QTL Mapping of Trichome Traits and Analysis of Candidate Genes in Leaves of Wheat (Triticum aestivum L.)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.1.1. Test Material
2.1.2. Sample Collection and Preparation
2.2. Methods
2.2.1. Trichome Trait Measurement and Data Analysis
2.2.2. QTL Mapping
2.2.3. Prediction of Candidate Genes
3. Results
3.1. Distribution of Trichomes on Wheat Leaves
3.2. Phenotypic Characteristics of Trichome-Related Traits in the DH Lines and Parents
3.3. Correlations of Trichome Traits in Wheat Leaves
3.4. QTL for Trichome Density in Wheat Leaves
3.5. QTL for Trichome Length in Wheat Leaves
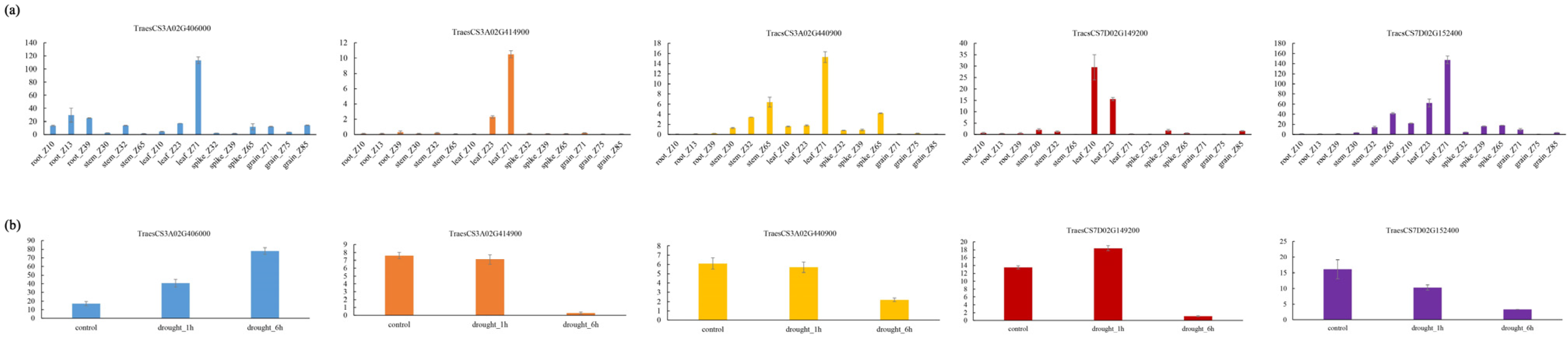
3.6. The Prediction of Candidate Genes
4. Discussion
4.1. Genetic Effects of Trichome-Related Traits
4.2. Linkage and Pleiotroty of QTL Related to Trichome Traits in Wheat
4.3. Prediction of Candidate Genes Associated with Trichome Traits
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Balkunde, R.; Pesch, M.; Hülskamp, M. Trichome patterning in Arabidopsis thaliana from genetic to molecular models. Curr. Top. Dev. Biol. 2010, 91, 299–321. [Google Scholar] [PubMed]
- Gao, Y.; Guo, J.Q.; Zhao, J.F. Molecular Mechanisms of Arabidopsis Trichome Development. Chin. Bull. Bot. 2011, 46, 119–127. [Google Scholar]
- Andrade, M.C.; Da Silva, A.A.; Neiva, I.P.; Oliveira, I.R.C.; De Castro, E.M.; Francis, D.M.; Maluf, W.R. Inheritance of type IV glandular trichome density and its association with whitefly resistance from Solanum galapagense accession LA1401. Euphytica 2017, 213, 52. [Google Scholar] [CrossRef]
- Houten, Y.M.; Glas, J.J.; Hoogerbrugge, H.; Rothe, J.; Bolckmans, K.J.F.; Simoni, S.; van Arkel, J.; Alba, J.M.; Kant, M.R.; Sabelis, M.W. Herbivory-associated degradation of tomato trichomes and its impact on biological control of Aculops lycopersici. Exp. Appl. Acarol. 2013, 60, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Kariyat, R.R.; Smith, J.D.; Stephenson, A.G.; De Moraes, C.M.; Mescher, M.C. Non-glandular trichomes of Solanum carolinense deter feeding by Manduca sexta caterpillars and cause damage to the gut peritrophic matrix. Proc. R. Soc. B 2017, 284, 20162323. [Google Scholar] [CrossRef] [PubMed]
- Steede, W.T.; Ma, J.M.; Eickholt, D.P.; Drake-Stowe, K.E.; Kernodle, S.P.; Shew, D.; Danehower, D.A.; Lewis, R.S. The tobacco trichome exudate Z-abienol and its relationship with plant resistance to Phytophthora nicotianae. Plant Dis. 2017, 101, 1214–1221. [Google Scholar] [CrossRef] [PubMed]
- Vendemiatti, E.; Zsögön, A.; Silva, G.F.F.E.; De Jesus, F.A.; Cutri, L.; Figueiredo, C.R.F.; Tanaka, F.A.O.; Nogueira, F.T.S.; Peres, L.E.P. Loss of type-IV glandular trichomes is a heterochronic trait in tomato and can be reverted by promoting juvenility. Plant Sci. 2017, 259, 35–47. [Google Scholar] [CrossRef]
- Mcdowell, E.T.; Kapteyn, J.; Schmidt, A.; Li, C.; Kang, J.H.; Descour, A.; Shi, F.; Larson, M.; Schilmiller, A.; An, L.L.; et al. Comparative functional genomic analysis of Solanum glandular trichome types. Plant Physiol. 2011, 155, 524–539. [Google Scholar] [CrossRef]
- Ma, Y.L.; Wang, L.; Liu, Y.-X.; Lan, H.-Y. Uptates on Stress Tolerance of Main Accessory Structures and Their Synergetic Interaction in Desert Plants. Plant Physiol. J. 2015, 51, 1821–1836. [Google Scholar]
