Metabolomic Profiling of Female Mink Serum during Early to Mid-Pregnancy to Reveal Metabolite Changes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Serum Collection
2.2. Sample Preparation
2.3. LC/MS Analysis
2.4. Data Processing and Pattern Recognition
3. Results
3.1. Original Chromatogram Based on LC/MS
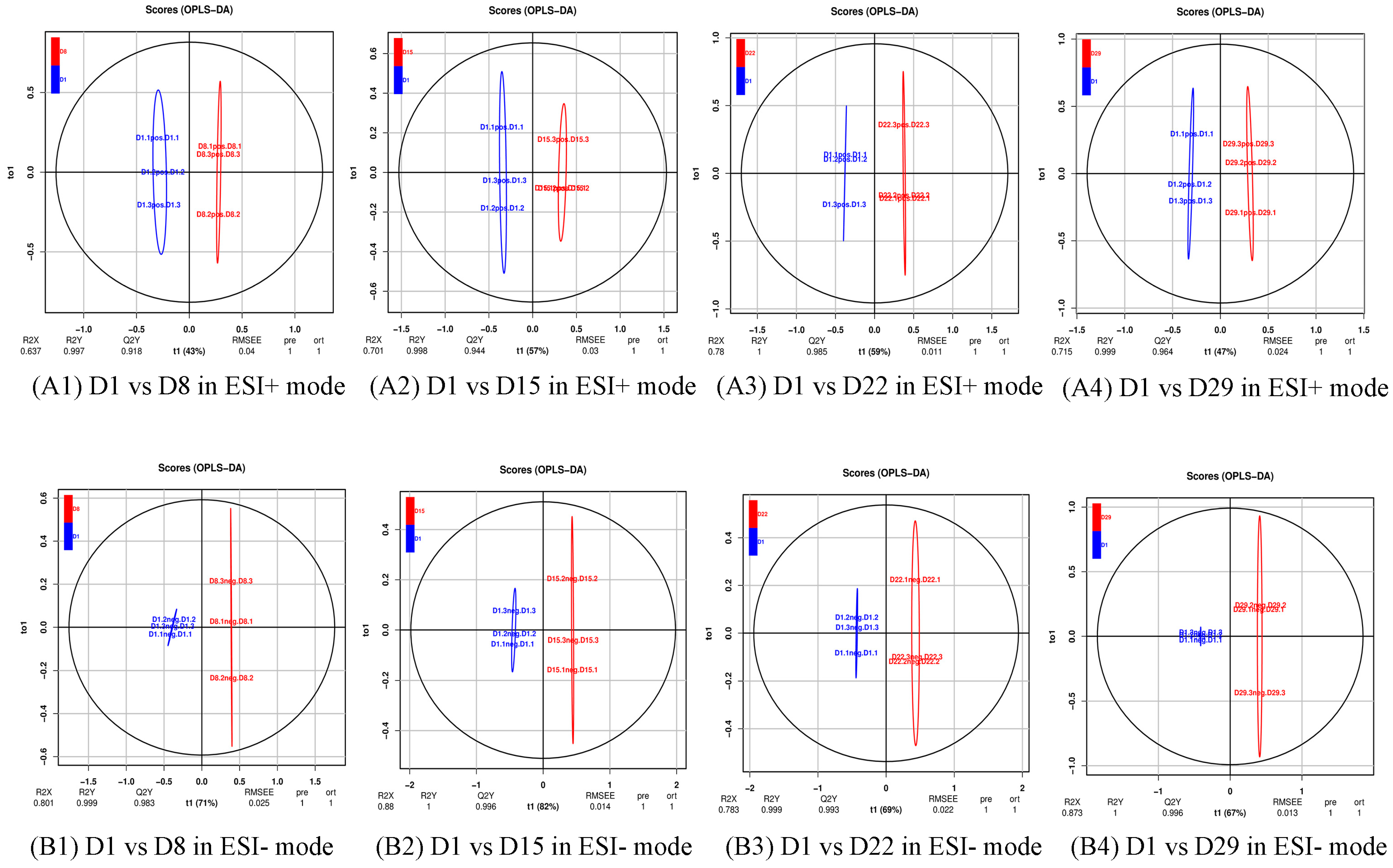
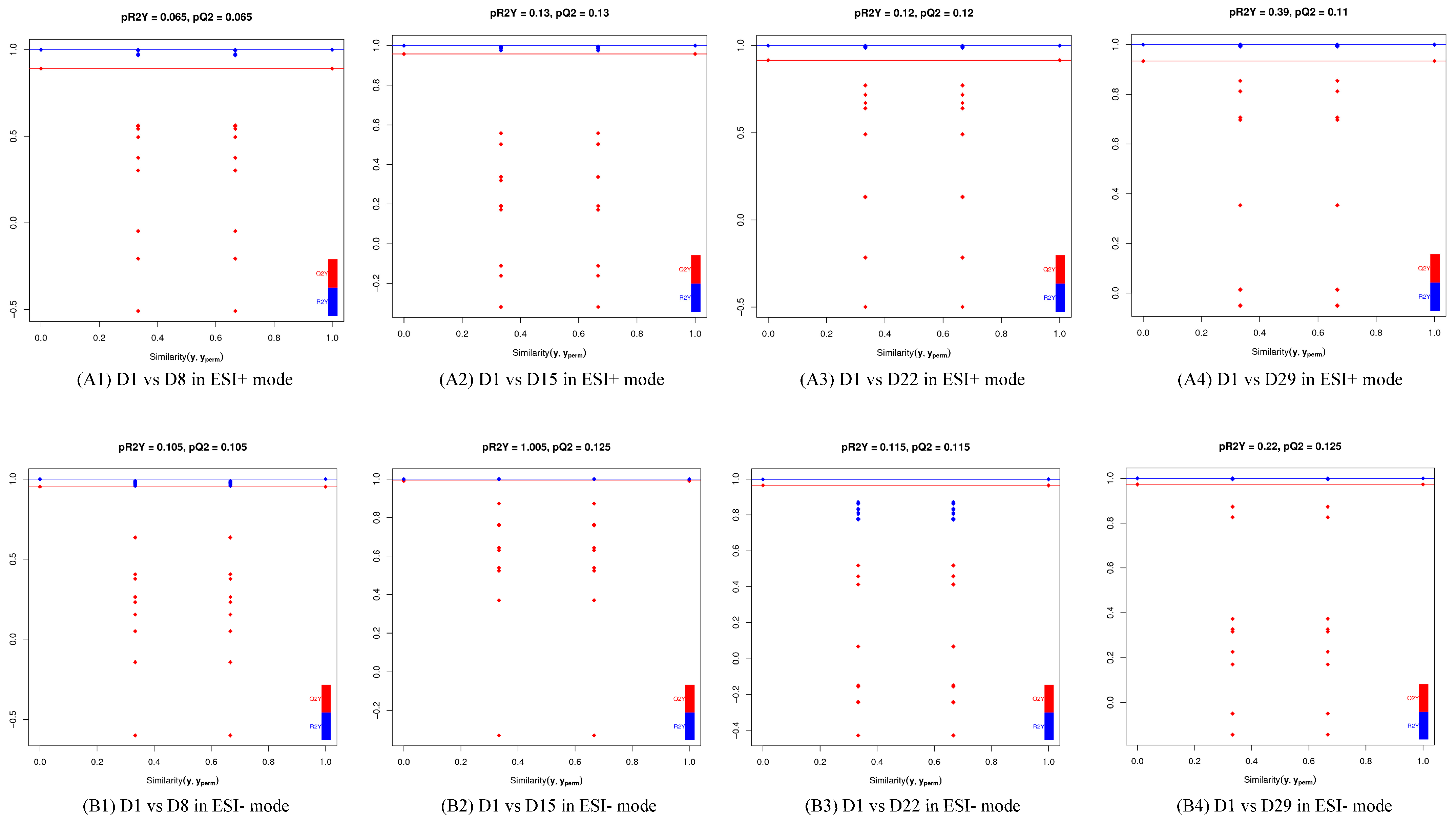
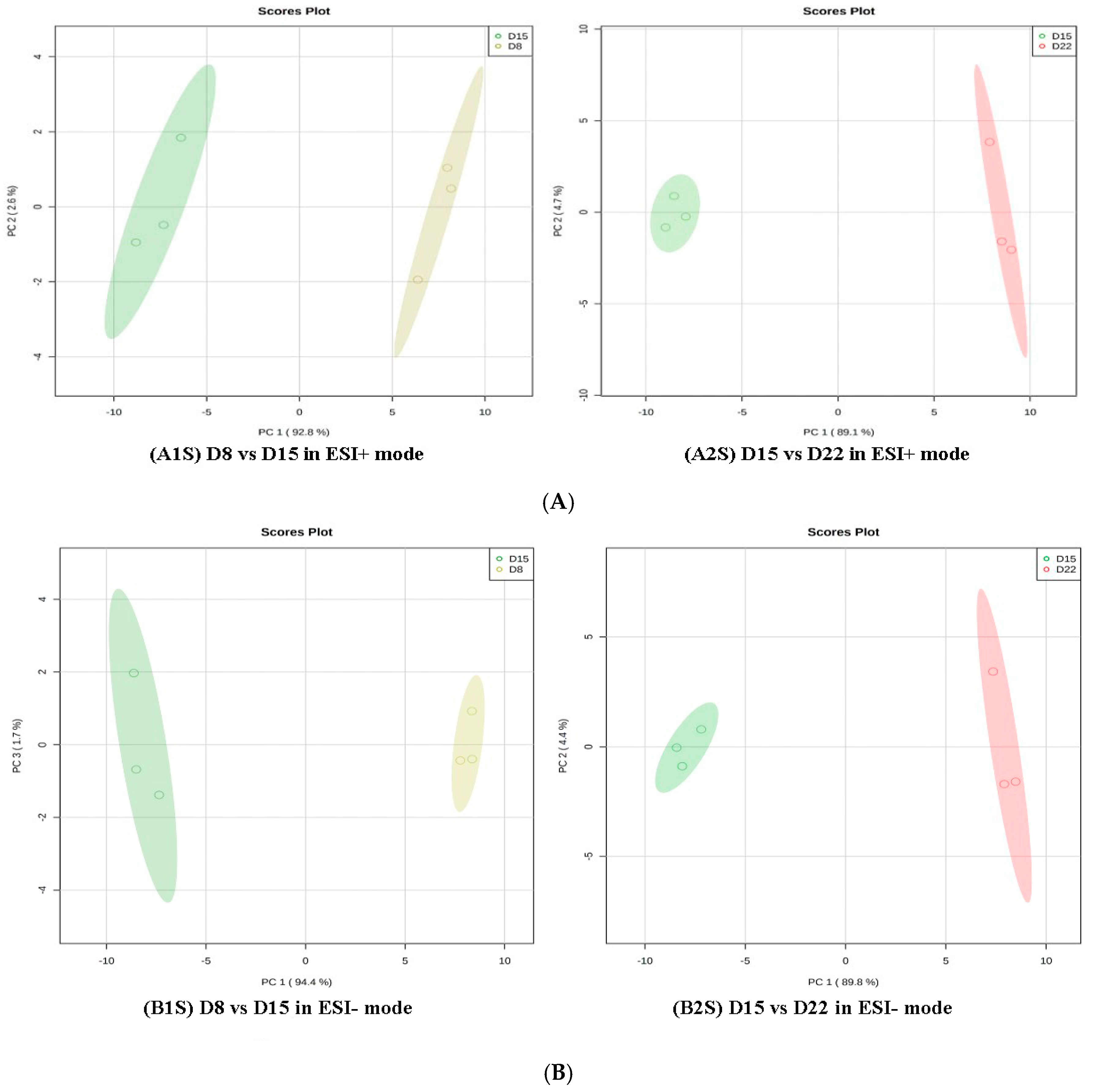
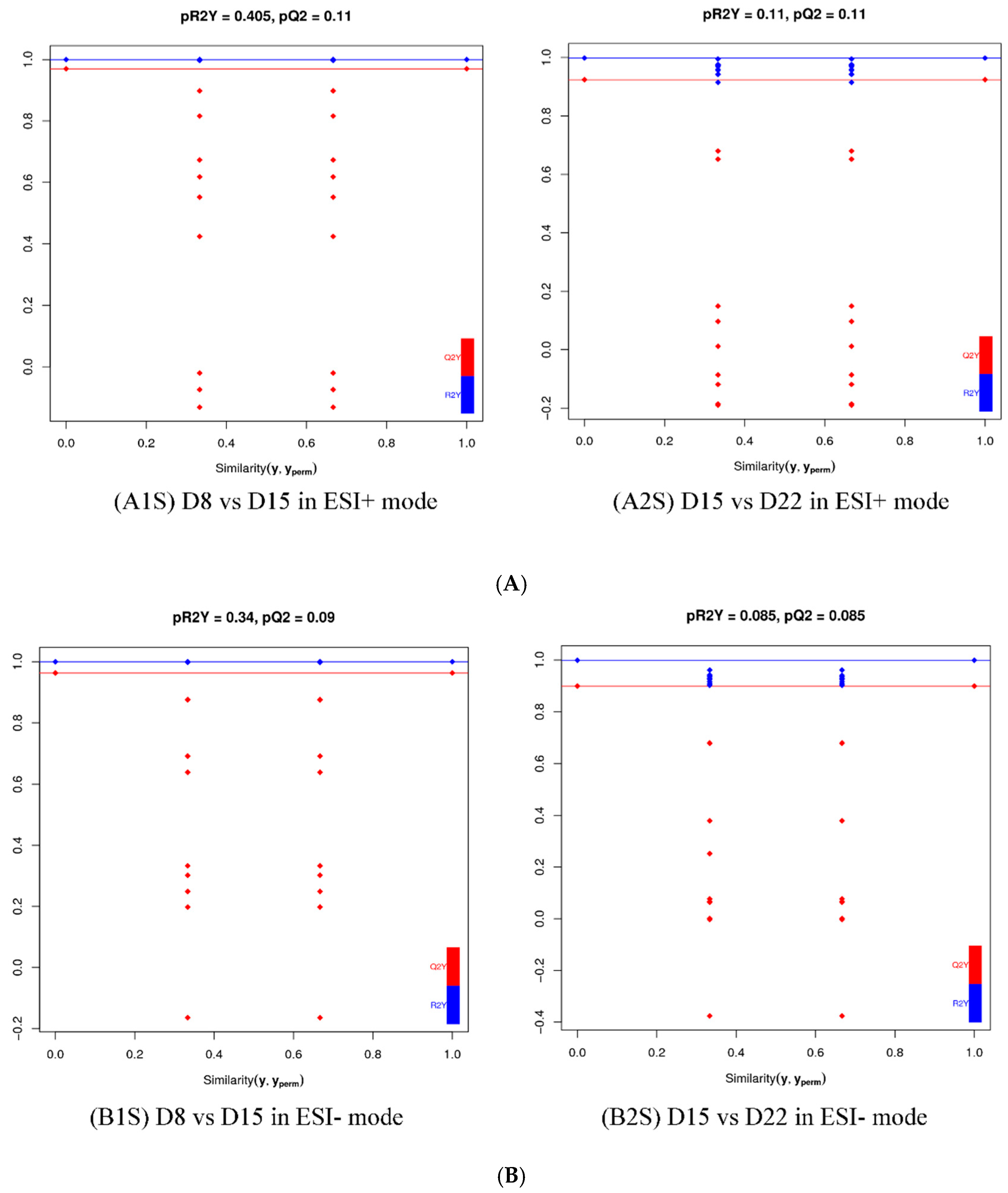
3.2. Multivariate Statistical Analysis
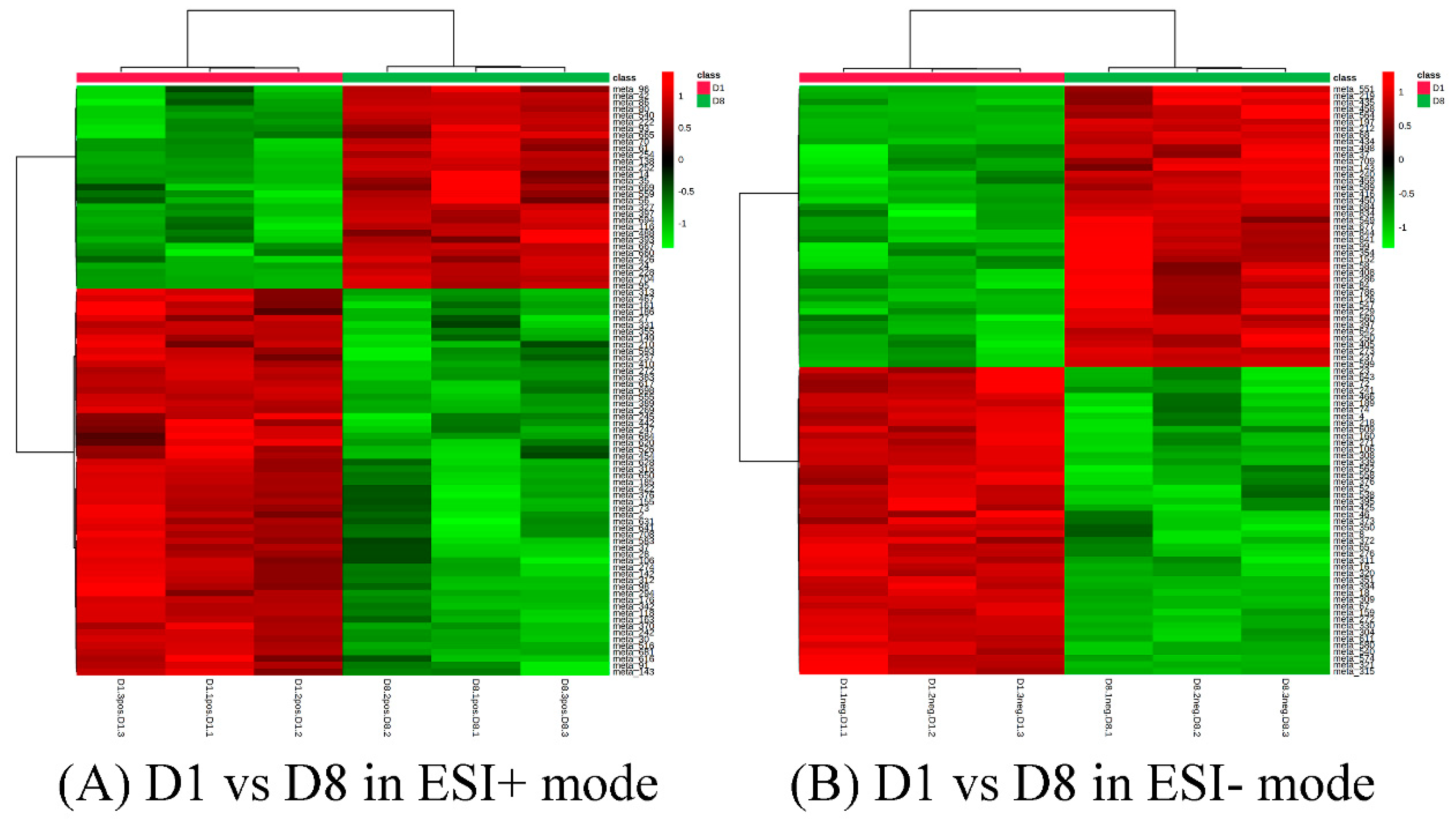
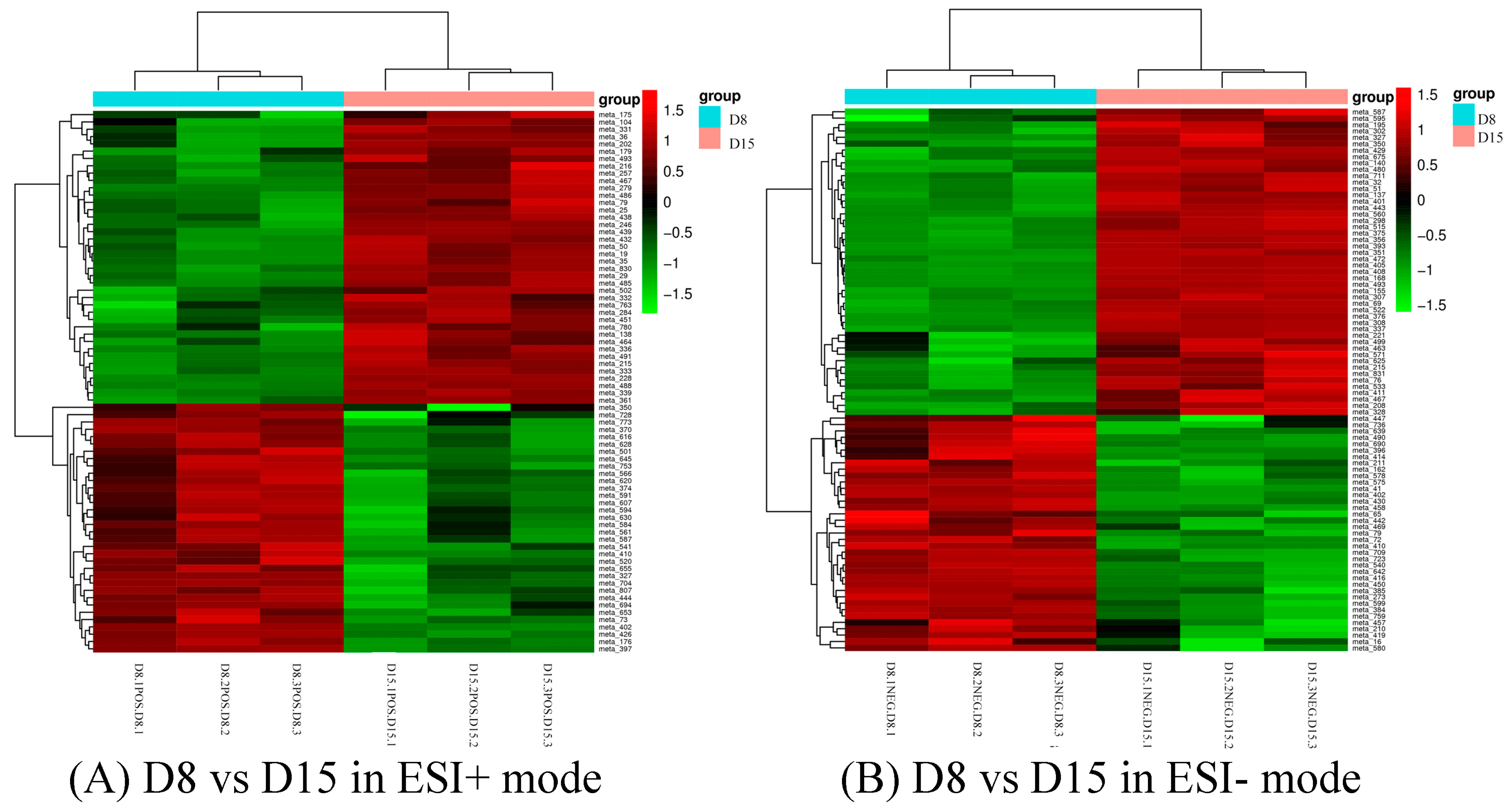
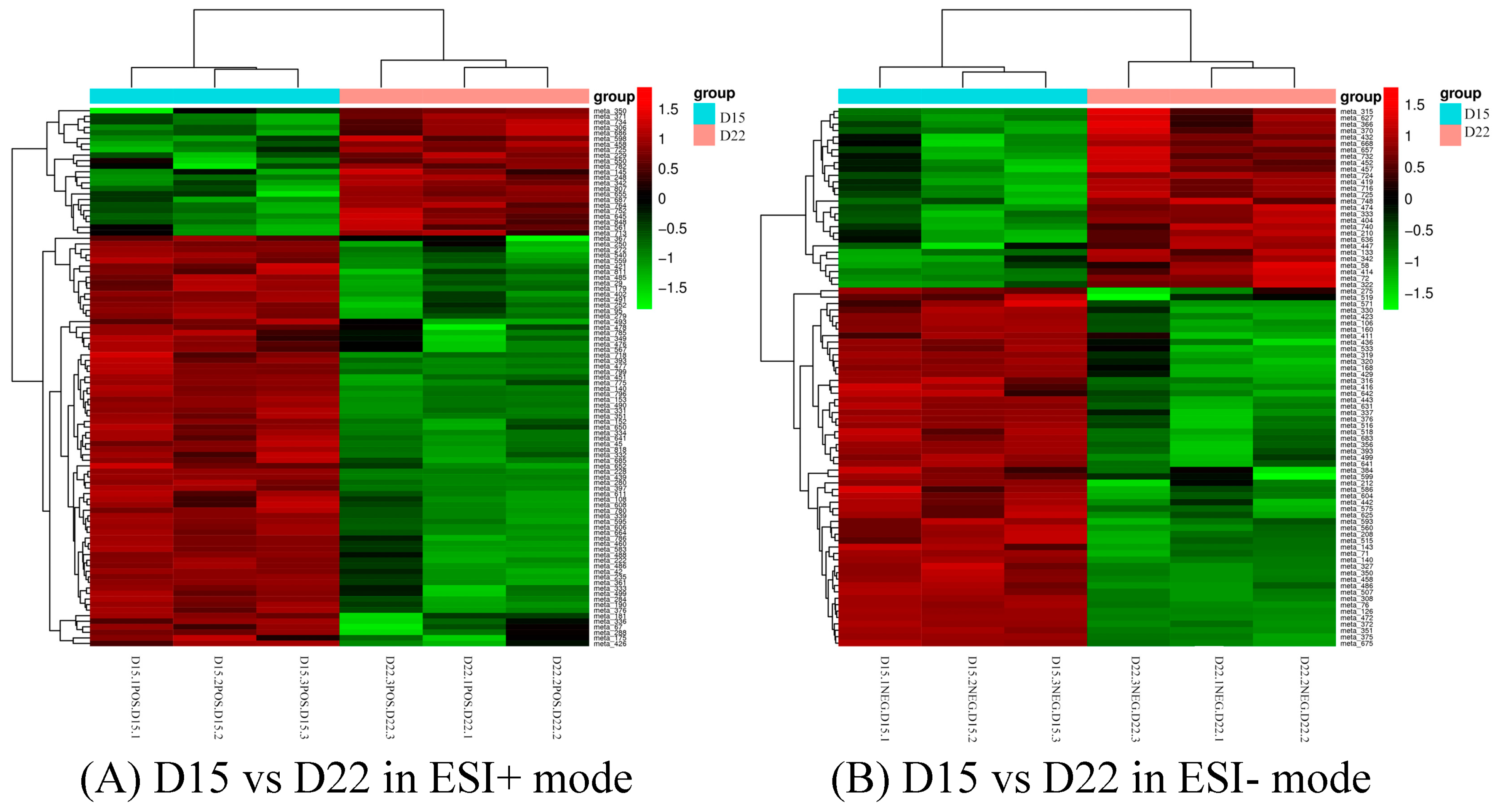
3.3. Identification of Differential Metabolites and Metabolic Pathways
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fan, B.; Han, Y.; Yang, Y.; Zhao, X.; Tang, Y.; Li, X.; Diao, Y.; Xu, B. Transcriptomic analysis of ovarian signaling at the emergence of the embryo from obligate diapause in the American mink (Neovison vison). Anim. Reprod. Sci. 2021, 232, 106823. [Google Scholar] [CrossRef] [PubMed]
- Murphy, B.D.; Di Gregorio, G.B.; Douglas, D.A.; González-Reyna, A. Interactions between melatonin and prolactin during gestation in mink (Mustela vison). J. Reprod. Fertil. 1990, 89, 423–429. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Li, C.; Chen, L.; Liu, Y.; Hou, R.; Zhou, X. Research advances on embryonic diapause in mammals. Anim. Reprod. Sci. 2018, 198, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Camilletti, M.A.; Abeledo-Machado, A.; Faraoni, E.Y.; Thomas, P.; Díaz-Torga, G. New insights into progesterone actions on prolactin secretion and prolactinoma development. Steroids 2019, 152, 108496. [Google Scholar] [CrossRef]
- Hirota, Y. Progesterone governs endometrial proliferation-differentiation switching and blastocyst implantation. Endocr. J. 2019, 66, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Papke, R.L.; Concannon, P.W.; Travis, H.F.; Hansel, W. Control of luteal function and implantation in the mink by prolactin. J. Anim. Sci. 1980, 50, 1102–1107. [Google Scholar] [CrossRef]
- Fenelon, J.C.; Banerjee, A.; Lefèvre, P.; Gratian, F.; Murphy, B.D. Polyamine-Mediated Effects of Prolactin Dictate Emergence from Mink Obligate Embryonic Diapause. Biol. Reprod. 2016, 95, 6. [Google Scholar] [CrossRef]
