Selection and Validation of Reference Genes for Gene Expression Studies in Euonymus japonicus Based on RNA Sequencing
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. RNA-Seq
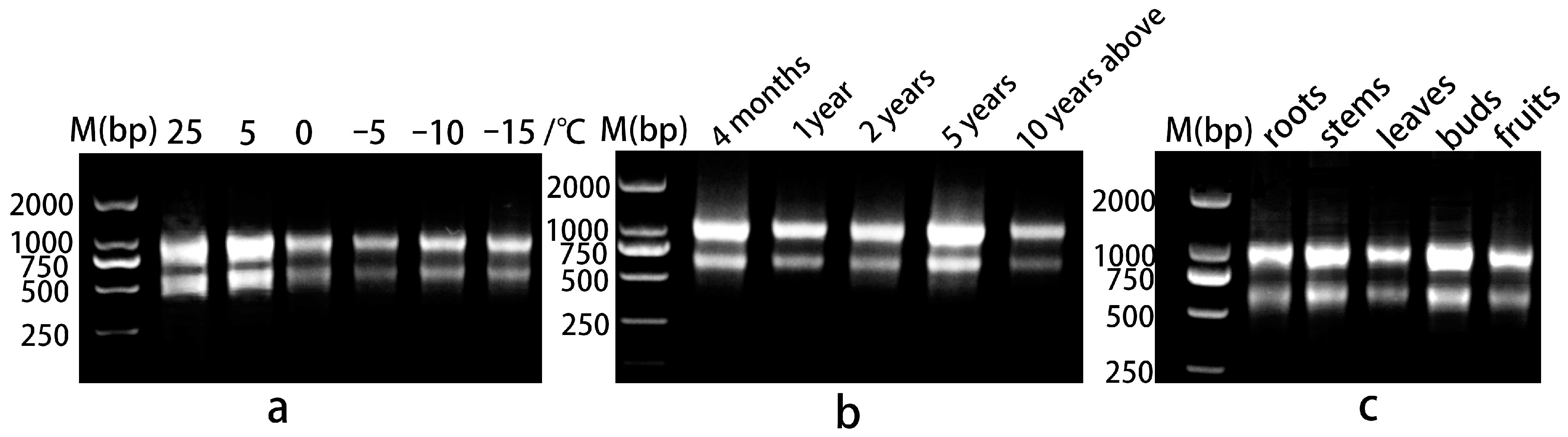
2.3. RNA Extraction and cDNA Reverse Transcription
2.4. Primer Design and PCR
2.5. Data Analysis
3. Results
3.1. Screening of Candidate RGs Based on the Results of RNA-Seq
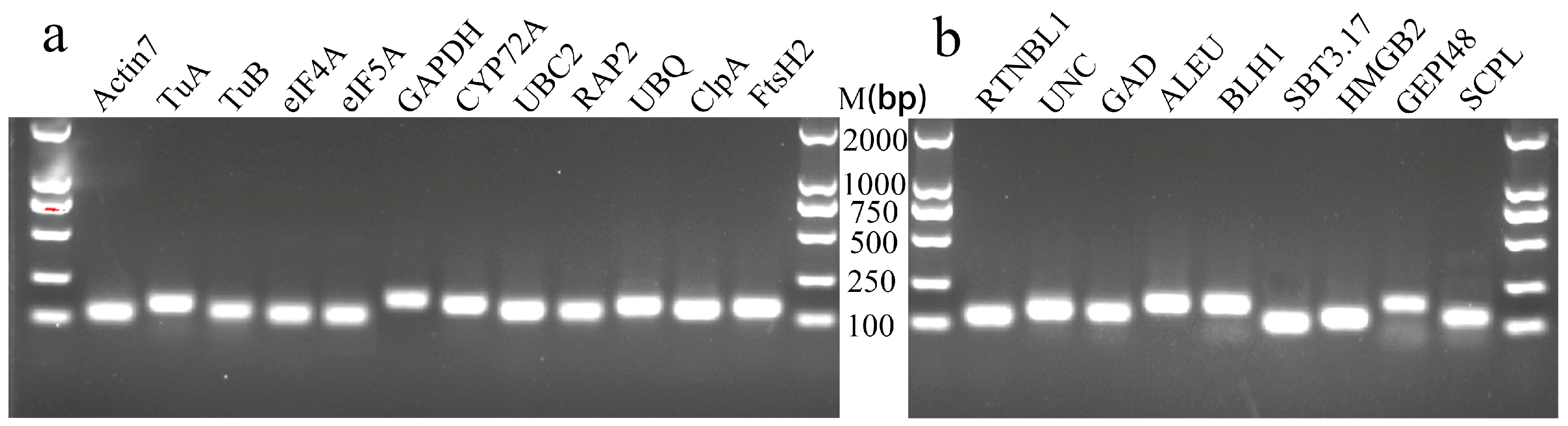
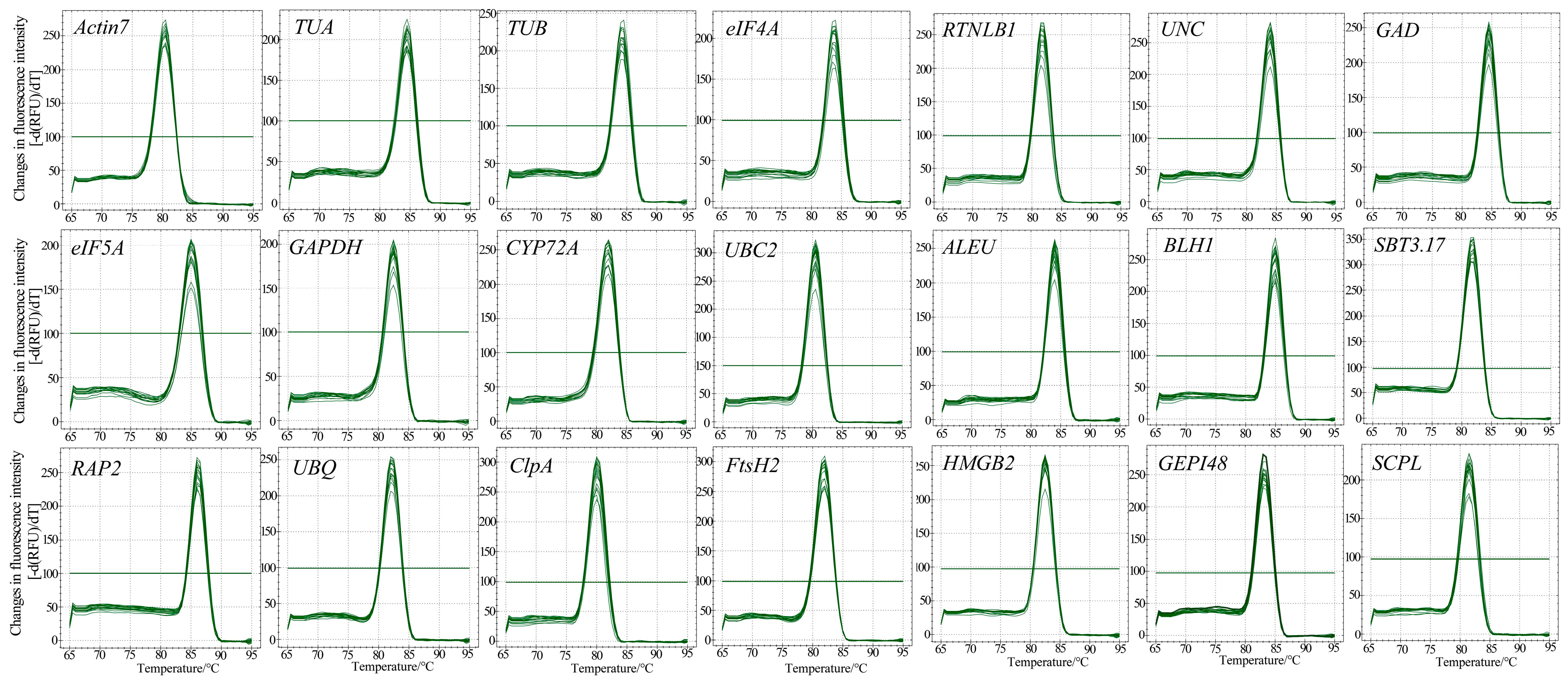
3.2. Primer Specificity and Amplification Efficiency of Candidate RGs
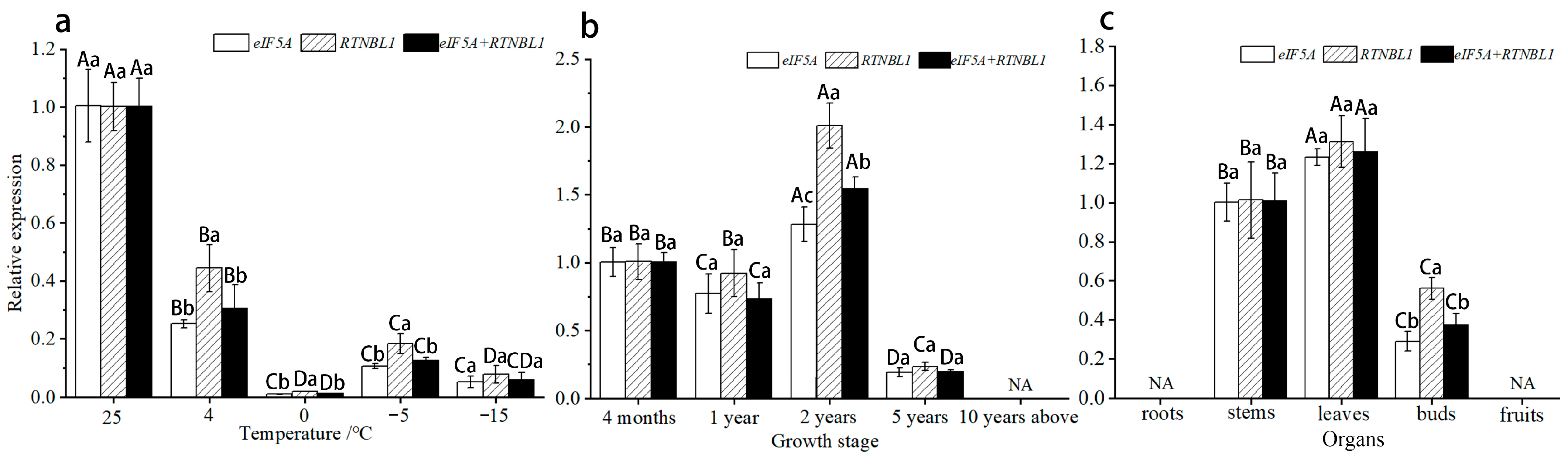
3.3. Expression Profiles of Candidate RGs
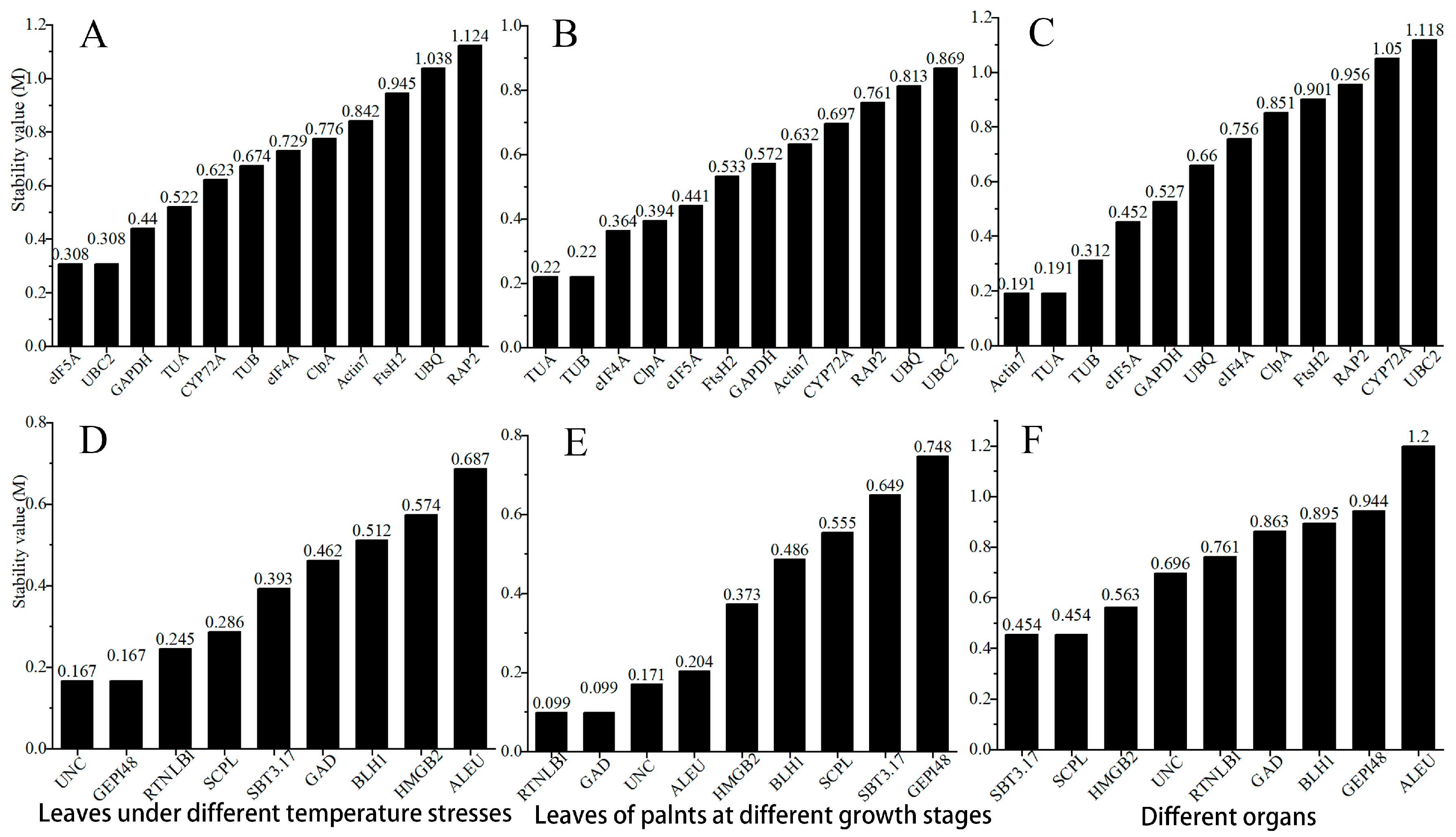
3.4. GeNorm Analysis
3.5. NormFinder Analysis
3.6. Bestkeeper Analysis
3.7. Comprehensive Analysis of RefFinder
3.8. Reference Gene Selection and Validation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xue, D. Multi-Omics Analysis of the Adaptation of Euonymus japonicus to Winter Freezing Stress. Master’s Thesis, Minzu University of China, Beijing, China, 2023. [Google Scholar] [CrossRef]
- Shiyuan, M.; Guiyun, L.; Mingzhong, Z.; Zhihao, Z.; Ye, W.; Huatian, W.; Xiumei, L. Physiolocical Response to Cold Stress and Evaluation of Cold Resistance for Five Species of Euonymus Linn. Acta Bot. Boreali Occident. Sin. 2020, 40, 624–634. [Google Scholar]
- Yankai, L.; Lijun, W.; Xiaohe, S. Survey and ldentification of Foliar Diseases on Euonymus japonicus. J. Yangtze Univ. Nat. Sci. Ed. 2017, 14, 5–8+3. [Google Scholar] [CrossRef]
- Huiwen, X. The Research on the Propagation Techniques of Euonymus japonicus cv. Microphyllus Butterscotch. Master’s Thesis, Central South University of Forestry and Technology, Changsha, China, 2011. [Google Scholar]
- Ma, L.; Wu, J.; Qi, W.; Coulter, J.A.; Fang, Y.; Li, X.; Liu, L.; Jin, J.; Niu, Z.; Yue, J.; et al. Screening and verification of reference genes for analysis of gene expression in winter rapeseed (Brassica rapa L.) under abiotic stress. PLoS ONE 2020, 15, e0236577. [Google Scholar] [CrossRef] [PubMed]
- Brunner, A.M.; Yakovlev, I.A.; Strauss, S.H. Validating internal controls for quantitative plant gene expression studies. BMC Plant Biol. 2004, 4, 14. [Google Scholar] [CrossRef] [PubMed]
- Huggett, J.; Dheda, K.; Bustin, S.; Zumla, A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005, 6, 279–284. [Google Scholar] [CrossRef]
- Udvardi, M.K.; Czechowski, T.; Scheible, W.-R.d. Eleven Golden Rules of Quantitative RT-PCR. Plant Cell 2008, 20, 1736–1737. [Google Scholar] [CrossRef]