- Hegebarth, D.; Buschhaus, C.; Wu, M.; Bird, D.; Jetter, R. The composition of surface wax on trichomes of Arabidopsis thaliana differs from wax on other epidermal cells. Plant J. 2016, 88, 762–774. [Google Scholar] [CrossRef]
- Rakha, M.; Bouba, N.; Ramasamy, S.; Regnard, J.L.; Hanson, P. Evaluation of wild tomato accessions (Solanum spp.) for resistance to two-spotted spider mite (Tetranychus urticae Koch) based on trichome type and acylsugar content. Genet Resour. Crop Evol. 2017, 64, 1011–1022. [Google Scholar] [CrossRef]
- Hu, B.; Wan, Y.; Li, X.; Zhang, F.; Yan, W.; Xie, J. Phenotypic characterization and genetic analysis of rice with pubescent leaves and glabrous hulls. Crop Sci. 2013, 53, 1878–1886. [Google Scholar] [CrossRef]
- Hendrick, M.F.; Finseth, F.R.; Mathiasson, M.E.; Palmer, K.A.; Broder, E.M.; Breigenzer, P.; Fishman, L. The genetics of extreme microgeographic adaptation: An integrated approach identifies a major gene underlying leaf trichome divergence in Yellowstone Mimulus guttatus. Mol. Ecol. 2016, 25, 5647–5662. [Google Scholar] [CrossRef] [PubMed]
- Mazie, A.R.; Baum, D.A. Clade-specific positive selection on a developmental gene: BRANCHLESS TRICHOME and the evolution of stellate trichomes in Physaria (Brassicaceae). Mol. Phylogenet. Evol. 2016, 100, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Ning, P.B.; Wang, J.H.; Zhou, Y.L.; Gao, L.F.; Wang, J.; Gong, C.M. Adaptional evolution of trichome in Caragana korshinskii to natural drought stress on the Loess Plateau, China. Ecol. Evol. 2016, 6, 3786–3795. [Google Scholar] [CrossRef] [PubMed]
- Oppenheimer, D.G.; Herman, P.L.; Sivakumaran, S.; Esch, J.; Marks, M.D. A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 1991, 67, 483–493. [Google Scholar] [CrossRef] [PubMed]
- Alahakoon, U.I.; Taheri, A.; Nayidu, N.K.; Epp, D.; Yu, M.; Parkin, I.; Hegedus, D.; Bonham-Smith, P.; Gruber, M.Y. Hairy canola (Brasssica napus) re-visited: Down-regulating TTG1 in an AtGL3-enhanced hairy leaf background improves growth, leaf trichome coverage, and metabolite gene expression diversity. BMC Plant Biol. 2016, 16, 12. [Google Scholar] [CrossRef] [PubMed]
- Ioannidi, E.; Rigas, S.; Tsitsekian, D.; Daras, G.; Alatzas, A.; Makris, A.; Tanou, G.; Argiriou, A.; Alexandrou, D.; Poethig, S.; et al. Trichome patterning control involves TTG1 interaction with SPL transcription factors. Plant Mol. Biol. 2016, 92, 675–687. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Q.; Shan, X.T.; Gao, R.F.; Yang, S.; Wang, S.C.; Gao, X.; Wang, L. Two IIIf clade-bHLHs from freesia hybrida play divergent roles in flavonoid biosynthesis and trichome formation when ectopically expressed in Arabidopsis. Sci. Rep. 2016, 6, 30514. [Google Scholar] [CrossRef]
- Gao, C.H.; Li, D.; Jin, C.Y.; Duan, S.W.; Qi, S.H.; Liu, K.G.; Wang, H.C.; Ma, H.L.; Hai, J.B.; Chen, M.X. Genome-wide identification of GLABRA3 downstream genes for anthocyanin biosynthesis and trichome formation in Arabidopsis. Biochem. Biophys. Res. Commun. 2017, 485, 360–365. [Google Scholar] [CrossRef]
- Dai, X.M.; Zhou, L.M.; Zhang, W.; Cai, L.; Guo, H.Y.; Tian, H.N.; Schiefelbein, J.; Wang, S.C. A single amino acid substitution in the R3 domain of GLABRA1 leads to inhibition of trichome formation in Arabidopsis without affecting its interaction with GLABRA3. Plant Cell Environ. 2016, 39, 897–907. [Google Scholar] [CrossRef]
