Aquaporins Transcripts with Potential Prognostic Value in Pancreatic Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethical Approval and Sample Characterization
2.2. RNA Isolation and RT-qPCR Analysis
2.3. Statistical Analysis
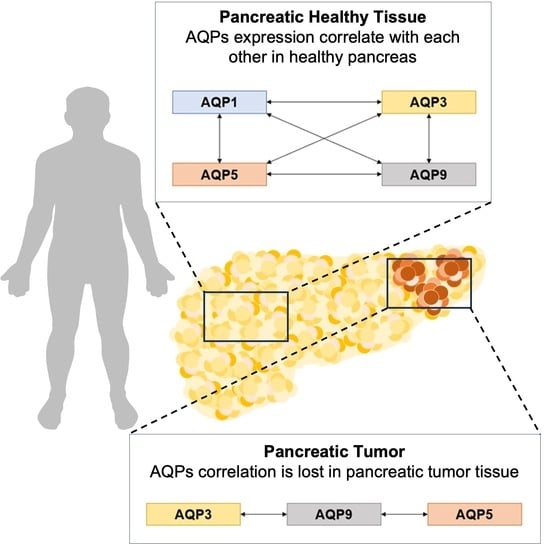
3. Results and Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Maitra, A.; Fukushima, N.; Takaori, K.; Hruban, R.H. Precursors to invasive pancreatic cancer. Adv. Anat. Pathol. 2005, 12, 81–91. [Google Scholar] [CrossRef]
- McGuigan, A.; Kelly, P.; Turkington, R.C.; Jones, C.; Coleman, H.G.; McCain, R.S. Pancreatic cancer: A review of clinical diagnosis, epidemiology, treatment and outcomes. World J. Gastroenterol. 2018, 24, 4846–4861. [Google Scholar] [CrossRef]
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef]
- Midha, S.; Chawla, S.; Garg, P.K. Modifiable and non-modifiable risk factors for pancreatic cancer: A review. Cancer Lett. 2016, 381, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Ilic, M.; Ilic, I. Epidemiology of pancreatic cancer. World J. Gastroenterol. 2016, 22, 9694–9705. [Google Scholar] [CrossRef]
- Ćwik, G.; Wallner, G.; Skoczylas, T.; Ciechański, A.; Zinkiewicz, K. Cancer antigens 19-9 and 125 in the differential diagnosis of pancreatic mass lesions. Arch. Surg. 2006, 141, 968–973. [Google Scholar] [CrossRef] [PubMed]
- Ballehaninna, U.K.; Chamberlain, R.S. The clinical utility of serum CA 19-9 in the diagnosis, prognosis and management of pancreatic adenocarcinoma: An evidence based appraisal. J. Gastrointest. Oncol. 2012, 3, 105–119. [Google Scholar] [CrossRef]
- Ballehaninna, U.K.; Chamberlain, R.S. Serum CA 19-9 as a biomarker for pancreatic cancer—A comprehensive review. Indian J. Surg. Oncol. 2011, 3, 88–100. [Google Scholar] [CrossRef]
- Verkman, A.S. Aquaporins in clinical medicine. Annu. Rev. Med. 2012, 63, 303–316. [Google Scholar] [CrossRef]
- Ishibashi, K.; Hara, S.; Kondo, S. Aquaporin water channels in mammals. Clin. Exp. Nephrol. 2009, 13, 107–117. [Google Scholar] [CrossRef]
- Papadopoulos, M.C.; Saadoun, S. Key roles of aquaporins in tumor biology. Biochim. Biophys. Acta 2015, 1848, 2576–2583. [Google Scholar] [CrossRef]
- Silva, P.M.; da Silva, I.V.; Sarmento, M.J.; Silva, I.C.; Carvalho, F.A.; Soveral, G.; Santos, N.C. Aquaporin-3 and aquaporin-5 facilitate migration and cell-cell adhesion in pancreatic cancer by modulating cell biomechanical properties. Cells 2022, 11, 1308. [Google Scholar] [CrossRef]
- Direito, I.; Madeira, A.; Brito, M.A.; Soveral, G. Aquaporin-5: From structure to function and dysfunction in cancer. Cell. Mol. Life Sci. CMLS 2016, 73, 1623–1640. [Google Scholar] [CrossRef] [PubMed]
- Marlar, S.; Jensen, H.H.; Login, F.H.; Nejsum, L.N. Aquaporin-3 in cancer. Int. J. Mol. Sci. 2017, 18, 2106. [Google Scholar] [CrossRef] [PubMed]
- Edamana, S.; Login, F.H.; Yamada, S.; Kwon, T.H.; Nejsum, L.N. Aquaporin water channels as regulators of cell-cell adhesion proteins. Am. J. Physiol. Cell Physiol. 2021, 320, C771–C777. [Google Scholar] [CrossRef]
- Moon, C.S.; Moon, D.; Kang, S.K. Aquaporins in cancer biology. Front. Oncol. 2022, 12, 782829. [Google Scholar] [CrossRef]
- Pimpão, C.; Wragg, D.; da Silva, I.V.; Casini, A.; Soveral, G. Aquaglyceroporin modulators as emergent pharmacological molecules for human diseases. Front. Mol. Biosci. 2022, 9, 845237. [Google Scholar] [CrossRef] [PubMed]
- Delporte, C. Aquaporins in salivary glands and pancreas. Biochim. Biophys. Acta 2014, 1840, 1524–1532. [Google Scholar] [CrossRef]
- Méndez-Giménez, L.; Ezquerro, S.; da Silva, I.V.; Soveral, G.; Frühbeck, G.; Rodríguez, A. Pancreatic aquaporin-7: A novel target for anti-diabetic drugs? Front. Chem. 2018, 6, 99. [Google Scholar] [CrossRef]