- Chen, Y.; He, Q.; Li, X.; Zhang, Y.; Li, J.; Zhang, L.; Yao, X.; Zhang, X.; Liu, C.; Wang, H. Identification of Accurate Reference Genes for qRT-PCR Analysis of Gene Expression in Eremochloa ophiuroides under Multiple Stresses of Phosphorus Deficiency and/or Aluminum Toxicity. Plants 2023, 12, 3751. [Google Scholar] [CrossRef]
- Kim, B.-R.; Nam, H.-Y.; Kim, S.-U.; Kim, S.-I.; Chang, Y.-J. Normalization of reverse transcription quantitative-PCR with housekeeping genes in rice. Biotechnol. Lett. 2003, 25, 1869–1872. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, C.; Harijati, N.; Diao, Y.; Liu, E.; Hu, Z. Selection and Evaluation of Reference Genes for RT-qPCR Analysis in Amorphophallus Konjac Based on Transcriptome Data. Genes 2023, 14, 1513. [Google Scholar] [CrossRef]
- Niu, J.-Z.; Dou, W.; Ding, T.-B.; Yang, L.-H.; Shen, G.-M.; Wang, J.-J. Evaluation of suitable reference genes for quantitative RT-PCR during development and abiotic stress in Panonychus citri (McGregor) (Acari: Tetranychidae). Mol. Biol. Rep. 2012, 39, 5841–5849. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, Y.; Ding, L.; Zhang, J.; Wei, J.; Wang, H. Validation of Reference Genes for Gene Expression by Quantitative Real-Time RT-PCR in Stem Segments Spanning Primary to Secondary Growth in Populus tomentosa. PLoS ONE 2016, 11, e0157370. [Google Scholar] [CrossRef] [PubMed]
- Nazari, F.; Parham, A.; Maleki, A.F. GAPDH, β-actin and β2-microglobulin, as three common reference genes, are not reliable for gene expression studies in equine adipose- and marrow-derived mesenchymal stem cells. J. Anim. Sci. Technol. 2015, 57, 18. [Google Scholar] [CrossRef] [PubMed]
- Rubie, C.; Kempf, K.; Hans, J.; Su, T.; Tilton, B.; Georg, T.; Brittner, B.; Ludwig, B.; Schilling, M. Housekeeping gene variability in normal and cancerous colorectal, pancreatic, esophageal, gastric and hepatic tissues. Mol. Cell. Probes 2005, 19, 101–109. [Google Scholar] [CrossRef]
- Tong, J.; Hu, M.; Han, B.; Ji, Y.; Wang, B.; Liang, H.; Liu, M.; Wu, Z.; Liu, N. Determination of reliable reference genes for gene expression studies in Chinese chive (Allium tuberosum) based on the transcriptome profiling. Sci. Rep. 2021, 11, 16558. [Google Scholar] [CrossRef] [PubMed]
- Yunxia, Q.; Yongbin, L.; Weiheng, R. RNA-Seq and its applications: A new technology for transcriptomics. Hereditas 2011, 33, 1191–1202. [Google Scholar]
- Haas, B.J.; Zody, M.C. Advancing RNA-Seq analysis. Nat. Biotechnol. 2010, 28, 421–423. [Google Scholar] [CrossRef] [PubMed]
- Martin, L.; Fei, Z.; Giovannoni, J.; Rose, J. Catalyzing plant science research with RNA-seq. Front. Plant Sci. 2013, 4, 66. [Google Scholar] [CrossRef]
