Cerebrospinal Fluid C1-Esterase Inhibitor and Tie-1 Levels Affect Cognitive Performance: Evidence from Proteome-Wide Mendelian Randomization
Abstract
1. Introduction
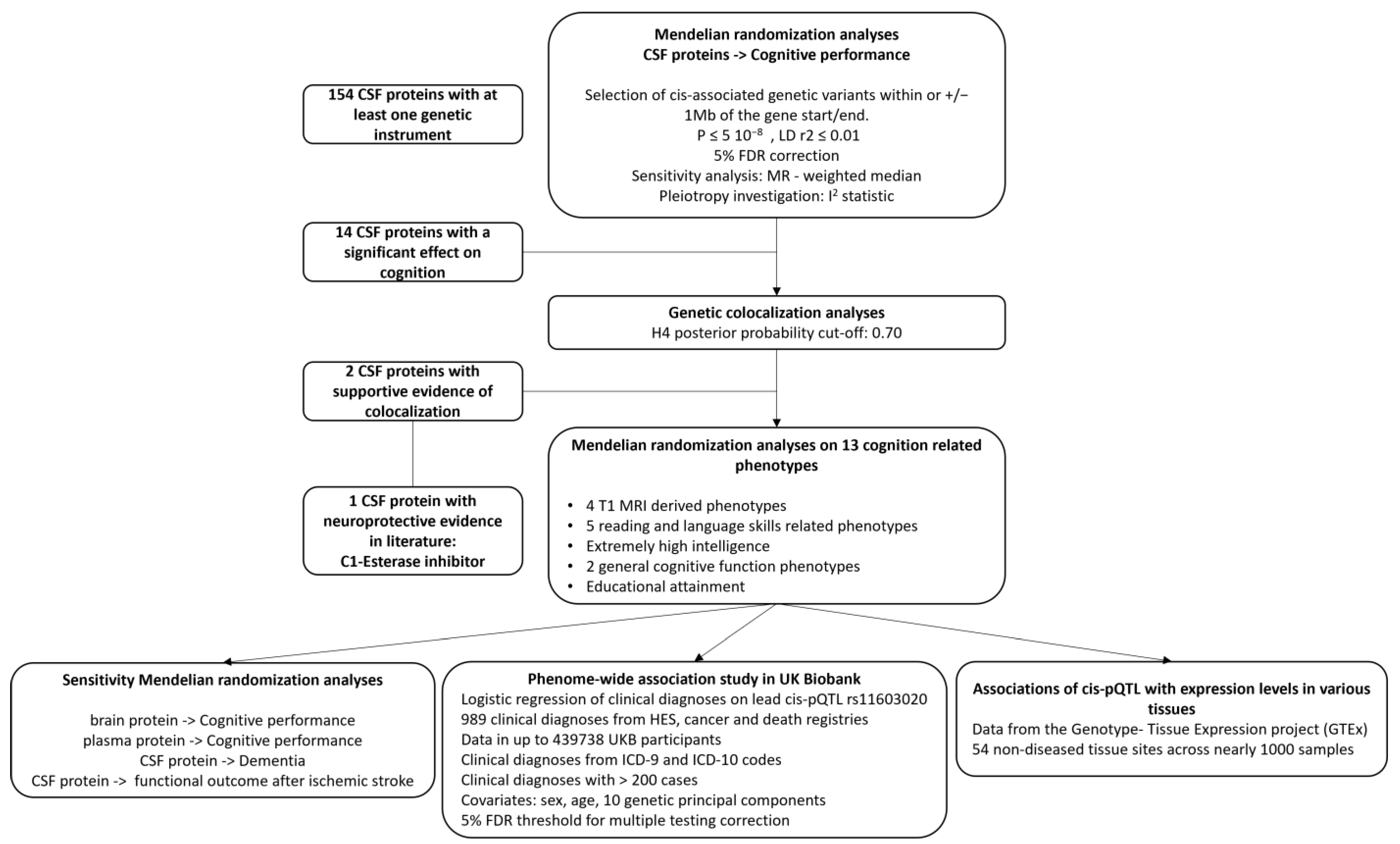
2. Materials and Methods
2.1. Study Design
2.2. Genetic Associations with Protein Levels in CSF and Other Tissues
