs242487. [Google Scholar] [CrossRef] [PubMed]
- Hallacli, E.; Kayatekin, C.; Nazeen, S.; Wang, X.H.; Sheinkopf, Z.; Sathyakumar, S.; Sarkar, S.; Jiang, X.; Dong, X.; Di Maio, R.; et al. The Parkinson’s disease protein α-synuclein is a modulator of processing bodies and mRNA stability. Cell 2022, 185, 2035–2056.e33. [Google Scholar] [CrossRef]
- Boccaccio, G.L.; Thomas, M.G.; García, C.C. Membraneless Organelles and Condensates Orchestrate Innate Immunity Against Viruses. J. Mol. Biol. 2023, 435, 167976. [Google Scholar] [CrossRef] [PubMed]
- Lavalée, M.; Curdy, N.; Laurent, C.; Fournié, J.-J.; Franchini, D.-M. Cancer cell adaptability: Turning ribonucleoprotein granules into targets. Trends Cancer 2021, 7, 902–915. [Google Scholar] [CrossRef] [PubMed]
- Rehwinkel, J.; Behm-Ansmant, I.; Gatfield, D.; Izaurralde, E. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA 2005, 11, 1640–1647. [Google Scholar] [CrossRef] [PubMed]
- Layana, C.; Vilardo, E.S.; Corujo, G.; Hernández, G.; Rivera-Pomar, R. Drosophila Me31B is a Dual eIF4E-Interacting Protein. J. Mol. Biol. 2023, 435, 167949. [Google Scholar] [CrossRef]
- Di Stefano, B.; Luo, E.-C.; Haggerty, C.; Aigner, S.; Charlton, J.; Brumbaugh, J.; Ji, F.; Rabano Jiménez, I.; Clowers, K.J.; Huebner, A.J.; et al. The RNA Helicase DDX6 Controls Cellular Plasticity by Modulating P-Body Homeostasis. Cell Stem Cell 2019, 25, 622–638.e13. [Google Scholar] [CrossRef]
- Kolaj, A.; Zahr, S.K.; Wang, B.S.; Krawec, T.; Kazan, H.; Yang, G.; Kaplan, D.R.; Miller, F.D. The P-body protein 4E-T represses translation to regulate the balance between cell genesis and establishment of the postnatal NSC pool. Cell Rep. 2023, 42, 112242. [Google Scholar] [CrossRef]
- Buddika, K.; Huang, Y.-T.; Ariyapala, I.S.; Butrum-Griffith, A.; Norrell, S.A.; O’Connor, A.M.; Patel, V.K.; Rector, S.A.; Slovan, M.; Sokolowski, M.; et al. Coordinated repression of pro-differentiation genes via P-bodies and transcription maintains Drosophila intestinal stem cell identity. Curr. Biol. 2022, 32, 386–397.e6. [Google Scholar] [CrossRef]
- Zabolotskaya, M.V.; Grima, D.P.; Lin, M.-D.; Chou, T.-B.; Newbury, S.F. The 5′-3′ exoribonuclease Pacman is required for normal male fertility and is dynamically localized in cytoplasmic particles in Drosophila testis cells. Biochem. J. 2008, 416, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Jensen, L.; Venkei, Z.G.; Watase, G.J.; Bisai, B.; Pletcher, S.; Lee, C.-Y.; Yamashita, Y.M. me31B regulates stem cell homeostasis by preventing excess dedifferentiation in the Drosophila male germline. J. Cell Sci. 2021, 134, jcs258757. [Google Scholar] [CrossRef] [PubMed]
- Sankaranarayanan, M.; Weil, T.T. Granule regulation by phase separation during Drosophila oogenesis. Emerg. Top. Life Sci. 2020, 4, 355–364. [Google Scholar]
- Wilsch-Bräuninger, M.; Schwarz, H.; Nüsslein-Volhard, C. A Sponge-like Structure Involved in the Association and Transport of Maternal Products during Drosophila Oogenesis. J. Cell Biol. 1997, 139, 817–829. [Google Scholar] [CrossRef] [PubMed]
- Snee, M.J.; Macdonald, P.M. Dynamic organization and plasticity of sponge bodies. Dev. Dyn. 2009, 238, 918–930. [Google Scholar] [CrossRef] [PubMed]
- McCambridge, A.; Solanki, D.; Olchawa, N.; Govani, N.; Trinidad, J.C.; Gao, M. Comparative Proteomics Reveal Me31B’s Interactome Dynamics, Expression Regulation, and Assembly Mechanism into Germ Granules during Drosophila Germline Development. Sci. Rep. 2020, 10, 564. [Google Scholar] [CrossRef]
- DeHaan, H.; McCambridge, A.; Armstrong, B.; Cruse, C.; Solanki, D.; Trinidad, J.C.; Arkov, A.L.; Gao, M. An in vivo proteomic analysis of the Me31B interactome in Drosophila germ granules. FEBS Lett. 2017, 591, 3536–3547. [Google Scholar] [CrossRef]
- Sankaranarayanan, M.; Emenecker, R.J.; Wilby, E.L.; Jahnel, M.; Trussina, I.R.E.A.; Wayland, M.; Alberti, S.; Holehouse, A.S.; Weil, T.T. Adaptable P body physical states differentially regulate bicoid mRNA storage during early Drosophila development. Dev. Cell 2021, 56, 2886–2901.e6. [Google Scholar] [CrossRef]
- Ferraiuolo, M.A.; Basak, S.; Dostie, J.; Murray, E.L.; Schoenberg, D.R.; Sonenberg, N. A role for the eIF4E-binding protein 4E-T in P-body formation and mRNA decay. J. Cell Biol. 2005, 170, 913–924. [Google Scholar] [CrossRef]
- Davidson, A.; Parton, R.M.; Rabouille, C.; Weil, T.T.; Davis, I. Localized Translation of gurken/TGF-α mRNA during Axis Specification Is Controlled by Access to Orb/CPEB on Processing Bodies. Cell Rep. 2016, 14, 2451–2462. [Google Scholar] [CrossRef]
- Fan, S.-J.; Marchand, V.; Ephrussi, A. Drosophila Ge-1 Promotes P Body Formation and oskar mRNA Localization. PLoS ONE 2011, 6, e20612. [Google Scholar] [CrossRef] [PubMed]
- Weil, T.T.; Parton, R.M.; Herpers, B.; Soetaert, J.; Veenendaal, T.; Xanthakis, D.; Dobbie, I.M.; Halstead, J.M.; Hayashi, R.; Rabouille, C.; et al. Drosophila patterning is established by differential association of mRNAs with P bodies. Nat. Cell Biol. 2012, 14, 1305–1313. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, A.; Amikura, R.; Hanyu, K.; Kobayashi, S. Me31B silences translation of oocyte-localizing RNAs through the formation of cytoplasmic RNP complex during Drosophila oogenesis. Development 2001, 128, 3233–3242. [Google Scholar] [CrossRef] [PubMed]
- Patel, P.H.; Barbee, S.A.; Blankenship, J.T. GW-Bodies and P-Bodies Constitute Two Separate Pools of Sequestered Non-Translating RNAs. PLoS ONE 2016, 11, e0150291. [Google Scholar] [CrossRef]
- Pérez-Vilaró, G.; Fernández-Carrillo, C.; Mensa, L.; Miquel, R.; Sanjuan, X.; Forns, X.; Pérez-del-Pulgar, S.; Díez, J. Hepatitis C virus infection inhibits P-body granule formation in human livers. J. Hepatol. 2015, 62, 785–790. [Google Scholar] [CrossRef]
- Derrick, C.J.; Weil, T.T. Translational control of gurken mRNA in Drosophila development. Cell Cycle 2017, 16, 23–32. [Google Scholar] [CrossRef]
- Shimada, Y.; Burn, K.M.; Niwa, R.; Cooley, L. Reversible response of protein localization and microtubule organization to nutrient stress during Drosophila early oogenesis. Dev. Biol. 2011, 355, 250–262. [Google Scholar] [CrossRef]