- Johnson, C.S.; Kolevski, B.; Smyth, D.R. TRANSPARENT TESTA GLABRA 2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 2002, 14, 1359–1375. [Google Scholar] [CrossRef]
- Zheng, K.J.; Tian, H.N.; Hu, Q.N.; Guo, H.Y.; Yang, L.; Cai, L.; Wang, X.T.; Liu, B.; Wang, S.C. Ectopic expression of R3 MYB transcription factor gene OsTCL1 in Arabidopsis, but not rice, affects trichome and root hair formation. Sci. Rep. 2016, 6, 19254. [Google Scholar] [CrossRef]
- Schellmann, S.; Schnittger, A.; Kirik, V.; Wada, T.; Okada, K.; Beermann, A.; Thumfahrt, J.; Jürgens, G.; Hülskamp, M. TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J. 2002, 21, 5036–5046. [Google Scholar] [CrossRef]
- Wada, T.; Tominaga-Wada, R. CAPRICE family genes control flowering time through both promoting and repressing CONSTANS and FLOWERING LOCUS T expression. Plant Sci. 2015, 241, 260–265. [Google Scholar] [CrossRef]
- Vernoud, V.; Laigle, G.; Rozier, F.; Meeley, R.B.; Perez, P.; Rogowsky, P.M. The HD-ZIP IV transcription factor OCL4 is necessary for trichome patterning and anther development in maize. Plant J. 2009, 59, 883–894. [Google Scholar] [CrossRef]
- Li, J.; Yuan, Y.; Lu, Z.; Yang, L.; Gao, R.; Lu, J.; Li, J.; Xiong, G. Glabrous Rice 1, encoding a homeodomain protein, regulates trichome development in rice. Rice 2012, 5, 32. [Google Scholar] [CrossRef]
- Li, C. Cloning and Function Analysis of Glabrous Leaf Gene GLR3, GL5 and Fine Mapping of Hairy Leaf Gene HL6 in Rice; China Agricultural University: Beijing, China, 2016. [Google Scholar]
- Zhang, H.; Wu, K.; Wang, Y.; Peng, Y.; Hu, F.; Wen, L.; Han, B.; Qian, Q.; Teng, S. A WUSCHEL-like homeobox gene, OsWOX3B responses to NUDA/GL-1 locus in rice. Rice 2012, 5, 30. [Google Scholar] [CrossRef]
- Angeles-Shim, R.B.; Asano, K.; Takashi, T.; Shim, J.; Kuroha, T.; Ayano, M.; Ashikari, M. A WUSCHEL-related homeobox 3B gene, Depilous(dep), confers glabrousness of rice leaves and glumes. Rice 2012, 5, 28–30. [Google Scholar] [CrossRef]
- Sun, W.; Gao, D.; Xiong, Y.; Tang, X.; Xiao, X.; Wang, C.; Yu, S. Hairy Leaf 6, an AP2/ERF transcription factor, interacts with OsWOX3B and regulates trichome formation in rice. Mol. Plant 2017, 10, 1417–1433. [Google Scholar] [CrossRef]
- Zeng, Y.; Zhu, Y.; Lian, L.; Xie, H.; Zhang, J.; Xie, H. Genetic analysis and fine mapping of the pubescence gene GL6 in rice (Oryza sativa L.). Chin. Sci. Bull. 2013, 58, 2992–2999. [Google Scholar] [CrossRef]
- Xie, Y.; Yu, X.; Jiang, S.; Xiao, K.; Wang, Y.; Li, L.; Wang, F.; He, W.; Cai, Q.; Xie, H.; et al. OsGL6, a conserved AP2 domain protein, promotes leaf trichome initiation in rice. Biochem. Biophys. Res. Commun. 2020, 522, 448–455. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, W.; Qin, P.; Huang, Y.; Ma, B.; Ouyang, X.; Chen, X.; Li, S. Characterization and fine mapping of GLABROUS RICE 2 in rice. J. Genet. Genom. 2013, 40, 579–582. [Google Scholar] [CrossRef] [PubMed]
- Song, H.-B.; Wang, B.; Chen, R.-J.; Zheng, X.-Y.; Yu, S.-B.; Lan, T. Genetic analysis and gene mapping of the glabrous leaf and hull mutant glr3 in rice (Oryza sativa L.). Hereditas 2016, 38, 1011–1018. [Google Scholar]
- Lan, T.; Zheng, Y.L.; Su, Z.L.; Yu, S.; Song, H.; Zheng, X.; Lin, G.; Wu, W. OsSPL10, a SBP-Box gene, plays a dual role in salt tolerance and trichome formation in rice (Oryza sativa L.). G3-Genes Genomes Genet. 2019, 9, 4107–4114. [Google Scholar] [CrossRef]