- Kurihara, S. Polyamine metabolism and transport in gut microbes. Biosci. Biotechnol. Biochem. 2022, 86, 957–966. [Google Scholar] [CrossRef]
- Shi, C.; Yan, Z.; Zhang, Y.; Qin, L.; Wu, W.; Gao, C.; Gao, L.; Liu, J.; Cui, Y. Effects of putrescine on the quality and epigenetic modification of mouse oocytes during in vitro maturation. Reprod. Fertil. Dev. 2022, 34, 957–970. [Google Scholar] [CrossRef]
- Hussain, T.; Tan, B.; Ren, W.; Rahu, N.; Kalhoro, D.H.; Yin, Y. Exploring polyamines: Functions in embryo/fetal development. Anim. Nutr. 2017, 3, 7–10. [Google Scholar] [CrossRef]
- Cui, L.; Lu, H.; Lee, Y.H. Challenges and emergent solutions for LC-MS/MS based untargeted metabolomics in diseases. Mass Spectrom. Rev. 2018, 37, 772–792. [Google Scholar] [CrossRef]
- Bian, Y.; Gao, C.; Kuster, B. On the potential of micro-flow LC-MS/MS in proteomics. Expert Rev. Proteom. 2022, 19, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Pang, Z.; Xia, J. Comprehensive investigation of pathway enrichment methods for functional interpretation of LC-MS global metabolomics data. Brief. Bioinform. 2023, 24, bbac553. [Google Scholar] [CrossRef] [PubMed]
- Okahashi, N.; Ueda, M.; Yasuda, S.; Tsugawa, H.; Arita, M. Global profiling of gut microbiota-associated lipid metabolites in antibiotic-treated mice by LC-MS/MS-based analyses. STAR Protoc. 2021, 2, 100492. [Google Scholar] [CrossRef] [PubMed]
- Hu, K.; Yu, Y. Metabolite availability as a window to view the early embryo microenvironment in vivo. Mol. Reprod. Dev. 2017, 84, 1027–1038. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wang, L.; Chen, C.; Qi, H.; Baker, P.N.; Liu, X.; Zhang, H.; Han, T.L. Metabolic Changes of Maternal Uterine Fluid, Uterus, and Plasma during the Peri-implantation Period of Early Pregnancy in Mice. Reprod. Sci. 2020, 27, 488–502. [Google Scholar] [CrossRef]
- Spindle, A.I.; Pedersen, R.A. Hatching, attachment, and outgrowth of mouse blastocysts in vitro: Fixed nitrogen requirements. J. Exp. Zool. 1973, 186, 305–318. [Google Scholar] [CrossRef]