- Burn, K.M.; Shimada, Y.; Ayers, K.; Vemuganti, S.; Lu, F.; Hudson, A.M.; Cooley, L. Somatic insulin signaling regulates a germline starvation response in Drosophila egg chambers. Dev. Biol. 2015, 398, 206–217. [Google Scholar] [CrossRef]
- Wippich, F.; Vaishali; Hennrich, M.L.; Ephrussi, A. Nutritional stress-induced regulation of microtubule organization and mRNP transport by HDAC1 controlled α-tubulin acetylation. Commun. Biol. 2023, 6, 776. [Google Scholar] [CrossRef]
- Courel, M.; Clément, Y.; Bossevain, C.; Foretek, D.; Vidal Cruchez, O.; Yi, Z.; Bénard, M.; Benassy, M.-N.; Kress, M.; Vindry, C.; et al. GC content shapes mRNA storage and decay in human cells. eLife 2019, 8, e49708. [Google Scholar] [CrossRef]
- Marcelo, A.; Koppenol, R.; de Almeida, L.P.; Matos, C.A.; Nóbrega, C. Stress granules, RNA-binding proteins and polyglutamine diseases: Too much aggregation? Cell Death Dis. 2021, 12, 592. [Google Scholar] [CrossRef] [PubMed]
- Horner, V.L.; Wolfner, M.F. Transitioning from egg to embryo: Triggers and mechanisms of egg activation. Dev. Dyn. 2008, 237, 527–544. [Google Scholar] [CrossRef] [PubMed]
- York-Andersen, A.H.; Parton, R.M.; Bi, C.J.; Bromley, C.L.; Davis, I.; Weil, T.T. A single and rapid calcium wave at egg activation in Drosophila. Biol. Open 2015, 4, 553–560. [Google Scholar] [CrossRef] [PubMed]
- Beadle, L.F.; Love, J.C.; Shapovalova, Y.; Artemev, A.; Rattray, M.; Ashe, H.L. Combined modelling of mRNA decay dynamics and single-molecule imaging in the Drosophila embryo uncovers a role for P-bodies in 5′ to 3′ degradation. PLOS Biol. 2023, 21, e3001956. [Google Scholar]
- Eichler, C.E.; Hakes, A.C.; Hull, B.; Gavis, E.R. Compartmentalized oskar degradation in the germ plasm safeguards germline development. eLife 2020, 9, e49988. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Ly, M.; Lugowski, A.; Laver, J.D.; Lipshitz, H.D.; Smibert, C.A.; Rissland, O.S. ME31B globally represses maternal mRNAs by two distinct mechanisms during the Drosophila maternal-to-zygotic transition. eLife 2017, 6, e27891. [Google Scholar] [CrossRef]
- Barbee, S.A.; Estes, P.S.; Cziko, A.-M.; Hillebrand, J.; Luedeman, R.A.; Coller, J.M.; Johnson, N.; Howlett, I.C.; Geng, C.; Ueda, R.; et al. Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron 2006, 52, 997–1009. [Google Scholar] [CrossRef]
- Vessey, J.P.; Vaccani, A.; Xie, Y.; Dahm, R.; Karra, D.; Kiebler, M.A.; Macchi, P. Dendritic Localization of the Translational Repressor Pumilio 2 and Its Contribution to Dendritic Stress Granules. J. Neurosci. 2006, 26, 6496–6508. [Google Scholar] [CrossRef]
- Miller, L.C.; Blandford, V.; McAdam, R.; Sanchez-Carbente, M.R.; Badeaux, F.; DesGroseillers, L.; Sossin, W.S. Combinations of DEAD box proteins distinguish distinct types of RNA: Protein complexes in neurons. Mol. Cell Neurosci. 2009, 40, 485–495. [Google Scholar] [CrossRef]
- Cougot, N.; Bhattacharyya, S.N.; Tapia-Arancibia, L.; Bordonné, R.; Filipowicz, W.; Bertrand, E.; Rage, F. Dendrites of Mammalian Neurons Contain Specialized P-Body-Like Structures That Respond to Neuronal Activation. J. Neurosci. 2008, 28, 13793–13804. [Google Scholar] [CrossRef]
- Formicola, N.; Vijayakumar, J.; Besse, F. Neuronal ribonucleoprotein granules: Dynamic sensors of localized signals. Traffic 2019, 20, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Formicola, N.; Heim, M.; Dufourt, J.; Lancelot, A.-S.; Nakamura, A.; Lagha, M.; Besse, F. Tyramine induces dynamic RNP granule remodeling and translation activation in the Drosophila brain. eLife 2021, 10, e65742. [Google Scholar] [CrossRef] [PubMed]
- Zeitelhofer, M.; Karra, D.; Macchi, P.; Tolino, M.; Thomas, S.; Schwarz, M.; Kiebler, M.; Dahm, R. Dynamic Interaction between P-Bodies and Transport Ribonucleoprotein Particles in Dendrites of Mature Hippocampal Neurons. J. Neurosci. 2008, 28, 7555–7562. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Banjade, S.; Cheng, H.-C.; Kim, S.; Chen, B.; Guo, L.; Llaguno, M.; Hollingsworth, J.V.; King, D.S.; Banani, S.F.; et al. Phase transitions in the assembly of multivalent signalling proteins. Nature 2012, 483, 336–340. [Google Scholar] [CrossRef]
- Chen, C.; Jia, H.; Nakamura, Y.; Kanekura, K.; Hayamizu, Y. Effect of Multivalency on Phase-Separated Droplets Consisting of Poly(PR) Dipeptide Repeats and RNA at the Solid/Liquid Interface. ACS Omega 2022, 7, 19280–19287. [Google Scholar] [CrossRef]
- Shapiro, D.M.; Ney, M.; Eghtesadi, S.A.; Chilkoti, A. Protein Phase Separation Arising from Intrinsic Disorder: First-Principles to Bespoke Applications. J. Phys. Chem. B 2021, 125, 6740–6759. [Google Scholar] [CrossRef]
- Martin, E.W.; Holehouse, A.S. Intrinsically disordered protein regions and phase separation: Sequence determinants of assembly or lack thereof. Emerg. Top. Life Sci. 2020, 4, 307–329. [Google Scholar]
- Posey, A.E.; Holehouse, A.S.; Pappu, R.V. Phase Separation of Intrinsically Disordered Proteins. Methods Enzymol. 2018, 611, 1–30. [Google Scholar]
- Molliex, A.; Temirov, J.; Lee, J.; Coughlin, M.; Kanagaraj, A.P.; Kim, H.J.; Mittag, T.; Taylor, J.P. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 2015, 163, 123–133. [Google Scholar] [CrossRef]
- Lee, J.; Cho, H.; Kwon, I. Phase separation of low-complexity domains in cellular function and disease. Exp. Mol. Med. 2022, 54, 1412–1422. [Google Scholar] [CrossRef]
- Kato, M.; Han, T.W.; Xie, S.; Shi, K.; Du, X.; Wu, L.C.; Mirzaei, H.; Goldsmith, E.J.; Longgood, J.; Pei, J.; et al. Cell-free Formation of RNA Granules: Low Complexity Sequence Domains Form Dynamic Fibers within Hydrogels. Cell 2012, 149, 753–767. [Google Scholar] [CrossRef] [PubMed]
- Brangwynne, C.P.; Tompa, P.; Pappu, R.V. Polymer physics of intracellular phase transitions. Nat. Phys. 2015, 11, 899–904. [Google Scholar] [CrossRef]
- Nott, T.J.; Petsalaki, E.; Farber, P.; Jervis, D.; Fussner, E.; Plochowietz, A.; Craggs, T.D.; Bazett-Jones, D.P.; Pawson, T.; Forman-Kay, J.D.; et al. Phase Transition of a Disordered Nuage Protein Generates Environmentally Responsive Membraneless Organelles. Mol. Cell 2015, 57, 936–947. [Google Scholar] [CrossRef]