- da Silva, I.V.; Cardoso, C.; Méndez-Giménez, L.; Camoes, S.P.; Frühbeck, G.; Rodríguez, A.; Miranda, J.P.; Soveral, G. Aquaporin-7 and aquaporin-12 modulate the inflammatory phenotype of endocrine pancreatic beta-cells. Arch. Biochem. Biophys. 2020, 691, 108481. [Google Scholar] [CrossRef] [PubMed]
- Bruun-Sørensen, A.S.; Edamana, S.; Login, F.H.; Borgquist, S.; Nejsum, L.N. Aquaporins in pancreatic ductal adenocarcinoma. APMIS 2021, 129, 700–705. [Google Scholar] [CrossRef]
- Arsenijevic, T.; Perret, J.; Van Laethem, J.; Delporte, C. Aquaporins involvement in pancreas physiology and in pancreatic diseases. Int. J. Mol. Sci. 2019, 20, 5052. [Google Scholar] [CrossRef] [PubMed]
- Direito, I.; Paulino, J.; Vigia, E.; Brito, M.A.; Soveral, G. Differential expression of aquaporin-3 and aquaporin-5 in pancreatic ductal adenocarcinoma. J. Surg. Oncol. 2017, 115, 980–996. [Google Scholar] [CrossRef] [PubMed]
- Edamana, S.; Pedersen, S.F.; Nejsum, L.N. Aquaporin water channels affect the response of conventional anticancer therapies of 3D grown breast cancer cells. Biochem. Biophys. Res. Commun. 2023, 639, 126–133. [Google Scholar] [CrossRef]
- Magouliotis, D.E.; Tasiopoulou, V.S.; Dimas, K.; Sakellaridis, N.; Svokos, K.A.; Svokos, A.A.; Zacharoulis, D. Transcriptomic analysis of the aquaporin (AQP) gene family interactome identifies a molecular panel of four prognostic markers in patients with pancreatic ductal adenocarcinoma. Pancreatology 2019, 19, 436–442. [Google Scholar] [CrossRef] [PubMed]
- World Medical Association. World Medical Association Declaration of Helsinki: Ethical principles for medical research involving human subjects. JAMA 2013, 310, 2191–2194. [Google Scholar] [CrossRef]
- da Silva, I.V.; Soares, B.P.; Pimpão, C.; Pinto, R.M.A.; Costa, T.; Freire, J.P.B.; Corrent, E.; Chalvon-Demersay, T.; Prates, J.A.M.; Lopes, P.A.; et al. Glutamine and cystine-enriched diets modulate aquaporins gene expression in the small intestine of piglets. PLoS ONE 2021, 16, e0245739. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Fleige, S.; Pfaffl, M.W. RNA integrity and the effect on the real-time qRT-PCR performance. Mol. Asp. Med. 2006, 27, 126–139. [Google Scholar] [CrossRef]
- SAS Institute. SAS/STAT User’s Guide, Version 9.1; SAS Institute: Cary, NC, USA, 2004. [Google Scholar]
- Zou, W.; Yang, Z.; Li, D.; Liu, Z.; Zou, Q.; Yuan, Y. AQP1 and AQP3 expression are associated with severe symptoms and poor-prognosis of the pancreatic ductal adenocarcinoma. Appl. Immunohistochem. Mol. Morphol. 2019, 27, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, C.; Pimpão, C.; Mósca, A.F.; Coxixo, A.S.; Lopes, D.; da Silva, I.V.; Pedersen, P.A.; Antunes, F.; Soveral, G. Human aquaporin-5 facilitates hydrogen peroxide permeation affecting adaption to oxidative stress and cancer cell migration. Cancers 2019, 11, 932. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, A.; Marinelli, R.A.; Tesse, A.; Fruhbeck, G.; Calamita, G. Sexual dimorphism of adipose and hepatic aquaglyceroporins in health and metabolic disorders. Front. Endocrinol. 2015, 6, 171. [Google Scholar] [CrossRef]
- Chen, J.; Wang, Z.; Xu, D.; Liu, Y.; Gao, Y. Aquaporin 3 promotes prostate cancer cell motility and invasion via extracellular signal-regulated kinase 1/2-mediated matrix metalloproteinase-3 secretion. Mol. Med. Rep. 2015, 11, 2882–2888. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.T.; Zhou, J.; Shi, S.; Xu, H.Y.; Qu, F.; Zhang, D.; Chen, Y.-D.; Yang, J.; Huang, H.-F.; Sheng, J.-Z. Identification of estrogen response element in aquaporin-3 gene that mediates estrogen-induced cell migration and invasion in estrogen receptor-positive breast cancer. Sci. Rep. 2015, 5, 12484. [Google Scholar] [CrossRef]