- Martin, P.M.; Sutherland, A.E. Exogenous amino acids regulate trophectoderm differentiation in the mouse blastocyst through an mTOR-dependent pathway. Dev. Biol. 2001, 240, 182–193. [Google Scholar] [CrossRef]
- Morris, M.B.; Ozsoy, S.; Zada, M.; Zada, M.; Zamfirescu, R.C.; Todorova, M.G.; Day, M.L. Selected Amino Acids Promote Mouse Pre-implantation Embryo Development in a Growth Factor-Like Manner. Front. Physiol. 2020, 11, 140. [Google Scholar] [CrossRef]
- Bauermeister, A.; Mannochio-Russo, H.; Costa-Lotufo, L.V.; Jarmusch, A.K.; Dorrestein, P.C. Mass spectrometry-based metabolomics in microbiome investigations. Nat. Rev. Microbiol. 2022, 20, 143–160. [Google Scholar] [CrossRef]
- Wang, S.; Blair, I.A.; Mesaros, C. Analytical Methods for Mass Spectrometry-Based Metabolomics Studies. Adv. Exp. Med. Biol. 2019, 1140, 635–647. [Google Scholar] [CrossRef] [PubMed]
- Desmarais, J.A.; Bordignon, V.; Lopes, F.L.; Smith, L.C.; Murphy, B.D. The escape of the mink embryo from obligate diapause. Biol. Reprod. 2004, 70, 662–670. [Google Scholar] [CrossRef] [PubMed]
- Martinet, L.; Allais, C.; Allain, D. The role of prolactin and LH in luteal function and blastocyst growth in mink (Mustela vison). J. Reprod. Fertil. Suppl. 1981, 29, 119–130. [Google Scholar] [PubMed]
- Fenelon, J.C.; Lefèvre, P.L.; Banerjee, A.; Murphy, B.D. Regulation of diapause in carnivores. Reprod. Domest. Anim. 2017, 52 (Suppl. S2), 12–17. [Google Scholar] [CrossRef] [PubMed]
- Grattan, D.R. 60 Years of Neuroendocrinology: The hypothalamo-prolactin axis. J. Endocrinol. 2015, 226, T101–T122. [Google Scholar] [CrossRef]
- Kirsch, P.; Kunadia, J.; Shah, S.; Agrawal, N. Metabolic effects of prolactin and the role of dopamine agonists: A review. Front. Endocrinol. 2022, 13, 1002320. [Google Scholar] [CrossRef]
- Zhu, W.; Mantione, K.J.; Shen, L.; Cadet, P.; Esch, T.; Goumon, Y.; Bianchi, E.; Sonetti, D.; Stefano, G.B. Tyrosine and tyramine increase endogenous ganglionic morphine and dopamine levels in vitro and in vivo: Cyp2d6 and tyrosine hydroxylase modulation demonstrates a dopamine coupling. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2005, 11, Br397–Br404. [Google Scholar]