- Pak, C.W.; Kosno, M.; Holehouse, A.S.; Padrick, S.B.; Mittal, A.; Ali, R.; Yunus, A.A.; Liu, D.R.; Pappu, R.V.; Rosen, M.K. Sequence Determinants of Intracellular Phase Separation by Complex Coacervation of a Disordered Protein. Mol. Cell 2016, 63, 72–85. [Google Scholar] [CrossRef]
- Riback, J.A.; Katanski, C.D.; Kear-Scott, J.L.; Pilipenko, E.V.; Rojek, A.E.; Sosnick, T.R.; Drummond, D.A. Stress-Triggered Phase Separation Is an Adaptive, Evolutionarily Tuned Response. Cell 2017, 168, 1028–1040.e19. [Google Scholar] [CrossRef] [PubMed]
- Murthy, A.C.; Dignon, G.L.; Kan, Y.; Zerze, G.H.; Parekh, S.H.; Mittal, J.; Fawzi, N.L. Molecular interactions underlying liquid-liquid phase separation of the FUS low complexity domain. Nat. Struct. Mol. Biol. 2019, 26, 637–648. [Google Scholar] [CrossRef]
- Dzuricky, M.; Rogers, B.A.; Shahid, A.; Cremer, P.S.; Chilkoti, A. De novo engineering of intracellular condensates using artificial disordered proteins. Nat. Chem. 2020, 12, 814–825. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.-H.; Yu, J.H.; Gulick, T.; Bloch, K.D.; Bloch, D.B. RNA-associated protein 55 (RAP55) localizes to mRNA processing bodies and stress granules. RNA 2006, 12, 547–554. [Google Scholar] [CrossRef] [PubMed]
- Ayache, J.; Bénard, M.; Ernoult-Lange, M.; Minshall, N.; Standart, N.; Kress, M.; Weil, D. P-body assembly requires DDX6 repression complexes rather than decay or Ataxin2/2L complexes. Mol. Biol. Cell 2015, 26, 2579–2595. [Google Scholar] [CrossRef]
- Serman, A.; Le Roy, F.; Aigueperse, C.; Kress, M.; Dautry, F.; Weil, D. GW body disassembly triggered by siRNAs independently of their silencing activity. Nucleic Acids Res. 2007, 35, 4715–4727. [Google Scholar] [CrossRef]
- Banani, S.F.; Rice, A.M.; Peeples, W.B.; Lin, Y.; Jain, S.; Parker, R.; Rosen, M.K. Compositional Control of Phase-Separated Cellular Bodies. Cell 2016, 166, 651–663. [Google Scholar] [CrossRef]
- Huch, S.; Nissan, T. An mRNA decapping mutant deficient in P body assembly limits mRNA stabilization in response to osmotic stress. Sci. Rep. 2017, 7, 44395. [Google Scholar] [CrossRef]
- Decker, C.J.; Teixeira, D.; Parker, R. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J. Cell Biol. 2007, 179, 437–449. [Google Scholar] [CrossRef]
- Kara, E.; McCambridge, A.; Proffer, M.; Dilts, C.; Pumnea, B.; Eshak, G.; Smith, K.; Fielder, L.; Doyle, D.; Ortega, B.; et al. An In Vivo Analysis of the Functional Motifs of DEAD-box RNA Helicase Me31B in Drosophila Fertility and Germline Development. bioRxiv. 2022. [CrossRef]
- Teixeira, D.; Parker, R. Analysis of P-Body Assembly in Saccharomyces cerevisiae. Mol. Biol. Cell 2007, 18, 2274–2287. [Google Scholar] [CrossRef] [PubMed]
- Rajyaguru, P.; She, M.; Parker, R. Scd6 targets eIF4G to repress translation: RGG-motif proteins as a class of eIF4G-binding proteins. Mol. Cell 2012, 45, 244–254. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.H.; Yang, W.-H.; Gulick, T.; Bloch, K.D.; Bloch, D.B. Ge-1 is a central component of the mammalian cytoplasmic mRNA processing body. RNA 2005, 11, 1795–1802. [Google Scholar] [CrossRef]
- Marnef, A.; Maldonado, M.; Bugaut, A.; Balasubramanian, S.; Kress, M.; Weil, D.; Standart, N. Distinct functions of maternal and somatic Pat1 protein paralogs. RNA 2010, 16, 2094–2107. [Google Scholar] [CrossRef]
- Ozgur, S.; Chekulaeva, M.; Stoecklin, G. Human Pat1b Connects Deadenylation with mRNA Decapping and Controls the Assembly of Processing Bodies. Mol. Cell Biol. 2010, 30, 4308–4323. [Google Scholar] [CrossRef]
- Yang, Z.; Jakymiw, A.; Wood, M.R.; Eystathioy, T.; Rubin, R.L.; Fritzler, M.J.; Chan, E.K.L. GW182 is critical for the stability of GW bodies expressed during the cell cycle and cell proliferation. J. Cell Sci. 2004, 117, 5567–5578. [Google Scholar] [CrossRef]
- Chekulaeva, M.; Hentze, M.W.; Ephrussi, A. Bruno acts as a dual repressor of oskar translation, promoting mRNA oligomerization and formation of silencing particles. Cell 2006, 124, 521–533. [Google Scholar] [CrossRef] [PubMed]
- Bose, M.; Lampe, M.; Mahamid, J.; Ephrussi, A. Liquid-to-solid phase transition of oskar ribonucleoprotein granules is essential for their function in Drosophila embryonic development. Cell 2022, 185, 1308–1324.e23. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Choi, J.-M.; Holehouse, A.S.; Lee, H.O.; Zhang, X.; Jahnel, M.; Maharana, S.; Lemaitre, R.; Pozniakovsky, A.; Drechsel, D.; et al. A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell 2018, 174, 688–699.e16. [Google Scholar] [CrossRef]
- Roden, C.; Gladfelter, A.S. RNA contributions to the form and function of biomolecular condensates. Nat. Rev. Mol. Cell Biol. 2021, 22, 183–195. [Google Scholar] [CrossRef]
- Delanoue, R.; Herpers, B.; Soetaert, J.; Davis, I.; Rabouille, C. Drosophila Squid/hnRNP helps Dynein switch from a gurken mRNA transport motor to an ultrastructural static anchor in sponge bodies. Dev. Cell 2007, 13, 523–538. [Google Scholar] [CrossRef] [PubMed]
- Strome, S.; Updike, D. Specifying and protecting germ cell fate. Nat. Rev. Mol. Cell Biol. 2015, 16, 406–416. [Google Scholar] [CrossRef]
- Hakes, A.C.; Gavis, E.R. Plasticity of Drosophila germ granules during germ cell development. PLOS Biol. 2023, 21, e3002069. [Google Scholar] [CrossRef]
- Thomson, T.; Liu, N.; Arkov, A.; Lehmann, R.; Lasko, P. Isolation of new polar granule components in Drosophila reveals P body and ER associated proteins. Mech. Dev. 2008, 125, 865–873. [Google Scholar] [CrossRef]
- Mahowald, A.P. Fine structure of pole cells and polar granules in Drosophila melanogaster. J. Exp. Zool. 1962, 151, 201–215. [Google Scholar] [CrossRef]
- Trcek, T.; Lehmann, R. Germ granules in Drosophila. Traffic 2019, 20, 650–660. [Google Scholar] [CrossRef]
- Trcek, T.; Douglas, T.E.; Grosch, M.; Yin, Y.; Eagle, W.V.I.; Gavis, E.R.; Shroff, H.; Rothenberg, E.; Lehmann, R. Sequence-Independent Self-Assembly of Germ Granule mRNAs into Homotypic Clusters. Mol. Cell 2020, 78, 941–950.e12. [Google Scholar] [CrossRef] [PubMed]