- Satooka, H.; Hara-Chikuma, M. Aquaporin-3 controls breast cancer cell migration by regulating hydrogen peroxide transport and its downstream cell signaling. Mol. Cell. Biol. 2016, 36, 1206–1218. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Huang, L.; Shao, M. Aquaporin 3 facilitates tumor growth in pancreatic cancer by modulating mTOR signaling. Biochem. Biophys. Res. Commun. 2017, 486, 1097–1102. [Google Scholar] [CrossRef] [PubMed]
- Costa, P.; Lemos, J.P.; Lopes, P.A.; Alfaia, C.M.; Costa, A.S.H.; Bessa, R.J.B.; Prates, J.A.M. Effect of low- and high-forage diets on meat quality and fatty acid composition of Alentejana and Barrosã beef breeds. Animal 2012, 6, 1187–1197. [Google Scholar] [CrossRef]
- da Silva, I.V.; Garra, S.; Calamita, G.; Soveral, G. The multifaceted role of aquaporin-9 in health and its potential as a clinical biomarker. Biomolecules 2022, 12, 897. [Google Scholar] [CrossRef]




| Patient | Gender | Age | Cancer Type | Aggressiveness Grade 1 | Invasiveness Grade 2 |
|---|---|---|---|---|---|
| 1 | M | 69 | Ampulla adenocarcinoma | Low | High |
| 2 | M | 55 | Kidney metastasis | Low | Low |
| 3 | F | 55 | Ductal adenocarcinoma | High | High |
| 4 | M | 63 | Ampulla adenocarcinoma | Low | High |
| 5 | M | 69 | Distal cholangiocarcinoma | High | High |
| 6 | M | 59 | Ductal adenocarcinoma | High | High |
| 7 | F | 69 | Non-invasive intraductal papillary mucinous neoplasm | Low | Low |
| 8 | F | 52 | Invasive intraductal papillary mucinous neoplasm | Low | High |
| 9 | F | 69 | Endocrine neoplasia | High | Low |
| 10 | F | 82 | Ductal adenocarcinoma | High | High |
| 11 | M | 58 | Distal cholangiocarcinoma | Low | High |
| 12 | F | 65 | Non-invasive intraductal papillary mucinous neoplasm | Low | Low |
| 13 | F | 65 | Ductal adenocarcinoma | High | High |
| 14 | M | 80 | Ductal adenocarcinoma | Low | High |
| 15 | F | 72 | Ampulla adenocarcinoma | Low | High |
| 16 | F | 60 | Ductal adenocarcinoma | High | High |
| 17 | M | 56 | Ampulla adenocarcinoma | Low | High |
| 18 | M | 56 | Cystic neuroendocrine tumor | Low | Low |
| 19 | M | 67 | Ductal adenocarcinoma | High | High |
| 20 | F | 78 | Ductal adenocarcinoma | High | High |
| 21 | M | 50 | Distal cholangiocarcinoma | Low | High |
| 22 | F | 80 | Invasive intraductal papillary mucinous neoplasm | Low | Low |
| 23 | M | 64 | Ductal adenocarcinoma | High | High |
| 24 | F | 51 | Cystic neuroendocrine tumor | Low | Low |
| Gene | Full Gene Name | TaqMan Gene Expression Assay |
|---|---|---|
| AQP1 | Aquaporin 1 | Hs01028916_m1 |
| AQP3 | Aquaporin 3 | Hs01105469_g1 |
| AQP5 | Aquaporin 5 | Hs00387048_m1 |
| AQP9 | Aquaporin 9 | Hs00175573_m1 |
| Housekeeping Gene | ||
| ACTB | β-actin | Hs99999903_m1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lopes, P.A.; Fonseca, E.; da Silva, I.V.; Vigia, E.; Paulino, J.; Soveral, G. Aquaporins Transcripts with Potential Prognostic Value in Pancreatic Cancer. Genes 2023, 14, 1694. https://doi.org/10.3390/genes14091694
Lopes PA, Fonseca E, da Silva IV, Vigia E, Paulino J, Soveral G. Aquaporins Transcripts with Potential Prognostic Value in Pancreatic Cancer. Genes. 2023; 14(9):1694. https://doi.org/10.3390/genes14091694
Chicago/Turabian StyleLopes, Paula A., Elisabete Fonseca, Inês V. da Silva, Emanuel Vigia, Jorge Paulino, and Graça Soveral. 2023. "Aquaporins Transcripts with Potential Prognostic Value in Pancreatic Cancer" Genes 14, no. 9: 1694. https://doi.org/10.3390/genes14091694
APA StyleLopes, P. A., Fonseca, E., da Silva, I. V., Vigia, E., Paulino, J., & Soveral, G. (2023). Aquaporins Transcripts with Potential Prognostic Value in Pancreatic Cancer. Genes, 14(9), 1694. https://doi.org/10.3390/genes14091694