- Nagatsu, T.; Nakashima, A.; Watanabe, H.; Ito, S.; Wakamatsu, K. Neuromelanin in Parkinson’s Disease: Tyrosine Hydroxylase and Tyrosinase. Int. J. Mol. Sci. 2022, 23, 4176. [Google Scholar] [CrossRef]
- Kapatos, G.; Zigmond, M. Dopamine biosynthesis from L-tyrosine and L-phenylalanine in rat brain synaptosomes: Preferential use of newly accumulated precursors. J. Neurochem. 1977, 28, 1109–1119. [Google Scholar] [CrossRef]
- Dong, Y.; Bai, Y.; Liu, G.; Wang, Z.; Cao, J.; Chen, Y.; Yang, H. The immunologic and antioxidant effects of L-phenylalanine on the uterine implantation of mice embryos during early pregnancy. Histol. Histopathol. 2014, 29, 1335–1342. [Google Scholar] [CrossRef]
- Gao, H. Amino Acids in Reproductive Nutrition and Health. Adv. Exp. Med. Biol. 2020, 1265, 111–131. [Google Scholar] [CrossRef]
- Chieffi Baccari, G.; Falvo, S.; Santillo, A.; Di Giacomo Russo, F.; Di Fiore, M.M. D-Amino acids in mammalian endocrine tissues. Amino Acids 2020, 52, 1263–1273. [Google Scholar] [CrossRef]
- Leese, H.J.; McKeegan, P.J.; Sturmey, R.G. Amino Acids and the Early Mammalian Embryo: Origin, Fate, Function and Life-Long Legacy. Int. J. Environ. Res. Public Health 2021, 18, 9874. [Google Scholar] [CrossRef]
- Zamfirescu, R.C.; Day, M.L.; Morris, M.B. mTORC1/2 signaling is downregulated by amino acid-free culture of mouse preimplantation embryos and is only partially restored by amino acid readdition. Am. J. Physiol. Cell Physiol. 2021, 320, C30–C44. [Google Scholar] [CrossRef]
- Špirková, A.; Kovaříková, V.; Šefčíková, Z.; Pisko, J.; Kšiňanová, M.; Koppel, J.; Fabian, D.; Čikoš, Š. Glutamate can act as a signaling molecule in mouse preimplantation embryos. Biol. Reprod. 2022, 107, 916–927. [Google Scholar] [CrossRef]
- Van Winkle, L.J. Amino acid transport regulation and early embryo development. Biol. Reprod. 2001, 64, 1–12. [Google Scholar] [CrossRef]
- Zhang, S.; Zeng, X.; Ren, M.; Mao, X.; Qiao, S. Novel metabolic and physiological functions of branched chain amino acids: A review. J. Anim. Sci. Biotechnol. 2017, 8, 10. [Google Scholar] [CrossRef]
- Chen, P.R.; Redel, B.K.; Spate, L.D.; Ji, T.; Salazar, S.R.; Prather, R.S. Glutamine supplementation enhances development of in vitro-produced porcine embryos and increases leucine consumption from the medium. Biol. Reprod. 2018, 99, 938–948. [Google Scholar] [CrossRef]