- Niepielko, M.G.; Eagle, W.V.I.; Gavis, E.R. Stochastic Seeding Coupled with mRNA Self-Recruitment Generates Heterogeneous Drosophila Germ Granules. Curr. Biol. 2018, 28, 1872–1881.e3. [Google Scholar] [CrossRef] [PubMed]
- Curnutte, H.A.; Lan, X.; Sargen, M.; Ieong, S.M.A.; Campbell, D.; Kim, H.; Liao, Y.; Lazar, S.B.; Trcek, T. Proteins rather than mRNAs regulate nucleation and persistence of Oskar germ granules in Drosophila. Cell Rep. 2023, 42, 112723. [Google Scholar] [CrossRef] [PubMed]
- Eagle, W.V.I.; Yeboah-Kordieh, D.K.; Niepielko, M.G.; Gavis, E.R. Distinct cis-acting elements mediate targeting and clustering of Drosophila polar granule mRNAs. Development 2018, 145, dev164657. [Google Scholar] [CrossRef]
- Little, S.C.; Sinsimer, K.S.; Lee, J.J.; Wieschaus, E.F.; Gavis, E.R. Independent and coordinate trafficking of single Drosophila germ plasm mRNAs. Nat. Cell Biol. 2015, 17, 558–568. [Google Scholar] [CrossRef]
- Trcek, T.; Grosch, M.; York, A.; Shroff, H.; Lionnet, T.; Lehmann, R. Drosophila germ granules are structured and contain homotypic mRNA clusters. Nat. Commun. 2015, 6, 7962. [Google Scholar] [CrossRef]
- Valentino, M.; Ortega, B.M.; Ulrich, B.; Doyle, D.A.; Farnum, E.D.; Joiner, D.A.; Gavis, E.R.; Niepielko, M.G. Computational modeling offers new insight into Drosophila germ granule development. Biophys. J. 2022, 121, 1465–1482. [Google Scholar] [CrossRef]
- Bratek-Skicki, A.; Pancsa, R.; Meszaros, B.; Van Lindt, J.; Tompa, P. A guide to regulation of the formation of biomolecular condensates. FEBS J. 2020, 287, 1924–1935. [Google Scholar] [CrossRef]
- Kaneuchi, T.; Sartain, C.V.; Takeo, S.; Horner, V.L.; Buehner, N.A.; Aigaki, T.; Wolfner, M.F. Calcium waves occur as Drosophila oocytes activate. Proc. Natl. Acad. Sci. USA 2015, 112, 791–796. [Google Scholar] [CrossRef]
- Alberti, S. Phase separation in biology. Curr. Biol. 2017, 27, R1097–R1102. [Google Scholar] [CrossRef]
- Zhang, Z.; Ahmed-Braimah, Y.H.; Goldberg, M.L.; Wolfner, M.F. Calcineurin-dependent Protein Phosphorylation Changes During Egg Activation in Drosophila melanogaster. Mol. Cell Proteom. 2019, 18, S145–S158. [Google Scholar] [CrossRef]
- Hara, M.; Lourido, S.; Petrova, B.; Lou, H.J.; Von Stetina, J.R.; Kashevsky, H.; Turk, B.E.; Orr-Weaver, T.L. Identification of PNG kinase substrates uncovers interactions with the translational repressor TRAL in the oocyte-to-embryo transition. eLife 2018, 7, e33150. [Google Scholar] [CrossRef] [PubMed]
- Doane, W.W. Completion of meiosis in uninseminated eggs of Drosophila melanogaster. Science 1960, 132, 677–678. [Google Scholar] [CrossRef] [PubMed]
- Page, A.W.; Orr-Weaver, T.L. Activation of the meiotic divisions in Drosophila oocytes. Dev. Biol. 1997, 183, 195–207. [Google Scholar] [CrossRef] [PubMed]
- Mahowald, A.P.; Goralski, T.J.; Caulton, J.H. In vitro activation of Drosophila eggs. Dev. Biol. 1983, 98, 437–445. [Google Scholar] [CrossRef]
- Heifetz, Y.; Yu, J.; Wolfner, M.F. Ovulation triggers activation of Drosophila oocytes. Dev. Biol. 2001, 234, 416–424. [Google Scholar] [CrossRef]
- York-Andersen, A.H.; Wood, B.W.; Wilby, E.L.; Berry, A.S.; Weil, T.T. Osmolarity-regulated swelling initiates egg activation in Drosophila. Open Biol. 2021, 11, 210067. [Google Scholar] [CrossRef]
- Mohapatra, S.; Wegmann, S. Biomolecular condensation involving the cytoskeleton. Brain Res. Bull. 2023, 194, 105–117. [Google Scholar] [CrossRef]
- Wiegand, T.; Hyman, A.A. Drops and fibers—How biomolecular condensates and cytoskeletal filaments influence each other. Emerg. Top. Life Sci. 2020, 4, 247–261. [Google Scholar]
- York-Andersen, A.H.; Hu, Q.; Wood, B.W.; Wolfner, M.F.; Weil, T.T. A calcium-mediated actin redistribution at egg activation in Drosophila. Mol. Reprod. Dev. 2020, 87, 293–304. [Google Scholar] [CrossRef]
- Avilés-Pagán, E.E.; Hara, M.; Orr-Weaver, T.L. The GNU subunit of PNG kinase, the developmental regulator of mRNA translation, binds BIC-C to localize to RNP granules. eLife 2021, 10, e67294. [Google Scholar] [CrossRef] [PubMed]
- Hara, M.; Petrova, B.; Orr-Weaver, T.L. Control of PNG kinase, a key regulator of mRNA translation, is coupled to meiosis completion at egg activation. eLife 2017, 6, e22219. [Google Scholar] [CrossRef] [PubMed]
- Brand, A.H.; Perrimon, N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 1993, 118, 401–415. [Google Scholar] [CrossRef] [PubMed]
- Perkins, L.A.; Holderbaum, L.; Tao, R.; Hu, Y.; Sopko, R.; McCall, K.; Yang-Zhou, D.; Flockhart, I.; Binari, R.; Shim, H.-S.; et al. The Transgenic RNAi Project at Harvard Medical School: Resources and Validation. Genetics 2015, 201, 843–852. [Google Scholar] [CrossRef] [PubMed]
- Zirin, J.; Hu, Y.; Liu, L.; Yang-Zhou, D.; Colbeth, R.; Yan, D.; Ewen-Campen, B.; Tao, R.; Vogt, E.; VanNest, S.; et al. Large-Scale Transgenic Drosophila Resource Collections for Loss- and Gain-of-Function Studies. Genetics 2020, 214, 755–767. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, E.; Chartrand, P.; Schaefer, M.; Shenoy, S.M.; Singer, R.H.; Long, R.M. Localization of ASH1 mRNA particles in living yeast. Mol. Cell 1998, 2, 437–445. [Google Scholar] [CrossRef]
- Forrest, K.M.; Gavis, E.R. Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila. Curr. Biol. 2003, 13, 1159–1168. [Google Scholar] [CrossRef]
- Tantale, K.; Mueller, F.; Kozulic-Pirher, A.; Lesne, A.; Victor, J.-M.; Robert, M.-C.; Capozi, S.; Chouaib, R.; Bäcker, V.; Mateos-Langerak, J.; et al. A single-molecule view of transcription reveals convoys of RNA polymerases and multi-scale bursting. Nat. Commun. 2016, 7, 12248. [Google Scholar] [CrossRef]
- Dufourt, J.; Bellec, M.; Trullo, A.; Dejean, M.; De Rossi, S.; Favard, C.; Lagha, M. Imaging translation dynamics in live embryos reveals spatial heterogeneities. Science 2021, 372, 840–844. [Google Scholar] [CrossRef]
- Vinter, D.J.; Hoppe, C.; Ashe, H.L. Live and fixed imaging of translation sites at single mRNA resolution in the Drosophila embryo. STAR Protoc. 2021, 2, 100812. [Google Scholar] [CrossRef]