- Bai, X.; Chen, T.; Wu, Y.; Tang, M.; Xu, Z.-F. Selection and Validation of Reference Genes for qRT-PCR Analysis in the Oil-Rich Tuber Crop Tiger Nut (Cyperus esculentus) Based on Transcriptome Data. Int. J. Mol. Sci. 2021, 22, 2569. [Google Scholar] [CrossRef]
- Liang, L.; He, Z.; Yu, H.; Wang, E.; Zhang, X.; Zhang, B.; Zhang, C.; Liang, Z. Selection and Validation of Reference Genes for Gene Expression Studies in Codonopsis pilosula Based on Transcriptome Sequence Data. Sci. Rep. 2020, 10, 1362. [Google Scholar] [CrossRef]
- Sabeh, M.; Duceppe, M.-O.; St-Arnaud, M.; Mimee, B. Transcriptome-wide selection of a reliable set of reference genes for gene expression studies in potato cyst nematodes (Globodera spp.). PLoS ONE 2018, 13, e0193840. [Google Scholar] [CrossRef]
- Zhao, J.; Yang, F.; Feng, J.; Wang, Y.; Lachenbruch, B.; Wang, J.; Wan, X. Genome-Wide Constitutively Expressed Gene Analysis and New Reference Gene Selection Based on Transcriptome Data: A Case Study from Poplar/Canker Disease Interaction. Front. Plant Sci. 2017, 8, 1876. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chen, Y.; Shi, C.; Huang, Z.; Zhang, Y.; Li, S.; Li, Y.; Ye, J.; Yu, C.; Li, Z.; et al. SOAPnuke: A MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. GigaScience 2018, 7, gix120. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.0031. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of Real-Time Quantitative Reverse Transcription-PCR Data: A Model-Based Variance Estimation Approach to Identify Genes Suited for Normalization, Applied to Bladder and Colon Cancer Data Sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Xie, F.; Wang, J.; Zhang, B. RefFinder: A web-based tool for comprehensively analyzing and identifying reference genes. Funct. Integr. Genom. 2023, 23, 125. [Google Scholar] [CrossRef]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef]
- Xu, H.; Bao, J.-D.; Dai, J.-S.; Li, Y.; Zhu, Y. Genome-Wide Identification of New Reference Genes for qRT-PCR Normalization under High Temperature Stress in Rice Endosperm. PLoS ONE 2015, 10, e0142015. [Google Scholar] [CrossRef] [PubMed]
- Ge, D.; Yang, H.; Lunlin, C.; Shuyu, L.; Laiqiang, S.; Jie, X. RNA-seg Based Discovery of New Reference Genes and Primers in Brassica napus under Aluminum Stress. Acta Agric. Boreali Sin. 2021, 36, 1–9. [Google Scholar]
- Fangmin, Z. Selection of Refercence Genes in Sweet Cherry. Master’s Thesis, Henan Agricultural University, Zhengzhou, China, 2014. [Google Scholar]
- Jianyang, W.; Bing, H.; Yujie, D.; Weicai, L.; Yongzan, W. Analysis Method of Systematically Evaluating Stability of Reference Genes Using geNorm, NormFinder and BestKeeper. Mod. Agric. Technol. 2017, 5, 278–281. [Google Scholar]