- González, I.M.; Martin, P.M.; Burdsal, C.; Sloan, J.L.; Mager, S.; Harris, T.; Sutherland, A.E. Leucine and arginine regulate trophoblast motility through mTOR-dependent and independent pathways in the preimplantation mouse embryo. Dev. Biol. 2012, 361, 286–300. [Google Scholar] [CrossRef]
- Tochitani, S. Functions of Maternally-Derived Taurine in Fetal and Neonatal Brain Development. Adv. Exp. Med. Biol. 2017, 975 Pt 1, 17–25. [Google Scholar] [CrossRef]
- Mu, T.; Feng, Y.; Che, Y.; Lv, Q.; Hu, J.; Yang, Q.; Yang, J. Taurine Promotes In-vitro Follicle Development, Oocyte Maturation, Fertilization and Cleavage of rats. Adv. Exp. Med. Biol. 2019, 1155, 197–203. [Google Scholar] [CrossRef]
- Sivashanmugam, M.; Jaidev, J.; Umashankar, V.; Sulochana, K.N. Ornithine and its role in metabolic diseases: An appraisal. Biomed. Pharmacother. Biomed. Pharmacother. 2017, 86, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Lenis, Y.Y.; Elmetwally, M.A.; Maldonado-Estrada, J.G.; Bazer, F.W. Physiological importance of polyamines. Zygote (Camb. Engl.) 2017, 25, 244–255. [Google Scholar] [CrossRef]
- Wu, G.; Bazer, F.W.; Hu, J.; Johnson, G.A.; Spencer, T.E. Polyamine synthesis from proline in the developing porcine placenta. Biol. Reprod. 2005, 72, 842–850. [Google Scholar] [CrossRef] [PubMed]
- Kwon, H.; Spencer, T.E.; Bazer, F.W.; Wu, G. Developmental changes of amino acids in ovine fetal fluids. Biol. Reprod. 2003, 68, 1813–1820. [Google Scholar] [CrossRef]
- Fenelon, J.C.; Murphy, B.D. Inhibition of polyamine synthesis causes entry of the mouse blastocyst into embryonic diapause. Biol. Reprod. 2017, 97, 119–132. [Google Scholar] [CrossRef] [PubMed]




| No. | Name | p-Value | VIP | Regulated | KEGG Pathway Annotation |
|---|---|---|---|---|---|
| 1 | L-proline | 0.003 | 1.300 | down | Metabolic pathways (ko01100); arginine and proline metabolism (ko00330) |
| 2 | L-valine | 0.004 | 1.294 | down | Metabolic pathways (ko01100); biosynthesis of amino acids (ko01230) |
| 3 | L-threonine | 0.015 | 1.225 | down | Valine, leucine, and isoleucine biosynthesis (ko00290) |