- Raj, A.; van den Bogaard, P.; Rifkin, S.A.; van Oudenaarden, A.; Tyagi, S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods 2008, 5, 877–879. [Google Scholar] [CrossRef] [PubMed]
- Kramer, S. Simultaneous detection of mRNA transcription and decay intermediates by dual colour single mRNA FISH on subcellular resolution. Nucleic Acids Res. 2017, 45, e49. [Google Scholar] [CrossRef] [PubMed]
- Halstead, J.M.; Wilbertz, J.H.; Wippich, F.; Lionnet, T.; Ephrussi, A.; Chao, J.A. Chapter Six—TRICK: A Single-Molecule Method for Imaging the First Round of Translation in Living Cells and Animals. In Methods in Enzymology; Filonov, G.S., Jaffrey, S.R., Eds.; Academic Press: Cambridge, MA, USA, 2016; Volume 572, pp. 123–157. [Google Scholar]
- Halstead, J.M.; Lionnet, T.; Wilbertz, J.H.; Wippich, F.; Ephrussi, A.; Singer, R.H.; Chao, J.A. Translation. An RNA biosensor for imaging the first round of translation from single cells to living animals. Science 2015, 347, 1367–1671. [Google Scholar] [CrossRef] [PubMed]
- Tanenbaum, M.E.; Gilbert, L.A.; Qi, L.S.; Weissman, J.S.; Vale, R.D. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 2014, 159, 635–646. [Google Scholar] [CrossRef]
- Yan, X.; Hoek, T.A.; Vale, R.D.; Tanenbaum, M.E. Dynamics of Translation of Single mRNA Molecules In Vivo. Cell 2016, 165, 976–989. [Google Scholar] [CrossRef] [PubMed]
- Pichon, X.; Bastide, A.; Safieddine, A.; Chouaib, R.; Samacoits, A.; Basyuk, E.; Peter, M.; Mueller, F.; Bertrand, E. Visualization of single endogenous polysomes reveals the dynamics of translation in live human cells. J. Cell Biol. 2016, 214, 769–781. [Google Scholar] [CrossRef]
- Wang, C.; Han, B.; Zhou, R.; Zhuang, X. Real-time imaging of translation on single mRNA transcripts in live cells. Cell 2016, 165, 990–1001. [Google Scholar] [CrossRef]
- Wu, B.; Eliscovich, C.; Yoon, Y.J.; Singer, R.H. Translation dynamics of single mRNAs in live cells and neurons. Science 2016, 352, 1430–1435. [Google Scholar] [CrossRef]
- Vinter, D.J.; Hoppe, C.; Minchington, T.G.; Sutcliffe, C.; Ashe, H.L. Dynamics of hunchback translation in real-time and at single-mRNA resolution in the Drosophila embryo. Development 2021, 148, dev196121. [Google Scholar] [CrossRef]
- Bellec, M.; Chen, R.; Dhayni, J.; Favard, C.; Trullo, A.; Lenden-Hasse, H.; Lehmann, R.; Bertrand, E.; Lagha, M.; Dufourt, J. Boosting the toolbox for live imaging of translation. BioRxiv 2023, preprint. [Google Scholar]
- Garcia, J.F.; Parker, R. MS2 coat proteins bound to yeast mRNAs block 5′ to 3′ degradation and trap mRNA decay products: Implications for the localization of mRNAs by MS2-MCP system. RNA 2015, 21, 1393–1395. [Google Scholar] [CrossRef] [PubMed]
- Pandey, U.B.; Nichols, C.D. Human disease models in Drosophila melanogaster and the role of the fly in therapeutic drug discovery. Pharmacol. Rev. 2011, 63, 411–436. [Google Scholar] [CrossRef] [PubMed]
- Arora, S.; Ligoxygakis, P. Beyond Host Defense: Deregulation of Drosophila Immunity and Age-Dependent Neurodegeneration. Front. Immunol. 2020, 11, 1574. [Google Scholar] [CrossRef]
- Verheyen, E.M. The power of Drosophila in modeling human disease mechanisms. Dis. Model. Mech. 2022, 15, dmm049549. [Google Scholar] [CrossRef]
- Cheng, L.; Baonza, A.; Grifoni, D. Drosophila Models of Human Disease. BioMed Res. Int. 2018, 2018, 7214974. [Google Scholar] [CrossRef]
- Tapia, A.; Giachello, C.N.; Palomino-Schätzlein, M.; Baines, R.A.; Galindo, M.I. Generation and Characterization of the Drosophila melanogaster paralytic Gene Knock-Out as a Model for Dravet Syndrome. Life 2021, 11, 1261. [Google Scholar] [CrossRef]
- Kim, K.; Lane, E.A.; Saftien, A.; Wang, H.; Xu, Y.; Wirtz-Peitz, F.; Perrimon, N. Drosophila as a model for studying cystic fibrosis pathophysiology of the gastrointestinal system. Proc. Natl. Acad. Sci. USA 2020, 117, 10357–10367. [Google Scholar] [CrossRef] [PubMed]
- Huggett, S.B.; Hatfield, J.S.; Walters, J.D.; McGeary, J.E.; Welsh, J.W.; Mackay, T.F.C.; Anholt, R.R.H.; Palmer, R.H.C. Ibrutinib as a potential therapeutic for cocaine use disorder. Transl. Psychiatry 2021, 11, 623. [Google Scholar] [CrossRef]
- Rouka, E.; Gourgoulianni, N.; Lüpold, S.; Hatzoglou, C.; Gourgoulianis, K.I.; Zarogiannis, S.G. Prediction and enrichment analyses of the Homo sapiens-Drosophila melanogaster COPD-related orthologs: Potential for modeling of human COPD genomic responses with the fruit fly. Am. J. Physiol. Integr. Comp. Physiol. 2022, 322, R77–R82. [Google Scholar] [CrossRef]
- Kotian, N.; Troike, K.M.; Curran, K.N.; Lathia, J.D.; McDonald, J.A. A Drosophila RNAi screen reveals conserved glioblastoma-related adhesion genes that regulate collective cell migration. G3 GenesGenomesGenetics 2021, 12, jkab356. [Google Scholar] [CrossRef]
- Belfer, S.J.; Bashaw, A.G.; Perlis, M.L.; Kayser, M.S. A Drosophila model of sleep restriction therapy for insomnia. Mol. Psychiatry 2021, 26, 492–507. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.-X.; Li, S.-S.; Li, A.-Q.; Liu, Z.-Y.; Neely, G.G.; Wang, Q.-P. dSec16 Acting in Insulin-like Peptide Producing Cells Controls Energy Homeostasis in Drosophila. Life 2022, 13, 81. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, T.; Matsukawa, K.; Watanabe, N.; Kishino, Y.; Kunugi, H.; Ihara, R.; Wakabayashi, T.; Hashimoto, T.; Iwatsubo, T. Self-assembly of FUS through its low-complexity domain contributes to neurodegeneration. Hum. Mol. Genet. 2018, 27, 1353–1365. [Google Scholar] [CrossRef]
- An, H.; Litscher, G.; Watanabe, N.; Wei, W.; Hashimoto, T.; Iwatsubo, T.; Buchman, V.L.; Shelkovnikova, T.A. ALS-linked cytoplasmic FUS assemblies are compositionally different from physiological stress granules and sequester hnRNPA3, a novel modifier of FUS toxicity. Neurobiol. Dis. 2022, 162, 105585. [Google Scholar] [CrossRef]
- Matsukawa, K.; Kukharsky, M.S.; Park, S.K.; Park, S.; Watanabe, N.; Iwatsubo, T.; Shelkovnikova, T.A. Long non-coding RNA NEAT1_1 ameliorates TDP-43 toxicity in in vivo models of TDP-43 proteinopathy. RNA Biol. 2021, 18, 1546–1554. [Google Scholar] [CrossRef] [PubMed]