- Li, W.; Wu, J.; Weng, S.; Zhang, D.; Zhang, Y.; Shi, C. Characterization and fine mapping of the glabrous leaf and hull mutants (gl1) in rice (Oryza sativa L.). Plant Cell Rep. 2010, 29, 617–627. [Google Scholar] [CrossRef]
- Hong, J.; Wang, Q.Z.; Fu, H.W.; Wu, D.-x.; Shu, Q.y. Fine mapping and candidate gene analysis of glabrous leaf and hull gene (gl1) in rice (Oryza sativa L.). J. Nucl. Agric. Sci. 2011, 25, 1088–1093+1190. [Google Scholar]
- DongChen, W.H.; Zhang, X.L.; Zhu, Q. Cloning and subcellular localization of a new glabrous-leaf mutant gene GLL in rice (Oryza sativa L.). Mol. Plant Breed. 2015, 13, 716–726. [Google Scholar]
- Li, Q.; Cao, C.X.; Zhang, C.J.; Zheng, S.S.; Wang, Z.H.; Wang, L.N.; Ren, Z.H. The identification of Cucumis sativus Glabrous 1 (CsGL1) required for the formation of trichomes uncovers a novel function for the homeodomain-leucine zipper I gene. J. Exp. Bot. 2015, 66, 2515–2526. [Google Scholar] [CrossRef]
- Ma, D.; Hu, Y.; Yang, C.Q.; Liu, B.L.; Fang, L.; Wan, Q.; Liang, W.H.; Mei, G.F.; Wang, L.J.; Wang, H.P.; et al. Genetic basis for glandular trichome formation in cotton. Nat. Commun. 2016, 7, 10456. [Google Scholar] [CrossRef]
- Wang, Y.L.; Nie, J.T.; Chen, H.M.; Guo, C.L.; Pan, J.; He, H.L.; Pan, J.S.; Cai, R. Identification and mapping of Tril, a homeodomain-leucine zipper gene involved in multicellular trichome initiation in Cucumis sativus. Theor. Appl. Genet. 2016, 129, 305–316. [Google Scholar] [CrossRef] [PubMed]
- Guan, X.Y.; Pang, M.X.; Nah, G.; Shi, X.L.; Ye, W.X.; Stelly, D.M.; Chen, Z.J. miRNA and miRNA regulate homoeologous MYB2 gene functions in Arabidopsis trichome and cotton fibre development. Nat. Commun. 2014, 5, 3050. [Google Scholar] [CrossRef] [PubMed]
- Du, W.J.; Yu, D.Y.; Fu, S.X. Analysis of QTLs the trichome density on the upper and downer surface of leaf blade in soybean [Glycine max (L.) Merr.]. Agric. Sci. China 2009, 8, 529–537. [Google Scholar] [CrossRef]
- Xing, G.-N.; Liu, Z.-X.-N.; Tan, L.-M.; Yue, H.; Wang, Y.-F.; Kim, H.-J.; Zhao, T.-J.; Gai, J.-Y. QTL Mapping of Pubescence Density and Length on Leaf Surface of Soybean. Acta Agron. Sin. 2013, 39, 12–20. [Google Scholar] [CrossRef]
- Ringlund, K.; Everson, E.H. Leaf pubescence in common wheat, Triticum aestivum L.; and resistance to the cereal leaf beetle, Oulema melanopus (L.). Crop Sci. 1968, 8, 705–710. [Google Scholar] [CrossRef]
- Maystrenko, O.I. Identification and location of genes controlling leaf hairing in young plants of common wheat. Genetika 1976, 12, 5–15. [Google Scholar]
- Arbuzova, V.S.; Efremova, T.T.; Laikova, L.I.; Maystrenko, O.I.; Popova, O.M.; Pshenichnikova, T.A. The development of precise genetic stocks in two wheat cultivars and their use in genetic analysis. Euphytica 1996, 89, 11–15. [Google Scholar] [CrossRef]
- Taketa, S.; Chang, C.L.; Ishii, M.; Takeda, K. Chromosome arm location of the gene controlling leaf pubescence of a Chinese local wheat cultivar ‘Hong-mang-mai’. Euphytica 2002, 125, 141–147. [Google Scholar] [CrossRef]
- Maystrenko, O.I. Using of cytogenetic methods in studies of common wheat ontogenesis. In Ontogenetics of Higher Plants; Shtiitsa: Kishinev, Moldova, 1992; pp. 98–114. [Google Scholar]