| 4 | Taurine | 0.049 | 1.201 | down | Taurine and hypotaurine metabolism (ko00430) |
| 5 | L-leucine | 0.021 | 1.204 | down | Metabolic pathways (ko01100); biosynthesis of amino acids (ko01230) |
| 6 | L-methionine | 0.036 | 1.249 | up | Cysteine and methionine metabolism (ko00270) |
| 7 | L-phenylalanine | 0.001 | 1.358 | up | Phenylalanine, tyrosine, and tryptophan biosynthesis (ko00400); phenylalanine metabolism (ko00360) |
| 8 | L-glutamine | 0.030 | 1.198 | down | Taurine and hypotaurine metabolism (ko00430); arginine and proline metabolism (ko00330) |
| 9 | Tyramine | 0.022 | 1.474 | up | Tyrosine metabolism (ko00350) |
| 10 | Dopamine | 0.000 | 1.543 | up | Metabolic pathways (ko01100); prolactin signaling pathway (ko04917) |
| 11 | D-ornithine | 0.004 | 1.487 | down | D-arginine and D-ornithine metabolism (ko00472); metabolic pathways (ko01100) |
| 12 | L-tyrosine | 0.000 | 1.535 | up | Phenylalanine metabolism (ko00360); dopaminergic synapse (ko04728); prolactin signaling pathway (ko04917) |
| 13 | L-lysine | 0.002 | 1.518 | down | Lysine degradation (ko00310); metabolic pathways (ko01100) |
| 14 | L-kynurenine | 0.000 | 1.533 | down | Tryptophan metabolism (ko00380) |
| No. | Name | p-Value | VIP | Regulated | KEGG Pathway Annotation |
|---|---|---|---|---|---|
| 1 | L-glutamate | 0.008 | 1.190 | down | Metabolic pathways (ko01100); biosynthesis of amino acids (ko01230) |
| 2 | L-methionine | 0.004 | 1.258 | up | Biosynthesis of amino acids (ko01230); cysteine and methionine metabolism (ko00270) |
| 3 | L-proline | 0.007 | 1.260 | down | Arginine and proline metabolism (ko00330); Metabolic pathways (ko01100) |
| 4 | L-threonine | 0.015 | 1.212 | down | Glycine, serine, and threonine metabolism (ko00260); metabolic pathways (ko01100) |
| 5 | L-phenylalanine | 0.004 | 1.270 | up | Biosynthesis of amino acids (ko01230); phenylalanine metabolism (ko00360) |
| 6 | L-tryptophan | 0.024 | 1.218 | down | Phenylalanine, tyrosine, and tryptophan biosynthesis (ko00400) |
| 7 | L-leucine | 0.024 | 1.182 | down | Valine, leucine, and isoleucine degradation (ko00280) |
| 8 | L-valine | 0.011 | 1.245 | down | Valine, leucine, and isoleucine biosynthesis (ko00290) |