- Markmiller, S.; Soltanieh, S.; Server, K.L.; Mak, R.; Jin, W.; Fang, M.Y.; Luo, E.-C.; Krach, F.; Yang, D.; Sen, A.; et al. Context-Dependent and Disease-Specific Diversity in Protein Interactions within Stress Granules. Cell 2018, 172, 590–604.e13. [Google Scholar] [CrossRef]
- Sharpe, J.L.; Harper, N.S.; Garner, D.R.; West, R.J.H. Modeling C9orf72-Related Frontotemporal Dementia and Amyotrophic Lateral Sclerosis in Drosophila. Front. Cell Neurosci. 2021, 15, 770937. [Google Scholar] [CrossRef]
- Sreedharan, J.; Blair, I.P.; Tripathi, V.B.; Hu, X.; Vance, C.; Rogelj, B.; Ackerley, S.; Durnall, J.C.; Williams, K.L.; Buratti, E.; et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science 2008, 319, 1668–1672. [Google Scholar] [CrossRef]
- Vance, C.; Rogelj, B.; Hortobágyi, T.; De Vos, K.J.; Nishimura, A.L.; Sreedharan, J.; Hu, X.; Smith, B.; Ruddy, D.; Wright, P.; et al. Mutations in FUS, an RNA Processing Protein, Cause Familial Amyotrophic Lateral Sclerosis Type 6. Science 2009, 323, 1208–1211. [Google Scholar] [CrossRef]
- Patel, A.; Lee, H.O.; Jawerth, L.; Maharana, S.; Jahnel, M.; Hein, M.Y.; Stoynov, S.; Mahamid, J.; Saha, S.; Franzmann, T.M.; et al. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell 2015, 162, 1066–1077. [Google Scholar] [CrossRef]
- Mackenzie, I.R.; Nicholson, A.M.; Sarkar, M.; Messing, J.; Purice, M.D.; Pottier, C.; Rademakers, R. TIA1 Mutations in Amyotrophic Lateral Sclerosis and Frontotemporal Dementia Promote Phase Separation and Alter Stress Granule Dynamics. Neuron 2017, 95, 808–816.e9. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Pu, J. α-Synuclein in Parkinson’s Disease: From Pathogenetic Dysfunction to Potential Clinical Application. Park. Dis. 2016, 2016, 1720621. [Google Scholar] [CrossRef] [PubMed]
- Olsen, A.L.; Feany, M.B. Glial α-synuclein promotes neurodegeneration characterized by a distinct transcriptional program in vivo. Glia 2019, 67, 1933–1957. [Google Scholar] [CrossRef] [PubMed]
- Feany, M.B.; Bender, W.W. A Drosophila model of Parkinson’s disease. Nature 2000, 404, 394–398. [Google Scholar] [CrossRef]
- Xiong, Y.; Yu, J. Modeling Parkinson’s Disease in Drosophila: What Have We Learned for Dominant Traits? Front. Neurol. 2018, 9, 228. [Google Scholar] [CrossRef]
- Balak, C.; Benard, M.; Schaefer, E.; Iqbal, S.; Ramsey, K.; Ernoult-Lange, M.; Mattioli, F.; Llaci, L.; Geoffroy, V.; Courel, M.; et al. Rare De Novo Missense Variants in RNA Helicase DDX6 Cause Intellectual Disability and Dysmorphic Features and Lead to P-Body Defects and RNA Dysregulation. Am. J. Hum. Genet. 2019, 105, 509–525. [Google Scholar] [CrossRef]
- Chan, H.Y.E.; Bonini, N.M. Drosophila models of human neurodegenerative disease. Cell Death Differ. 2000, 7, 1075–1080. [Google Scholar] [CrossRef]
- Hardy, S.D.; Shinde, A.; Wang, W.-H.; Wendt, M.K.; Geahlen, R.L. Regulation of epithelial-mesenchymal transition and metastasis by TGF-β, P-bodies, and autophagy. Oncotarget 2017, 8, 103302–103314. [Google Scholar] [CrossRef]
- Tang, L.; Zhao, P.; Kong, D. Muscleblind-like 1 destabilizes Snail mRNA and suppresses the metastasis of colorectal cancer cells via the Snail/E-cadherin axis. Int. J. Oncol. 2019, 54, 955–965. [Google Scholar] [CrossRef]
- Corcoran, J.A.; Khaperskyy, D.A.; Johnston, B.P.; King, C.A.; Cyr, D.P.; Olsthoorn, A.V.; McCormick, C. Kaposi’s sarcoma-associated herpesvirus G-protein-coupled receptor prevents AU-rich-element-mediated mRNA decay. J. Virol. 2012, 86, 8859–8871. [Google Scholar] [CrossRef]
- Hopkins, K.C.; McLane, L.M.; Maqbool, T.; Panda, D.; Gordesky-Gold, B.; Cherry, S. A genome-wide RNAi screen reveals that mRNA decapping restricts bunyaviral replication by limiting the pools of Dcp2-accessible targets for cap-snatching. Genes Dev. 2013, 27, 1511–1525. [Google Scholar] [CrossRef]
- Khong, A.; Jan, E. Modulation of Stress Granules and P Bodies during Dicistrovirus Infection. J. Virol. 2011, 85, 1439–1451. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.; Magwire, M.M.; Bayer, F.; Jiggins, F.M. A Polymorphism in the Processing Body Component Ge-1 Controls Resistance to a Naturally Occurring Rhabdovirus in Drosophila. PLOS Pathog. 2016, 12, e1005387. [Google Scholar]
- Göertz, G.P.; van Bree, J.W.M.; Hiralal, A.; Fernhout, B.M.; Steffens, C.; Boeren, S.; Visser, T.M.; Vogels, C.B.F.; Abbo, S.R.; Fros, J.J.; et al. Subgenomic flavivirus RNA binds the mosquito DEAD/H-box helicase ME31B and determines Zika virus transmission by Aedes aegypti. Proc. Natl. Acad. Sci. USA 2019, 116, 19136–19144. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Hu, S.; Xu, F.; Mei, S.; Liu, X.; Yin, L.; Zhao, F.; Zhao, X.; Sun, H.; Xiong, Z.; et al. MOV10 sequesters the RNP of influenza A virus in the cytoplasm and is antagonized by viral NS1 protein. Biochem. J. 2019, 476, 467–481. [Google Scholar] [CrossRef]
- Liu, Y.-C.; Mok, B.W.-Y.; Wang, P.; Kuo, R.-L.; Chen, H.; Shih, S.-R. Cellular 5′-3′ mRNA Exoribonuclease XRN1 Inhibits Interferon β Activation and Facilitates Influenza A Virus Replication. mBio 2021, 12, e0094521. [Google Scholar] [CrossRef]
- Welke, R.-W.; Sperber, H.S.; Bergmann, R.; Koikkarah, A.; Menke, L.; Sieben, C.; Krüger, D.H.; Chiantia, S.; Herrmann, A.; Schwarzer, R. Characterization of Hantavirus N Protein Intracellular Dynamics and Localization. Viruses 2022, 14, 457. [Google Scholar] [CrossRef]
- Oceguera, A.; Peralta, A.V.; Martínez-Delgado, G.; Arias, C.F.; López, S. Rotavirus RNAs sponge host cell RNA binding proteins and interfere with their subcellular localization. Virology 2018, 525, 96–105. [Google Scholar] [CrossRef]
- Kuroshima, T.; Matsuda, A.Y.; Hossain, E.; Yasuda, M.; Kitamura, T.; Kitagawa, Y.; Higashino, F. Adenovirus infection controls processing bodies to stabilize AU-rich element-containing mRNA. Virology 2022, 573, 124–130. [Google Scholar] [CrossRef]
- Fan, S.; Xu, Z.; Liu, P.; Qin, Y.; Chen, M. Enterovirus 71 2A Protease Inhibits P-Body Formation to Promote Viral RNA Synthesis. J. Virol. 2021, 95, e0092221. [Google Scholar] [CrossRef]
- Perez-Pepe, M.; Fernández-Alvarez, A.J.; Boccaccio, G.L. Life and Work of Stress Granules and Processing Bodies: New Insights into Their Formation and Function. Biochemistry 2018, 57, 2488–2498. [Google Scholar] [CrossRef]