- Blanco, A.; Bellomo, M.P.; Cenci, A.; De Giovanni, C.; D’Ovidio, R.; Iacono, E.; Laddomada, B.; Pagnotta, M.A.; Porceddu, E.; Sciancalepore, A.; et al. A genetic linkage map of durum wheat. Theor. Appl. Genet. 1998, 97, 721–728. [Google Scholar] [CrossRef]
- Khestkina, E.K.; Pestsovo, E.G.; Salina, E.; Röder, M.S. Molecular mapping and tagging of wheat genes using RAPD, STS and SSR markers. Cell. Mol. Biol. Lett. 2002, 7, 795–802. [Google Scholar]
- Love, H.H.; Craig, W.T. The inheritance of pubescent nodes in a cross between two varieties of wheat. J. Agric. Res. 1924, 28, 841–844. [Google Scholar]
- Gaines, E.F.; Carstens, A. The linkage of pubescent node and beard factors as evidenced by a cross between two varieties of wheat. J. Agric. Res. 1926, 33, 753–755. [Google Scholar]
- Sourdille, P.; Cadalen, T.; Gay, G.; Gill, B.; Bernard, M. Molecular and physical mapping of genes affecting awning in wheat. Plant Breed. 2002, 121, 320–324. [Google Scholar] [CrossRef]
- Wan, H.; Yang, Y.; Li, J.; Zhang, Z.; Yang, W. Mapping a major QTL for hairy leafsheath introgressed from Aegilops tauschii and its association with enhanced grain yield in bread wheat. Euphytica 2015, 205, 275–285. [Google Scholar] [CrossRef]
- Luo, W.; Liu, J.; Ding, P.; Li, C.; Liu, H.; Mu, Y.; Tang, H.; Jiang, Q.; Liu, Y.; Chen, G.; et al. Transcriptome analysis of near-isogenic lines for glume hairiness of wheat. Gene 2020, 739, 144517. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zheng, Z.; Luo, W.; Zhou, H.; Ying, Z.; Liu, C. Detection of a major QTL conditioning trichome length and density on chromosome arm 4BL and development of near isogenic lines targeting this locus in bread wheat. Mol. Breed. 2021, 41, 10. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Guan, W.; Shi, Y.; Wang, S.; Fan, H.; Yang, J.; Chen, W.; Zhang, W.; Sun, D.; Jing, R. QTL mapping and candidate gene analysis of seed vigor-related traits during artificial aging in wheat (Triticum aestivum). Sci. Rep. 2020, 10, 22060. [Google Scholar] [CrossRef]
- Li, L.; Peng, Z.; Mao, X.; Wang, J.; Li, C.; Chang, X.; Jing, R. Genetic insights into natural variation underlying salt tolerance in wheat. J. Exp. Bot. 2021, 72, 1135–1150. [Google Scholar] [CrossRef]
- Ma, S.W.; Wang, M.; Wu, J.H.; Guo, W.L.; Chen, Y.M.; Li, G.W.; Wang, Y.P.; Shi, W.M.; Xia, G.M.; Fu, D.L.; et al. WheatOmics: A platform combining multiple omics data to accelerate functional genomics studies in wheat. Mol. Plant 2021, 14, 1965–1968. [Google Scholar] [CrossRef]
- Tanksley, S.; Nelson, D.; Nelson, J.C. Advanced backcross QTL analysis: A method for the simultaneous discovery and transfer of valuable QTLs from unadaoted germlasm into elite breeding lines. Theor. Appl. Genet. 1996, 92, 191–203. [Google Scholar] [CrossRef]
- Fracheboud, Y.; Ribaut, J.M.; Vargas, M.; Messmer, R.; Stamp, P. Identification of quantitative trait loci for cold-tolerance of photosynthesis in maize (Zea mays L.). J. Exp. Bot. 2002, 53, 1967–1977. [Google Scholar] [CrossRef] [PubMed]
- Tuberosa, R.; Salvi, S.; Sanguineti, M.C.; Landi, P.; Maccaferri, M.; Conti, S. Mapping QTLs regulating morpho-physiological traits and yield: Case studies, shortcomings and perspectives in drought-stressed maize. Ann. Bot. 2002, 89, 941–963. [Google Scholar] [CrossRef] [PubMed]