| 9 | L-lysine | 0.041 | 1.063 | down | Lysine biosynthesis (ko00300); metabolic pathways (ko01100) |
| 10 | Taurine | 0.047 | 1.252 | down | Metabolic pathways (ko01100); taurine and hypotaurine metabolism (ko00430) |
| 11 | L-lysine | 0.011 | 1.338 | down | Lysine biosynthesis (ko00300); metabolic pathways (ko01100) |
| 12 | L-arginine | 0.006 | 1.383 | down | Biosynthesis of amino acids (ko01230); metabolic pathways (ko01100) |
| 13 | L-glutamate | 0.012 | 1.340 | down | Alanine, aspartate, and glutamate metabolism (ko00250) |
| No. | Name | p-Value | VIP | Regulated | KEGG Pathway Annotation |
|---|---|---|---|---|---|
| 1 | L-valine | 0.006 | 1.207 | down | Valine, leucine, and isoleucine degradation (ko00280) |
| 2 | L-glutamine | 0.008 | 1.200 | down | Biosynthesis of amino acids (ko01230); metabolic pathways (ko01100) |
| 3 | L-phenylalanine | 0.001 | 1.270 | up | Phenylalanine metabolism (ko00360); biosynthesis of amino acids (ko01230) |
| 4 | L-tryptophan | 0.022 | 1.132 | down | Phenylalanine, tyrosine, and tryptophan biosynthesis (ko00400) |
| 5 | L-threonine | 0.011 | 1.196 | down | Biosynthesis of amino acids (ko01230); metabolic pathways (ko01100) |
| 6 | L-methionine | 0.008 | 1.241 | up | Metabolic pathways (ko01100); cysteine and methionine metabolism (ko00270) |
| 7 | L-proline | 0.002 | 1.256 | down | Arginine and proline metabolism (ko00330); metabolic pathways (ko01100) |
| 8 | L-leucine | 0.005 | 1.217 | down | Biosynthesis of amino acids (ko01230); metabolic pathways (ko01100) |
| 9 | Dopamine | 0.000 | 1.543 | up | Metabolic pathways (ko01100); prolactin signaling pathway (ko04917) |
| 10 | D-ornithine | 0.004 | 1.487 | down | D-arginine and D-ornithine metabolism (ko00472); metabolic pathways (ko01100) |
| 11 | L-tyrosine | 0.000 | 1.535 | up | Phenylalanine metabolism (ko00360); dopaminergic synapse (ko04728); prolactin signaling pathway (ko04917) |
| 12 | L-lysine | 0.002 | 1.518 | down | Lysine degradation (ko00310); metabolic pathways (ko01100) |
| No. | Name | p-Value | VIP | Regulated | KEGG Pathway Annotation |
|---|---|---|---|---|---|