- Xiaozhe, Y.; Lan, W.; Li, X.; Mengyue, W.; Shilin, C.; Yuhua, S.; Xia, L. Screening of reference genes for quantitative real-time PCR in Artemisia argyi. China J. Chin. Mater. Medica 2022, 47, 659–667. [Google Scholar] [CrossRef]
- Huang, L.; Yan, H.; Jiang, X.; Zhang, Y.; Zhang, X.; Ji, Y.; Zeng, B.; Xu, B.; Yin, G.; Lee, S.; et al. Reference gene selection for quantitative real-time reverse-transcriptase PCR in orchardgrass subjected to various abiotic stresses. Gene 2014, 553, 158–165. [Google Scholar] [CrossRef]
- Lin, Y.; Zhang, C.; Lan, H.; Gao, S.; Liu, H.; Liu, J.; Cao, M.; Pan, G.; Rong, T.; Zhang, S. Validation of Potential Reference Genes for qPCR in Maize across Abiotic Stresses, Hormone Treatments, and Tissue Types. PLoS ONE 2014, 9, e95445. [Google Scholar] [CrossRef] [PubMed]
- Xiaojuan, S.; Baoguo, F.; Lichai, Y.; Xiena, C.; Shanfa, L. Selection and Validation of Reference Genes for Ouantitative RT-PCR Analvsis of Gene Expression in Populus trichocarpa. J. Integr. Plant Biol. 2013, 48, 507–518. [Google Scholar]
- Wenzhe, L.; Mingyue, N.; Xiuyun, L.; Erpei, L.; Huahong, H.; Zaikang, T. The Selection of Reference Genes for Quantitative PCR in Betula luminifera. Sci. Silvae Sin. 2016, 52, 29–37. [Google Scholar]
- Silveira, G.O.; Amaral, M.S.; Coelho, H.S.; Maciel, L.F.; Pereira, A.S.A.; Olberg, G.G.O.; Miyasato, P.A.; Nakano, E.; Verjovski-Almeida, S. Assessment of reference genes at six different developmental stages of Schistosoma mansoni for quantitative RT-PCR. Sci. Rep. 2021, 11, 16816. [Google Scholar] [CrossRef]
- Yin, H.; Yin, D.; Zhang, M.; Gao, Z.; Tuluhong, M.; Li, X.; Li, J.; Li, B.; Cui, G. Validation of Appropriate Reference Genes for qRT–PCR Normalization in Oat (Avena sativa L.) under UV-B and High-Light Stresses. Int. J. Mol. Sci. 2022, 23, 11187. [Google Scholar] [CrossRef]
- Mandal, A.; Mandal, S.; Park, M.H. Genome-Wide Analyses and Functional Classification of Proline Repeat-Rich Proteins: Potential Role of eIF5A in Eukaryotic Evolution. PLoS ONE 2014, 9, e111800. [Google Scholar] [CrossRef] [PubMed]
- Dever, T.E.; Gutierrez, E.; Shin, B.-S. The hypusine-containing translation factor eIF5A. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 413–425. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.-S.; Liu, L.-q.; Xu, C.; Zhao, Y.-h.; Zhu, X.-d.; Huang, S.-Z. Reference Gene Selection for Quantitative Real-Time RT-PCR Normalization in Iris. lactea var. chinensis Roots under Cadmium, Lead, and Salt Stress Conditions. Sci. World J. 2014, 2014, 532713. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-Y.; Chen, W.-Z.; Yang, S.-H.; Xu, C.-L.; Huang, X.; Chen, C.; Xie, H. Screening of reference genes in real-time PCR for Radopholus similis. PeerJ 2019, 7, e6253. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, T.T.; Nishimura, D.S.; De Nadai, F.B.; Figueira, A.; Latado, R.R. Selection of reference genes for expression analyses of red-fleshed sweet orange (Citrus sinensis). Genet. Mol. Res. 2015, 14, 18440–18451. [Google Scholar] [CrossRef]
- Roebroek, A.J.; van de Velde, H.J.; Van Bokhoven, A.; Broers, J.L.; Ramaekers, F.C.; Van de Ven, W.J. Cloning and expression of alternative transcripts of a novel neuroendocrine-specific gene and identification of its 135-kDa translational product. J. Biol. Chem. 1993, 268, 13439–13447. [Google Scholar] [CrossRef]
- Steiner, P.; Kulangara, K.; Sarria, J.C.F.; Glauser, L.; Regazzi, R.; Hirling, H. Reticulon 1-C/neuroendocrine-specific protein-C interacts with SNARE proteins. J. Neurochem. 2004, 89, 569–580. [Google Scholar] [CrossRef]
- Wakana, Y.; Koyama, S.; Nakajima, K.-i.; Hatsuzawa, K.; Nagahama, M.; Tani, K.; Hauri, H.-P.; Melançon, P.; Tagaya, M. Reticulon 3 is involved in membrane trafficking between the endoplasmic reticulum and Golgi. Biochem. Biophys. Res. Commun. 2005, 334, 1198–1205. [Google Scholar] [CrossRef]

| Category | Number | Gene | Gene Description | Homologous Sequence | Identities |
|---|---|---|---|---|---|
| Candidate traditional housekeeping genes | 1 | Actin7 | actin-7 | XM_038865290.1 | 96% |
| 2 | TuA | tubulin α-5 chain | XM_038840525.1 | 96% | |
| 3 | TuB | tubulin β chain-like | XM_038823732.1 | 95% | |
| 4 | eIF4A | eukaryotic initiation factor 4A-3 | XM_038845408.1 | 92% | |
| 5 | eIF5A | eukaryotic translation initiation factor 5A | XM_038856304.1 | 94% | |
| 6 | GAPDH | glyceraldehyde-3-phosphate dehydrogenase, cytosolic | XM_038838191.1 | 91% | |
| 7 | CYP72A | cytochrome P450 72A765 | MN738192.1 | 90% | |
| 8 | UBC2 | ubiquitin-conjugating enzyme E2 2 | XM_038844679.1 | 95% | |
| 9 | RAP2 | ethylene-responsive transcription factor RAP2-4 | XM_038860091.1 | 90% | |
| 10 | UBQ | polyubiquitin | XM_038834479.1 | 91% | |
| 11 | ClpA | ATP-dependent Clp protease ATP-binding subunit ClpA | XM_038830531.1 | 96% | |
| 12 | FtsH2 | ATP-dependent zinc metalloprotease FTSH 2 | XM_038853108.1 | 94% | |
| Candidate new RGs | 1 | RTNLB1 | reticulon-like protein B1 | XM_038853888.1 | 94% |
| 2 | UNC | uncharacterized | XM_038867966.1 | 91% | |
| 3 | GAD | glutamate decarboxylase | XM_038864250.1 | 92% | |
| 4 | ALEU | thiol protease aleurain-like | XM_038855394.1 | 92% | |