- Moncada, P.; Martínez, C.; Borrero, J.; Chatel, M.; Gauch, H., Jr.; Guimaraes, E.; Tohme, J.; McCouch, S.R. Quantitative trait loci for yield and yield components in an Oryza sativa×Oryza rufipogon BC2F2 population evaluated in an upland environment. Theor. Appl. Genet. 2001, 102, 41–52. [Google Scholar] [CrossRef]
- Wen, T.; Wu, M.; Shen, C.; Gao, B.; Zhu, D.; Zhang, X.; You, C.; Lin, Z. Linkage and association mapping reveals the genetic basis of brown fibre (Gossypium hirsutum). Plant Biotechnol. J. 2018, 16, 1654–1666. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Koizuka, N.; Martin, R.C.; Nonogaki, H. The BME3 (Blue Micropylar end 3) GATA zinc finger transcription factor is a positive regulator of Arabidopsis seed germination. Plant J. 2005, 44, 960–971. [Google Scholar] [CrossRef] [PubMed]
- Mao, H.; Li, S.; Chen, B.; Jian, C.; Mei, F.; Zhang, Y.; Li, F.; Chen, N.; Li, T.; Du, L.; et al. Variation in cis-regulation of a NAC transcription factor contributes to drought tolerance in wheat. Mol. Plant 2022, 15, 276–292. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tian, H.; Chen, D.; Zhang, H.; Sun, M.; Chen, S.; Qin, Z.; Ding, Z.; Dai, S. Cysteine-rich receptor-like protein kinases: Emerging regulators of plant stress responses. Trends Plant Sci. 2023, 28, 776–794. [Google Scholar] [CrossRef] [PubMed]
- Arellano-Villagómez, F.C.; Guevara-Olvera, L.; Zuñiga-Mayo, V.M.; Cerbantez-Bueno, V.E.; Verdugo-Perales, M.; Medina, H.R.; De Folter, S.; Acosta-García, G. Arabidopsis cysteine-rich receptor-like protein kinase CRK33 affects stomatal density and drought tolerance. Plant Signal. Behav. 2021, 16, 1905335. [Google Scholar] [CrossRef]
- Mousley, C.J.; Tyeryar, K.R.; Ryan, M.M.; Bankaitis, V.A. Sec14p-like proteins regulate phosphoinositide homoeostasis and intracellular protein and lipid trafficking in yeast. Biochem. Soc. Trans. 2006, 34 Pt 3, 346–350. [Google Scholar] [CrossRef]
- Meng, Q.; Kim, S.J.; Costa, M.A.; Moinuddin, S.G.A.; Celoy, R.M.; Smith, C.A.; Cort, J.R.; Davin, L.B.; Lewis, N.G. Dirigent protein subfamily function and structure in terrestrial plant phenol metabolism. Methods Enzymol. 2023, 683, 101–150. [Google Scholar]
- Handy, D.E.; Loscalzo, J. The role of glutathione peroxidase-1 in health and disease. Free Radic Biol. Med. 2022, 188, 146–161. [Google Scholar] [CrossRef]
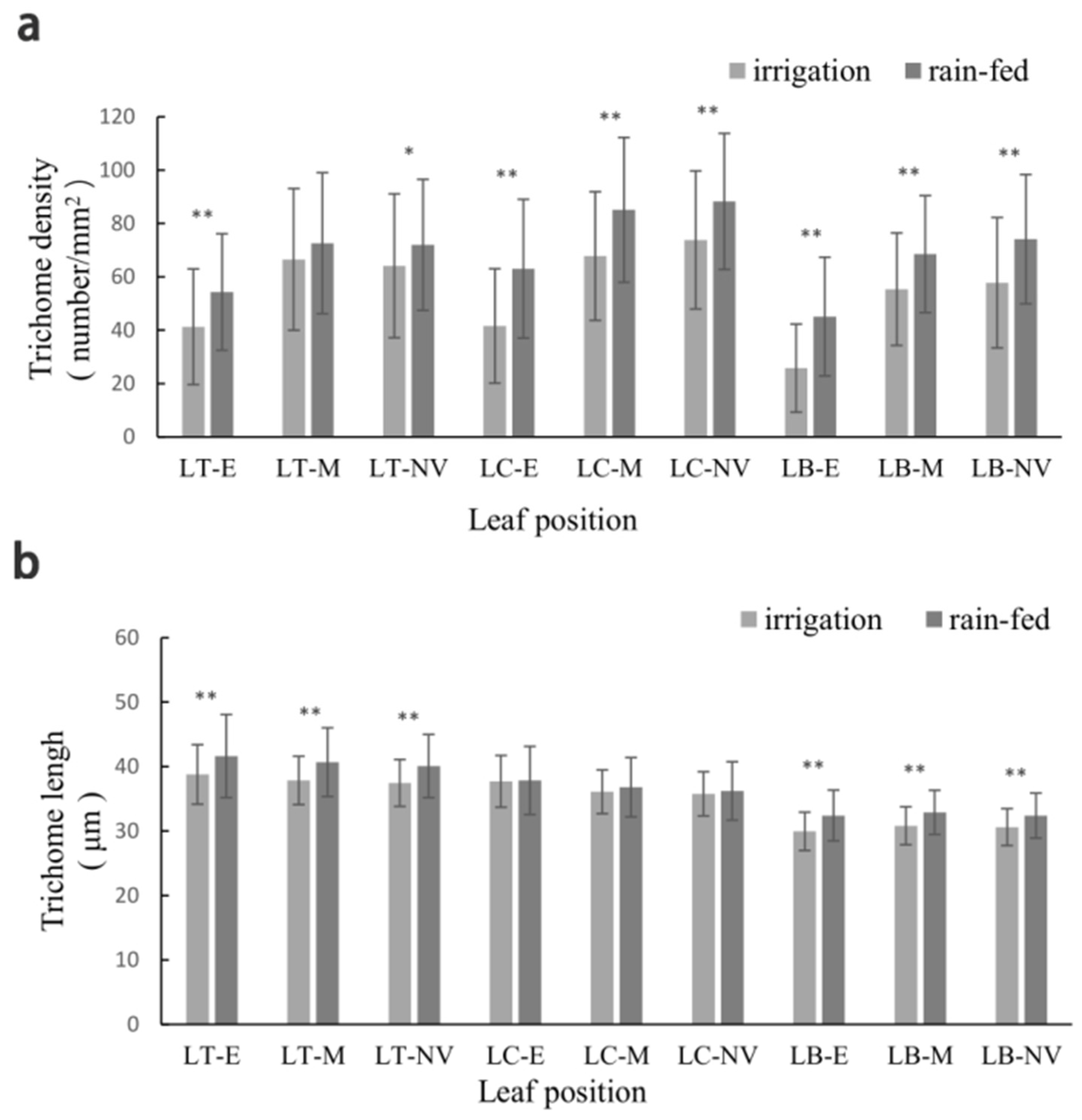


| Part | TD | TL | ||
|---|---|---|---|---|
| Irrigation | Rain-Fed | Irrigation | Rain-Fed | |
| LT | 57.076 Aa | 61.745 Bb | 37.967 Aa | 40.754 ** Aa |
| LC | 60.813 Aa | 72.973 ** Aa | 36.49 2 Bb | 36.843 Bb |
| LB | 46.218 Bb | 55.680 ** Bb | 30.276 Cc | 32.634 ** Cc |
| E | 35.785 Bb | 50.231 ** Bb | 35.380 Aa | 37.264 ** Aa |
| M | 62.535 Aa | 70.075 ** Aa | 34.816 Aab | 36.717 ** Aab |
| NV | 64.930 Aa | 72.653 ** Aa | 34.540 Ab | 36.257 ** Ab |
| QTL | Environment | Part | Positon (1) | Flanking Marker | LOD (2) | PVE (100%) (3) | Add (4) |
|---|---|---|---|---|---|---|---|
| Qtd-2A-1 | Rain-fed | LT-E | 38 | AX-110366793-AX-111505145 | 9.75 | 17.94 | −10.09 |
| LT-NV | 5.45 | 14.97 | −9.63 | ||||
| Qtd-2A-2 | Irrigation | LT-E | 29 | Xgwm275-Xwmc51 | 7.58 | 15.66 | 9.36 |
| Qtd-2A-3 | Irrigation | LC-E | 34 | AX-94695451-AX-95104154 | 8.84 | 14.929 | −11.36 |
| LC-M | 15.65 | 21.83 | −16.71 | ||||
| LC-NV | 22.32 | 27.993 | −17.94 | ||||
| Qtd-2A-4 | Irrigation | LC-E | 101 | AX-111172526-AX-111745616 | 4.84 | 7.689 | 8.15 |