| 1 | L-phenylalanine | 0.046 | 1.200 | up | Phenylalanine, tyrosine, and tryptophan biosynthesis (ko00400) |
| 2 | L-glutamate | 0.016 | 1.190 | down | Arginine biosynthesis (ko00220) |
| 3 | L-tryptophan | 0.031 | 1.288 | down | Phenylalanine, tyrosine, and tryptophan biosynthesis (ko00400) |
| 4 | L-phenylalanine | 0.022 | 1.291 | up | Phenylalanine, tyrosine, and tryptophan biosynthesis (ko00400) |
| 5 | L-alanine | 0.025 | 1.340 | down | Metabolic pathways (ko01100); cysteine and methionine metabolism (ko00270) |
| 6 | L-glutamate | 0.048 | 1.254 | down | Arginine and proline metabolism (ko00330); metabolic pathways (ko01100) |
| 7 | L-lysine | 0.031 | 1.339 | down | Biosynthesis of amino acids (ko01230); metabolic pathways (ko01100) |
| No. | Name | D1 vs. D8 | D8 vs. D15 | D15 vs. D22 | D22 vs. D29 |
|---|---|---|---|---|---|
| 1 | Dopamine | up | up | down | down |
| 2 | Tyramine | up | up | down | down |
| 3 | L-phenylalanine | up | up | down | down |
| 4 | L-tyrosine | up | up | down | down |
| 5 | Tyrosine | up | up | down | down |
| 6 | L-kynurenine | down | up | down | down |
| 7 | L-lysine | down | up | down | down |
| 8 | L-arginine | down | up | down | down |
| 9 | D-ornithine | down | down | down | down |
| 10 | Leucine | down | up | down | down |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, Y.; Huang, Y.; Deng, L.; Li, Z.; Li, C. Metabolomic Profiling of Female Mink Serum during Early to Mid-Pregnancy to Reveal Metabolite Changes. Genes 2023, 14, 1759. https://doi.org/10.3390/genes14091759
Luo Y, Huang Y, Deng L, Li Z, Li C. Metabolomic Profiling of Female Mink Serum during Early to Mid-Pregnancy to Reveal Metabolite Changes. Genes. 2023; 14(9):1759. https://doi.org/10.3390/genes14091759
Chicago/Turabian StyleLuo, Yuxin, Yiqiu Huang, Liang Deng, Zheng Li, and Chunjin Li. 2023. "Metabolomic Profiling of Female Mink Serum during Early to Mid-Pregnancy to Reveal Metabolite Changes" Genes 14, no. 9: 1759. https://doi.org/10.3390/genes14091759
APA StyleLuo, Y., Huang, Y., Deng, L., Li, Z., & Li, C. (2023). Metabolomic Profiling of Female Mink Serum during Early to Mid-Pregnancy to Reveal Metabolite Changes. Genes, 14(9), 1759. https://doi.org/10.3390/genes14091759