- Fernández-Carrillo, C.; Pérez-Vilaró, G.; Díez, J.; Pérez-del-Pulgar, S. Hepatitis C virus plays with fire and yet avoids getting burned. A review for clinicians on processing bodies and stress granules. Liver Int. 2018, 38, 388–398. [Google Scholar] [CrossRef] [PubMed]
- Ariumi, Y. Host Cellular RNA Helicases Regulate SARS-CoV-2 Infection. J. Virol. 2022, 96, e00002-22. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, N.M. Challenges and opportunities in controlling mosquito-borne infections. Nature 2018, 559, 490–497. [Google Scholar] [CrossRef] [PubMed]
- Kwaśnik, M.; Rożek, W.; Rola, J. Rift Valley Fever—A Growing Threat to Humans and Animals. J. Vet. Res. 2021, 65, 7–14. [Google Scholar] [CrossRef]
- Liu, L.; Weiss, E.; Panas, M.D.; Götte, B.; Sellberg, S.; Thaa, B.; McInerney, G.M. RNA processing bodies are disassembled during Old World alphavirus infection. J. Gen. Virol. 2019, 100, 1375–1389. [Google Scholar] [CrossRef]
- Morrison, T.E. Reemergence of Chikungunya Virus. J. Virol. 2014, 88, 11644–11647. [Google Scholar] [CrossRef]
- Hopkins, K.; Cherry, S. Bunyaviral cap-snatching vs. decapping. Cell Cycle 2013, 12, 3711–3712. [Google Scholar] [CrossRef]
- Harnish, J.M.; Link, N.; Yamamoto, S. Drosophila as a Model for Infectious Diseases. Int. J. Mol. Sci. 2021, 22, 2724. [Google Scholar] [CrossRef]
- Hedges, S.B.; Dudley, J.; Kumar, S. TimeTree: A public knowledge-base of divergence times among organisms. Bioinformatics 2006, 22, 2971–2972. [Google Scholar] [CrossRef]
- Rubin, G.M.; Yandell, M.D.; Wortman, J.R.; Gabor, G.L.; Miklos, G.L.G.; Nelson, C.R.; Hariharan, I.K.; Fortini, M.E.; Li, P.W.; Apweiler, R.; et al. Comparative genomics of the eukaryotes. Science 2000, 287, 2204–2215. [Google Scholar] [CrossRef] [PubMed]
- Reiter, L.T.; Potocki, L.; Chien, S.; Gribskov, M.; Bier, E. A Systematic Analysis of Human Disease-Associated Gene Sequences in Drosophila melanogaster. Genome Res. 2001, 11, 1114–1125. [Google Scholar] [CrossRef] [PubMed]
| Protein | Role | Localisation to P bodies | Human Orthologue | Yeast Orthologue | |
|---|---|---|---|---|---|
| Translational regulators | eIF4E- Transporter (4E-T) | Involved in the negative regulation of the eukaryotic translation initiation factor 4F complex assembly, through competitive binding with eIF4G for eIF4E | S2 cells [39] | 4E-T | NC |
| Eukaryotic translation initiation factor 4E1 (eIF4E1) | Part of the eukaryotic translational initiation factor 4F complex, which is capable of binding to the 5′ mRNA cap | S2 cells and egg chamber [13,40] | eIF4E | eIF4Ep | |
| Maternal expression at 31B (Me31B) | A DEAD box RNA helicase that plays a critical role in translational repression and mRNA decapping | S2 cells and egg chamber [13,40] | DDX6 | Dhh1p | |
| Trailer Hitch (Tral) | An sm-like protein with a variety of roles including translational repression and the ability to bind to DEAD box helices | S2 cells and egg chamber [13,40] | Lsm14A | Scd6p | |
| Staufen (Stau) | A double-stranded RNA binding protein involved in multiple mRNAs’ localisation | S2 cells and egg chamber [13,40] | Stau1 | NC | |
| Degradation machinery | Decapping protein 1 (DCP1) | A subunit of the mRNA decay holoenzyme, also involved in RNA localisation | S2 cells and egg chamber [13,14] | Dcp1 | Dcp1p |
| Decapping protein 2 (DCP2) | The catalytic subunit of the mRNA decay holoenzyme, an m7G(5′)pppN diphosphatase responsible for removal of the 5′ cap | S2 cells and egg chamber [13,14] | Dcp2 | Dcp2p | |
| Enhancer of decapping 3 (Edc3) | Promotes the efficient removal of the 5′ cap from mRNA | S2 cells and egg chamber [13,14] | Edc3 | Edc3p | |
| Ge-1 | A decapping activator that couples mRNA deadenylation to decapping and may act as a scaffold to physically connect these two processes | S2 cells and egg chamber [13,41] | Edc4 | NC | |
| Like Sm 1 (LSm1) | An sm-like protein that is part of the Lsm1-7-Pat1 complex, thought to enable RNA cap binding | S2 cells [13] | Lsm1 | Lsm1p | |
| NOT1 | Part of the CAF-1CCR4-NOT complex that degrades the mRNA poly(A) tail | S2 cells [13,14] | CNOT1 | CDC39p | |
| Pacman (Pcm) | A 5′ to 3′ exoribonuclease that degrades decapped mRNA | S2 cells and egg chamber [13,14] | XRN1 | XRN1p | |
| Protein associated with topo II related-1 (HPat) | A decapping activator that couples mRNA deadenylation and decapping | S2 cells and egg chamber [13,14] | Pat1A | Pat1p | |
| Twin (CCR4) | Part of the CAF-1CCR4-NOT complex that degrades the mRNA poly(A) tail | S2 cells [13,14] | CCR4 | CCR4 | |
| miRNA machinery | Argonaute 2 (AGO2) | Interacts with small interfering RNAs (siRNAs) to form RNA-induced silencing complexes (RISCs) | S2 cells [13] | Ago2 | NC |
| Dicer-1 (Dcr-1) | Cleaves double-stranded RNA and is involved in the production of mature miRNAs | S2 cells [13] | Dicer-1 | NC | |
| Dicer-2 (Dcr-2) | Cleaves double-stranded RNA and is involved in the production of mature miRNAs | S2 cells [13] | Dicer-2 | NC | |
| Drosha | Cleaves double-stranded RNA and is involved in the production of mature miRNAs | S2 cells [13] | Drosha | NC | |
| miRNA machinery (cont.) | Gawky (GW) | Required for gene silencing by micro-RNAs and promotes both deadenylation and decapping through the recruitment of the CCR4-NOT and the DCP1–DCP2 complexes | S2 cells [13] | GW182 | NC |
| Partner of Drosha (Pasha) | Cleaves double-stranded RNA and is involved in the production of mature miRNAs | S2 cells [13] | Pasha | NC | |
| Egg chamber specific components | Bruno 1 (Bru1) | An RNA binding protein that is involved in multiple aspects of post-transcriptional gene regulation, including localisation, translational repression, and activation of translation | Egg chamber [35] | CELF1 /CELF2 | WHI3p(?) |
| Cup | Involved in translational repression in eIF4E dependent and independent mechanisms | Egg chamber [40] | 4E-T | NC | |
| Exuperantia (Exu) | Involved in bcd mRNA localisation to the anterior of the oocyte in mid-oogenesis | Egg chamber [35,40,42,43] | NC | NC | |