| Qtd-2A-5 | Irrigation | LC-M | 20 | Xcwm138.2-AX-110686688 | 5.64 | 6.92 | 9.52 |
| Qtd-2A-6 | Irrigation | LC-M | 115 | AX-109532624-AX-111046111 | 3.85 | 4.43 | 7.54 |
| Qtd-2A-7 | Irrigation | LC-NV | 28 | Xgwm425-Xpsp3088 | 8.79 | 9.923 | 10.68 |
| Qtd-3A-1 | Rain-fed | LT-M | 96 | AX-95653062-AX-95235020 | 4.69 | 12.37 | −8.94 |
| LB-E | 99 | 2.98 | 9.06 | −5.05 | |||
| LB-M | 3.30 | 9.24 | −6.43 | ||||
| Irrigation | LT-NV | 96 | 5.10 | 11.93 | −8.25 | ||
| Qtd-3A-2 | Rain-fed | LT-NV | 91 | AX-95207768-AX-95679334 | 6.18 | 16.67 | −10.22 |
| Irrigation | LT-E | 92 | 2.90 | 5.34 | −5.52 | ||
| LC-NV | 5.12 | 4.69 | −7.40 | ||||
| Qtd-3A-3 | Rain-fed | LC-E | 90 | AX-95232910-AX-95207768 | 4.31 | 9.20 | −6.55 |
| LB-NV | 4.427 | 10.12 | −8.19 | ||||
| Irrigation | LB-M | 89 | 4.73 | 11.17 | −8.16 | ||
| Qtd-3A-4 | Rain-fed | LC-M | 100 | AX-95235020-AX-95658735 | 4.09 | 10.15 | −6.67 |
| LC-NV | 3.76 | 10.39 | −7.78 | ||||
| Irrigation | LT-M | 4.97 | 10.37 | −8.26 | |||
| Qtd-3A-5 | Irrigation | LB-E | 83 | AX-110617585-AX-95072294 | 4.92 | 11.68 | −8.07 |
| Qtd-5A-1 | Irrigation | LT-M | 0 | AX-109541070-AX-95630232 | 3.65 | 7.29 | −6.95 |
| Qtd-5A-2 | Irrigation | LT-NV | 20 | AX-95659236-AX-109921026 | 3.07 | 6.89 | −6.37 |
| Qtd-6A-1 | Rain-fed | LB-M | 63 | AX-110543147-Xpsp3071 | 2.57 | 7.34 | 5.90 |
| Qtd-6A-2 | Irrigation | LC-E | 33 | Xcwm162-AX-111619297 | 5.39 | 8.98 | −9.28 |
| Qtd-2B-1 | Rain-fed | LC-NV | 3 | Xcwm529-AX-94481482 | 4.09 | 11.28 | 8.37 |
| Qtd-2B-2 | Irrigation | LT-M | 2 | AX-110555744-Xcwm529 | 2.66 | 5.86 | 6.39 |
| Qtd-3B | Rain-fed | LT-E | 47 | AX-108735878-Xwmc231 | 4.93 | 9.56 | −7.359 |
| Qtd-5B-1 | Rain-fed | LB-NV | 99 | Xgwm408-Xgwm604 | 2.577 | 5.97 | 6.27 |
| Qtd-5B-2 | Irrigation | LC-NV | 133 | AX-110945396-AX-109581522 | 2.65 | 2.41 | 5.30 |
| Qtd-5B-3 | Irrigation | LB-NV | 249 | AX-109455033-AX-108853192 | 2.76 | 8.60 | 7.08 |
| Qtd-6B | Rain-fed | LT-E | 75 | EST138.1-Xwmc341 | 3.18 | 5.19 | 6.559 |
| Qtd-7B | Rain-fed | LT-E | 66 | Xwmc269.1-Xgwm297 | 4.63 | 7.92 | 6.70 |
| Qtd-1D | Irrigation | LC-NV | 86 | Xgwm337-AX-110521547 | 3.19 | 3.223 | −6.08 |
| Qtd-2D-1 | Rain-fed | LC-E | 95 | AX-109879970-AX-111066402 | 6.93 | 15.24 | −8.42 |
| Qtd-2D-2 | Rain-fed | LB-NV | 89 | AX-110744761-Xwmc453.1 | 4.47 | 10.64 | −8.45 |
| Qtd-2D-3 | Irrigation | LB-M | 116 | AX-94735076-AX-111914966 | 3.53 | 8.13 | −6.94 |
| Qtd-3D | Rain-fed | LC-M | 216 | AX-109367171-AX-111568509 | 3.09 | 7.55 | 5.76 |
| LC-NV | 3.51 | 9.60 | 7.49 | ||||
| Qtd-4D-1 | Irrigation | LC-E | 5 | AX-111475478-AX-89654830 | 4.94 | 7.86 | −8.25 |
| LC-M | 7 | 5.77 | 6.913 | −9.39 | |||
| LC-NV | 8 | 6.67 | 6.42 | −8.58 | |||
| Qtd-4D-2 | Irrigation | LB-E | 2 | AX-95659047-AX-109516054 | 4.27 | 9.99 | −7.47 |
| Qtd-6D | Rain-fed | LT-E | 64 | AX-108849732-AX-109779203 | 2.60 | 4.07 | 4.80 |
| Qtd-7D-1 | Rain-fed | LT-M | 108 | AX-111529990-AX-95631292 | 2.57 | 6.24 | 6.35 |
| Qtd-7D-2 | Rain-fed | LC-M | 101 | AX-95014724-AX-109507404 | 5.31 | 13.49 | 7.77 |
| QTL | Environment | Part | Position (1) | Flanking Marker | LOD (2) | PVE (100%) (3) | Add (4) |
|---|---|---|---|---|---|---|---|
| Qtl-1A | Rain-fed | LC-E | 73 | Xcwm517-Xcwm516 | 2.50 | 6.03 | −1.07 |
| Qtl-2A-1 | Rain-fed | LT-E | 45 | AX-94738148-Xgwm328 | 4.48 | 12.54 | 1.588 |