| 5 | BLH1 | BEL1-like homeodomain protein 1 | XM_038829031.1 | 88% | |
| 6 | SBT3.17 | subtilisin-like protease SBT3.17 | XM_038863645.1 | 92% | |
| 7 | HMGB2 | high mobility group B protein 2-like | XM_038849099.1 | 92% | |
| 8 | GEPI48 | UDP-glucose 4-epimerase GEPI48-like | XM_038869380.1 | 96% | |
| 9 | SCPL | serine carboxypeptidase-like | XM_038834759.1 | 94% |
| Category | Genes | Primer Sequence (5′-3′) Forward | Primer Sequence (5′-3′) Reverse | Product (bp *) | R2 * | E * (%) |
|---|---|---|---|---|---|---|
| Candidate traditional house-keeping genes | Actin7 | GTGTCATGGTTGGGATGGGT | ACGATACCGTGTTCGATGGG | 150 | 0.9975 | 103 |
| TUA | ACTGGCTTCAAGTGCGGTAT | GATCGATGCGTCCAAACACC | 133 | 0.9962 | 104 | |
| TUB | GCCGGACAATTTCGTCTTCG | TTCCTCACGACATCGAGCAC | 108 | 0.9988 | 104 | |
| eIF4A | CCGGCAAGACCTCCATGATT | TTCCCTCGTAGGCGACACTA | 95 | 1.0000 | 108 | |
| eIF5A | GAGGAGCACCACTTCGAGTC | GGGCGATTCTTGGTGACGAT | 130 | 0.9985 | 102 | |
| GAPDH | GGCTTGAGAAGGAGGCTACC | ACCAGCCTTGGCGTCAAATA | 158 | 0.9991 | 110 | |
| CYP72A | GACGCCGAGTGTGACGATAA | GGTCCATTTCTCGCCCTCAA | 142 | 0.9961 | 101 | |
| UBC2 | GCTCTGGAACGCTGTCATTT | CGAAGCGAACTGTAGGAGGC | 116 | 0.9995 | 109 | |
| RAP2 | GCTTTACCGAGGAGTCAGGC | CCTCGGCGGTATCAAAGGTA | 107 | 0.9921 | 105 | |
| UBQ | CCCTTGAAGTGGAAAGCAGTG | ATCAGCCAGAGTCCTTCCATC | 134 | 0.9992 | 104 | |
| ClpA | TGCTGGAACCAAGTACCGTG | TGCCCCAGCTCCAATTAAGG | 124 | 0.9990 | 105 | |
| FtsH2 | GGAGCTGATCTTGCCAACCT | CCCTCCATTCCAGCCACAAT | 122 | 0.9967 | 104 | |
| Candidate new RGs | RTNLB1 | CGGAGCATACTGGTGAGCAT | ATCGGAATCGGAAGACGACG | 104 | 0.9981 | 98 |
| UNC | TGGTACTTCGGGTTTGCAGC | TTGATGGCGTGCGAAGGTAT | 135 | 0.9906 | 104 | |
| GAD | TCAGTCCACTCCACTTTCGC | TTCCCGTCCAACATCAGCTC | 128 | 0.9988 | 110 | |
| ALEU | GTCGGCAACACTCATAACGC | ACAGCGAGCTTGTAAGGCAA | 164 | 0.9989 | 105 | |
| BLH1 | CCACCGCACTCCAACCTAAT | ATGAGTCCGTGCAAAGCAGA | 155 | 0.9933 | 104 | |
| SBT3.17 | GAGGTTGACGCAATCGTTGT | ATGTGGACCTTCGATTCGGG | 99 | 0.9994 | 103 | |
| HMGB2 | CCAAGGATCCGAACAAGCCT | CCAACAACGGCAACGGATTT | 115 | 0.9992 | 103 | |
| GEPI48 | CTGGCATTGGTTGTGAGGTG | TTCACGTTCTGCCTTGTCCG | 173 | 0.9942 | 105 | |
| SCPL | GTGGGCATTCCTGCTCTTCT | GACCAGACCACTTCATGGCA | 121 | 0.9968 | 103 |
| Category | Rank | Leaves under Different Temperature Stresses | Leaves of Plants at Different Growth Stages | Different Organs | |||
|---|---|---|---|---|---|---|---|
| Gene | Stability Value | Gene | Stability Value | Gene | Stability Value | ||
| Candidate traditional housekeeping genes | 1 | TUA | 0.25 | eIF5A | 0.135 | eIF5A | 0.308 |
| 2 | eIF5A | 0.335 | TUB | 0.2 | GAPDH | 0.438 | |