| Heterogenous nuclear ribonucleoprotein at 27C (Hrb27C) | A heterogenous nuclear ribonucleoprotein, an RNA binding protein, involved in the localisation and translational regulation of mRNA | Egg chamber [40] | DAZAP1 | HRP1p(?) | |
| Lost | Involved in mRNA localisation to the posterior of the oocyte in late oogenesis, present in multiple RNP complexes, and likely has a broader role in RNA metabolism | Egg chamber [40] | MTHFSD | Fau1p(?) | |
| Oo18 RNA binding protein (Orb) | Involved in mRNA polyadenylation, promoting translation (but may also act as a deadenylator and translational repressor dependent on its phosphorylation status) | Egg chamber [40] | CPEB | NC | |
| Squid (Sqd) | A heterogenous nuclear ribonucleoprotein A (hnRNPA), an RNA binding protein, involved in the localisation and translational regulation of grk mRNA | Egg chamber [40,42] | HNRNPAB/HNRNPD | HRP1(?) | |
| Ypsilon Schachtel (Yps) | RNA binding protein involved in various processes, such as translational repression and RNA stabilisation | Egg chamber [40,43] | YBX1 | NC |
| Protein Disrupted | Drosophila S2 Cells | Intestinal Stem Cells | Nurse Cells | Oocyte | Testes | Human (Adapted from [19]) | Budding Yeast | |
|---|---|---|---|---|---|---|---|---|
| Translational regulators | eIF4E- Transporter (4E-T) | - | No effect (Tral) [30] | - | - | - | Diffuse (DDX6, eIF4E, CCR4, Lsm1, Lsm14A), cannot be reinduced under stress [17,79] | Not conserved |
| Eukaryotic translation initiation factor E1 (eIF4E1) | Diffuse (Me31B) [27] | No effect (Tral) [30] | - | - | - | - | - | |
| Maternal expression at 31B (Me31B) | Diffuse (Tral, Ge-1) [13] | Smaller (Pat1) [30] | No effect (Tral and Cup) [84] | - | - | Diffuse (Lsm1, eIF4E, CCR4, 4E-T, Edc4, Dcp1a), cannot be re-induced under stress [17,79,80] | Smaller (under starvation) (Dcp1, Dcp2, Edc3, Xrn1, Dhh1, Pat1) [85] | |
| Trailer Hitch (Tral) | No effect (Ge-1) [13] | Smaller (Pat1) [30] | Diffuse (Me31B) [36] | Shape altered (Me31B) [38] | - | Diffuse (Edc4, Dcp1a), cannot be re-induced under stress [78,79] | Smaller (under starvation) (Dcp2) [86] | |
| Staufen (Stau) | - | No effect (Tral) [30] | - | - | - | - | Not conserved | |
| Degradation machinery | Decapping protein 1 (DCP1) | No effect (Tral, Ge1) [13] | - | Larger (Pcm) [14] | - | - | - | Larger (unstressed) (Ccr4, Dhh1, Pat1, Lsm1, Xrn1, Dcp2, Edc3) [85] |
| Decapping protein 2 (DCP2) | Larger (Tral, Ge1) [13] | No effect (Tral) [30] | Larger (Dcp1) [14] | - | - | Larger (LSm1, DDX6, eIF4E, CCR4) [17] No effect (Ge-1) [87] | Smaller (under starvation) (Ccr4, Dhh1, Pat1, Lsm1, Xrn1, Edc3) [85] | |
| Enhancer of decapping 3 (Edc3) | No effect (Tral, Ge1) [13] | Larger (Tral) [30] | - | - | - | No effect (Edc4) [79] | Smaller (under starvation) (Dhh1, Pat1, Lsm1, Dcp1, Dcp2, Xrn1) [83] | |
| Degradation machinery (cont.) | Ge-1 | Diffuse (Tral) [13] | Diffuse (Tral) [30] | - | Diffuse [41] | - | Smaller/fewer/diffuse, can be re-induced by stress (DDX6, Lsm14A, Dcp1a) [79,87] | Not conserved |
| Like Sm 1 (LSm1) | Diffuse (Tral, Ge1) [13] | Larger (Tral) [30] | - | - | - | Diffuse (DDX6, eIF4E, CCR4, 4E-T) [17] | Larger (unstressed) (Dcp1, Dcp2, Edc3, Xrn1, Dhh1, Pat1) [85] | |
| Not1 | Diffuse (Tral, Ge1) [13] | Diffuse (Tral) [30] | Not localised to P bodies [14] | - | - | - | - | |
| Pacman (Pcm) | Larger (Tral, Ge1) [13] | Larger (Tral) [30] | Larger (Dcp1, Dcp2) [14] | - | Larger (Dcp1) [31] | Larger (Dcp2) [16] | Larger (unstressed) (Ccr4, Dhh1, Pat1, Lsm1, Dcp1, Dcp2, Edc3) [85] | |
| Protein associated with topo II related-1 (HPat) | Diffuse (Tral, Ge1) [13] | Diffuse (Tral) [30] | - | - | - | Smaller/fewer/diffuse, can be re-induced by stress (Edc4) [76,88,89] | Smaller (Dcp1, Dcp2, Edc3, Xrn1, Dhh1, Pat1) [85] | |
| Twin (CCR4) | - | No effect (Tral) [30] | Not localised to P bodies [14] | - | - | Diffuse (DDX6, eIF4E, Lsm1, 4E-T) [17] | Smaller (under starvation) (Dcp2, Edc3, Dhh1, Pat1, Lsm1, Xrn1, Dcp1) [85] | |
| miRNA machinery | Argonaute 2 (AGO2) | Diffuse (Tral, Ge1) [13] | No effect (Tral) [30] | - | - | - | - | Not conserved |
| Dicer-1 (Dcr-1) | Diffuse (Tral, Ge1) [13] | No effect (Tral) [30] | - | - | - | - | Not conserved | |
| Dicer-2 (Dcr-2) | Diffuse (Tral, Ge1) [13] | No effect (Tral) [30] | - | - | - | - | Not conserved | |
| Drosha | Diffuse (Tral, Ge1) [13] | No effect (Tral) [30] | - | - | - | - | Not conserved | |
| Gawky (GW) | Diffuse (Tral, Ge1) [13] | No effect (Tral) [30] | - | - | - | Diffuse, can be re-induced by stress (Dcp1a, Lsm4) [80,90] | Not conserved | |
| Partner of Drosha (Pasha) | Diffuse (Tral, Ge1) [13] | No effect (Tral) [30] | - | - | - | - | Not conserved |
| In Vitro Mechanisms for Disassembling Condensates | Comparative Mechanisms for Disassembling Condensates at Egg Activation |
|---|---|
| Changes in the ionic concentration [108] | An increase in the intracellular calcium level [53,109] |
| Changes to post-translational modifications [110] | Phosphorylation of P body components [111,112] |
| Changes to the protein concentration [108,110] | Swelling and increase in volume [113,114,115,116,117], lowering of the cytoplasmic concentrations of P body proteins |
| Changes to the cytoskeletal architecture [118,119] | Multiple instances of actin cytoskeleton remodelling [120] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wilby, E.L.; Weil, T.T. Relating the Biogenesis and Function of P Bodies in Drosophila to Human Disease. Genes 2023, 14, 1675. https://doi.org/10.3390/genes14091675
Wilby EL, Weil TT. Relating the Biogenesis and Function of P Bodies in Drosophila to Human Disease. Genes. 2023; 14(9):1675. https://doi.org/10.3390/genes14091675
Chicago/Turabian StyleWilby, Elise L., and Timothy T. Weil. 2023. "Relating the Biogenesis and Function of P Bodies in Drosophila to Human Disease" Genes 14, no. 9: 1675. https://doi.org/10.3390/genes14091675
APA StyleWilby, E. L., & Weil, T. T. (2023). Relating the Biogenesis and Function of P Bodies in Drosophila to Human Disease. Genes, 14(9), 1675. https://doi.org/10.3390/genes14091675