| LT-M | 46 | 4.66 | 12.23 | 1.29 | |||
| Qtl-2A-2 | Rain-fed | LT-NV | 37 | AX-95684521-AX-111808444 | 3.18 | 8.53 | 1.05 |
| Qtl-2A-3 | Irrigation | LT-E | 43 | AX-94664242-AX-94738148 | 7.53 | 18.46 | 2.83 |
| LT-M | 44 | 3.56 | 9.53 | 1.65 | |||
| Qtl-2A-4 | Irrigation | LT-NV | 31 | Xgwm249-Xwmc63 | 2.78 | 7.21 | 1.29 |
| Qtl-2A-5 | Irrigation | LB-E | 95 | AX-110402534-AX-94450655 | 2.57 | 6.66 | 1.04 |
| Qtl-4A | Rain-fed | LC-M | 53 | AX-95145339-Xgwm610 | 3.68 | 8.09 | 1.00 |
| Qtl-1B-1 | Rain-fed | LT-E | 118 | Xwmc44-AX-108745931 | 2.95 | 8.16 | −1.28 |
| Qtl-1B-2 | Rain-fed | LC-NV | 42 | AX-94658630-AX-94935185 | 2.54 | 9.18 | −0.89 |
| Qtl-2B-1 | Rain-fed | LC-E | 54 | AX-108725943-AX-94940181 | 2.90 | 6.51 | 1.12 |
| Qtl-2B-2 | Rain-fed | LC-M | 58 | AX-108744217-AX-108728331 | 2.57 | 5.52 | 0.84 |
| Qtl-3B | Irrigation | LT-M | 34 | AX-108749246-AX-111641535 | 2.91 | 7.23 | 1.44 |
| Qtl-6B | Irrigation | LT-E | 128 | AX-111565328-AX-110521824 | 3.13 | 6.63 | 1.70 |
| Qtl-7B-1 | Rain-fed | LC-E | 74 | Xwmc269.1-Xgwm297 | 4.02 | 10.37 | −1.39 |
| Qtl-7B-2 | Irrigation | LB-E | 165 | Xcwm466-AX-111065705 | 4.13 | 11.30 | 1.35 |
| Qtl-7B-3 | Irrigation | LB-NV | 145 | AX-108748008-AX-108801907 | 3.50 | 11.24 | 1.03 |
| Qtl-2D | Irrigation | LC-M | 90 | Xwmc453.1-AX-109261081 | 3.39 | 10.79 | −1.48 |
| Qtl-3D | Irrigation | LB-M | 161 | AX-111086016-AX-111082209 | 3.96 | 8.49 | 1.08 |
| Qtl-6D | Irrigation | LB-M | 0 | AX-111540806-AX-109555484 | 3.46 | 7.24 | 1.01 |
| Qtl-7D-1 | Rain-fed | LT-M | 96 | AX-95119219-Xgwm44 | 6.09 | 16.53 | 1.51 |
| LB-M | 3.09 | 9.48 | 1.20 | ||||
| Irrigation | LT-M | 96 | 4.27 | 10.94 | 1.78 | ||
| LC-NV | 95 | 5.66 | 16.01 | 1.83 | |||
| Qtl-7D-2 | Rain-fed | LT-NV | 108 | AX-111529990-AX-95631292 | 3.91 | 10.18 | 1.15 |
| Qtl-7D-3 | Rain-fed | LC-M | 98 | AX-109879968-AX-95014724 | 7.48 | 17.43 | 1.48 |
| Irrigation | LT-NV | 100 | 6.43 | 17.70 | 2.05 | ||
| LB-NV | 98 | 4.53 | 14.77 | 1.19 | |||
| Qtl-7D-4 | Rain-fed | LC-NV | 94 | AX-94850949-AX-95119219 | 3.88 | 14.14 | 1.12 |
| Irrigation | LT-E | 93 | 4.81 | 10.60 | 2.17 | ||
| Qtl-7D-5 | Rain-fed | LB-E | 103 | AX-95014724-AX-109507404 | 2.67 | 8.34 | 1.09 |
| Irrigation | LC-E | 101 | 2.99 | 9.13 | 1.60 | ||
| LB-M | 103 | 4.94 | 10.65 | 1.21 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, H.; Xu, J.; Ao, D.; Jia, T.; Shi, Y.; Li, N.; Jing, R.; Sun, D. QTL Mapping of Trichome Traits and Analysis of Candidate Genes in Leaves of Wheat (Triticum aestivum L.). Genes 2024, 15, 42. https://doi.org/10.3390/genes15010042
Fan H, Xu J, Ao D, Jia T, Shi Y, Li N, Jing R, Sun D. QTL Mapping of Trichome Traits and Analysis of Candidate Genes in Leaves of Wheat (Triticum aestivum L.). Genes. 2024; 15(1):42. https://doi.org/10.3390/genes15010042
Chicago/Turabian StyleFan, Hua, Jianchao Xu, Dan Ao, Tianxiang Jia, Yugang Shi, Ning Li, Ruilian Jing, and Daizhen Sun. 2024. "QTL Mapping of Trichome Traits and Analysis of Candidate Genes in Leaves of Wheat (Triticum aestivum L.)" Genes 15, no. 1: 42. https://doi.org/10.3390/genes15010042
APA StyleFan, H., Xu, J., Ao, D., Jia, T., Shi, Y., Li, N., Jing, R., & Sun, D. (2024). QTL Mapping of Trichome Traits and Analysis of Candidate Genes in Leaves of Wheat (Triticum aestivum L.). Genes, 15(1), 42. https://doi.org/10.3390/genes15010042