| 3 | CYP72A | 0.408 | GAPDH | 0.304 | ClpA | 0.573 | |
| 4 | UBC2 | 0.545 | FtsH2 | 0.4 | FtsH2 | 0.658 | |
| 5 | GAPDH | 0.603 | TUA | 0.411 | RAP2 | 0.745 | |
| 6 | ClpA | 0.635 | ClpA | 0.685 | TUA | 0.778 | |
| 7 | TUB | 0.642 | eIF4A | 0.689 | TUB | 0.796 | |
| 8 | eIF4A | 0.682 | CYP72A | 0.748 | Actin7 | 0.842 | |
| 9 | Actin7 | 1.071 | Actin7 | 0.778 | eIF4A | 0.924 | |
| 10 | UBQ | 1.231 | UBQ | 0.824 | UBQ | 1.019 | |
| 11 | FtsH2 | 1.389 | RAP2 | 0.943 | CYP72A | 1.245 | |
| 12 | RAP2 | 1.418 | UBC2 | 1.038 | UBC2 | 1.301 | |
| Candidate new RGs | 1 | SCPL | 0.102 | ALEU | 0.237 | HMGB2 | 0.508 |
| 2 | RTNLB1 | 0.217 | HMGB2 | 0.249 | RTNLB1 | 0.518 | |
| 3 | SBT3.17 | 0.292 | SCPL | 0.433 | UNC | 0.535 | |
| 4 | GEPI48 | 0.373 | RTNLB1 | 0.451 | GAD | 0.608 | |
| 5 | UNC | 0.467 | UNC | 0.507 | BLH1 | 0.703 | |
| 6 | GAD | 0.474 | GAD | 0.531 | SCPL | 0.743 | |
| 7 | BLH1 | 0.526 | BLH1 | 0.55 | SBT3.17 | 0.855 | |
| 8 | HMGB2 | 0.7 | SBT3.17 | 0.802 | GEPI48 | 0.945 | |
| 9 | ALEU | 1.004 | GEPI48 | 1.008 | ALEU | 1.996 | |
| Category | Rank | Leaves under Different Temperature Stresses | Leaves of Plants at Different Growth Stages | Different Organs | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | SD * | CV * | Gene | SD | CV | Gene | SD | CV | ||
| Candidate traditional house-keeping genes | 1 | eIF5A | 0.32 | 1.31 | TUB | 0.33 | 1.29 | Actin7 | 0.52 | 2.07 |
| 2 | UBC2 | 0.36 | 1.26 | GAPDH | 0.33 | 1.4 | eIF5A | 0.52 | 2.22 | |
| 3 | GAPDH | 0.36 | 1.37 | eIF5A | 0.34 | 1.53 | UBQ | 0.61 | 2.31 | |
| 4 | TUA | 0.45 | 1.61 | TUA | 0.43 | 1.68 | TUA | 0.67 | 2.59 | |
| 5 | UBQ | 0.61 | 2.65 | FtsH2 | 0.49 | 2.02 | TUB | 0.82 | 3.11 | |
| 6 | TUB | 0.67 | 2.35 | ClpA | 0.5 | 2.48 | GAPDH | 0.85 | 3.32 | |
| 7 | CYP72A | 0.75 | 2.77 | CYP72A | 0.53 | 2.21 | eIF4A | 0.91 | 3.49 | |
| 8 | ClpA | 0.8 | 3.7 | UBQ | 0.53 | 2.5 | FtsH2 | 1 | 3.56 | |
| 9 | RAP2 | 0.9 | 3.16 | eIF4A | 0.57 | 2.3 | ClpA | 1.09 | 4.63 | |
| 10 | eIF4A | 0.9 | 3.41 | Actin7 | 0.7 | 2.84 | RAP2 | 1.25 | 4.4 | |
| 11 | FtsH2 | 1.19 | 4.53 | RAP2 | 0.8 | 2.97 | UBC2 | 1.29 | 4.6 | |
| 12 | Actin7 | 1.24 | 4.89 | UBC2 | 0.81 | 3.05 | CYP72A | 1.44 | 5.3 | |
| Candidate new RGs | 1 | HMGB2 | 0.65 | 2.64 | HMGB2 | 0.38 | 1.63 | SCPL | 0.5 | 1.9 |
| 2 | SBT3.17 | 0.74 | 2.89 | RTNLB1 | 0.42 | 1.77 | SBT3.17 | 0.57 | 2.23 | |
| 3 | SCPL | 0.79 | 3.02 | ALEU | 0.42 | 1.81 | HMGB2 | 0.71 | 2.9 | |
| 4 | GAD | 0.84 | 3.27 | GAD | 0.46 | 1.97 | GAD | 0.77 | 2.89 | |
| 5 | RTNLB1 | 0.91 | 3.55 | UNC | 0.5 | 1.97 | RTNLB1 | 0.85 | 3.23 | |
| 6 | BLH1 | 0.92 | 3.69 | BLH1 | 0.56 | 2.39 | BLH1 | 0.88 | 3.31 | |
| 7 | GEPI48 | 1.06 | 4.16 | SCPL | 0.6 | 2.35 | UNC | 0.9 | 3.27 | |
| 8 | UNC | 1.11 | 4.11 | SBT3.17 | 0.69 | 2.78 | GEPI48 | 0.9 | 3.42 | |
| 9 | ALEU | 1.15 | 4.58 | GEPI48 | 0.73 | 3.01 | ALEU | 1.47 | 5.56 | |
| Category | Rank | Leaves under Different Temperature Stresses | Leaves of Plants at Different Growth Stages | Different Organs | |||
|---|---|---|---|---|---|---|---|
| Gene | Stability Value | Gene | Stability Value | Gene | Stability Value | ||
| Candidate traditional house-keeping genes | 1 | eIF5A | 1.41 | TUB | 1.41 | eIF5A | 1.68 |
| 2 | TUA | 2 | eIF5A | 1.97 | Actin7 | 2.63 | |
| 3 | UBC2 | 2.38 | TUA | 2.99 | TUA | 3.13 | |
| 4 | GAPDH | 3.87 | GAPDH | 3.35 | GAPDH | 3.31 | |
| 5 | CYP72A | 4.21 | FtsH2 | 4.95 | ClpA | 5.05 | |
| 6 | TUB | 6.48 | ClpA | 5.63 | TUB | 5.21 | |
| 7 | ClpA | 7.44 | eIF4A | 5.8 | FtsH2 | 6.16 | |
| 8 | eIF4A | 7.61 | CYP72A | 7.97 | UBQ | 6.51 | |
| 9 | UBQ | 8.61 | Actin7 | 8.97 | eIF4A | 7.94 | |
| 10 | Actin7 | 9.67 | UBQ | 9.69 | RAP2 | 7.95 | |
| 11 | FtsH2 | 10.74 | RAP2 | 10.74 | CYP72A | 11.24 | |
| 12 | RAP2 | 11.17 | UBC2 | 12 | UBC2 | 11.74 | |
| Candidate new RGs | 1 | SCPL | 1.86 | ALEU | 1.86 | HMGB2 | 1.73 |
| 2 | RTNLB1 | 2.78 | RTNLB1 | 2 | SCPL | 2.21 | |
| 3 | GEPI48 | 3.03 | HMGB2 | 2.34 | SBT3.17 | 3.15 | |
| 4 | SBT3.17 | 3.31 | GAD | 3.31 | RTNLB1 | 3.5 | |
| 5 | UNC | 3.76 | UNC | 4.16 | UNC | 3.6 | |
| 6 | HMGB2 | 4.76 | SCPL | 5.45 | GAD | 4.68 | |
| 7 | GAD | 5.42 | BLH1 | 6.48 | BLH1 | 5.96 | |
| 8 | BLH1 | 6.74 | SBT3.17 | 8 | GEPI48 | 8 | |
| 9 | ALEU | 9 | GEPI48 | 9 | ALEU | 9 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, W.; Yang, Y.; Ma, B.; Wang, W.; Hu, Z.; Leng, P. Selection and Validation of Reference Genes for Gene Expression Studies in Euonymus japonicus Based on RNA Sequencing. Genes 2024, 15, 131. https://doi.org/10.3390/genes15010131
Guo W, Yang Y, Ma B, Wang W, Hu Z, Leng P. Selection and Validation of Reference Genes for Gene Expression Studies in Euonymus japonicus Based on RNA Sequencing. Genes. 2024; 15(1):131. https://doi.org/10.3390/genes15010131
Chicago/Turabian StyleGuo, Wei, Yihui Yang, Bo Ma, Wenbo Wang, Zenghui Hu, and Pingsheng Leng. 2024. "Selection and Validation of Reference Genes for Gene Expression Studies in Euonymus japonicus Based on RNA Sequencing" Genes 15, no. 1: 131. https://doi.org/10.3390/genes15010131
APA StyleGuo, W., Yang, Y., Ma, B., Wang, W., Hu, Z., & Leng, P. (2024). Selection and Validation of Reference Genes for Gene Expression Studies in Euonymus japonicus Based on RNA Sequencing. Genes, 15(1), 131. https://doi.org/10.3390/genes15010131