2.3. Genetic Associations with the Outcomes
2.4. Selection of Genetic Proxies for CSF Protein Levels
2.5. Statistical Analysis
2.5.1. Mendelian Randomization Analysis
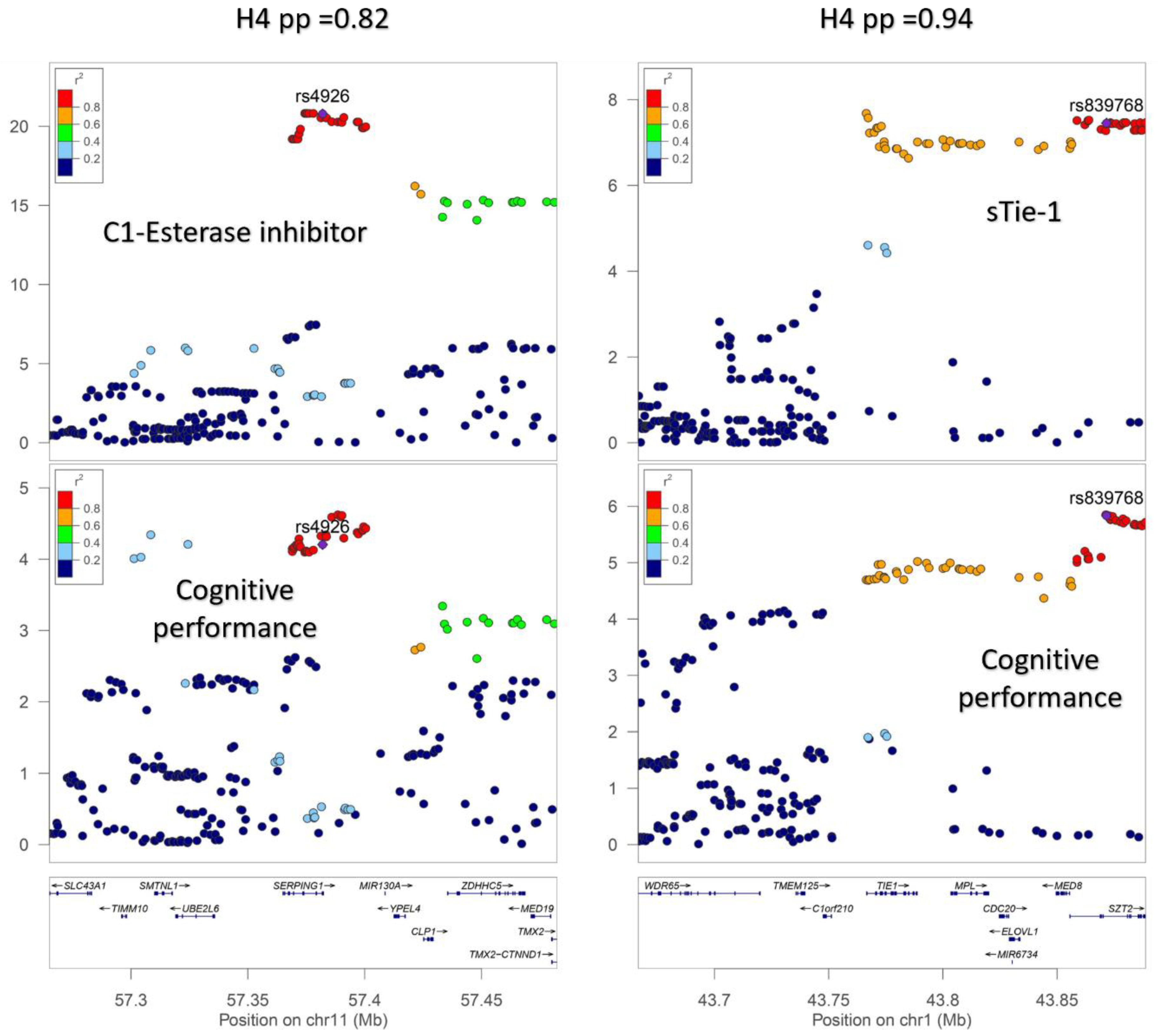
2.5.2. Genetic Colocalization Analysis
2.5.3. Complementary Analysis for C1-Esterase Inhibitor
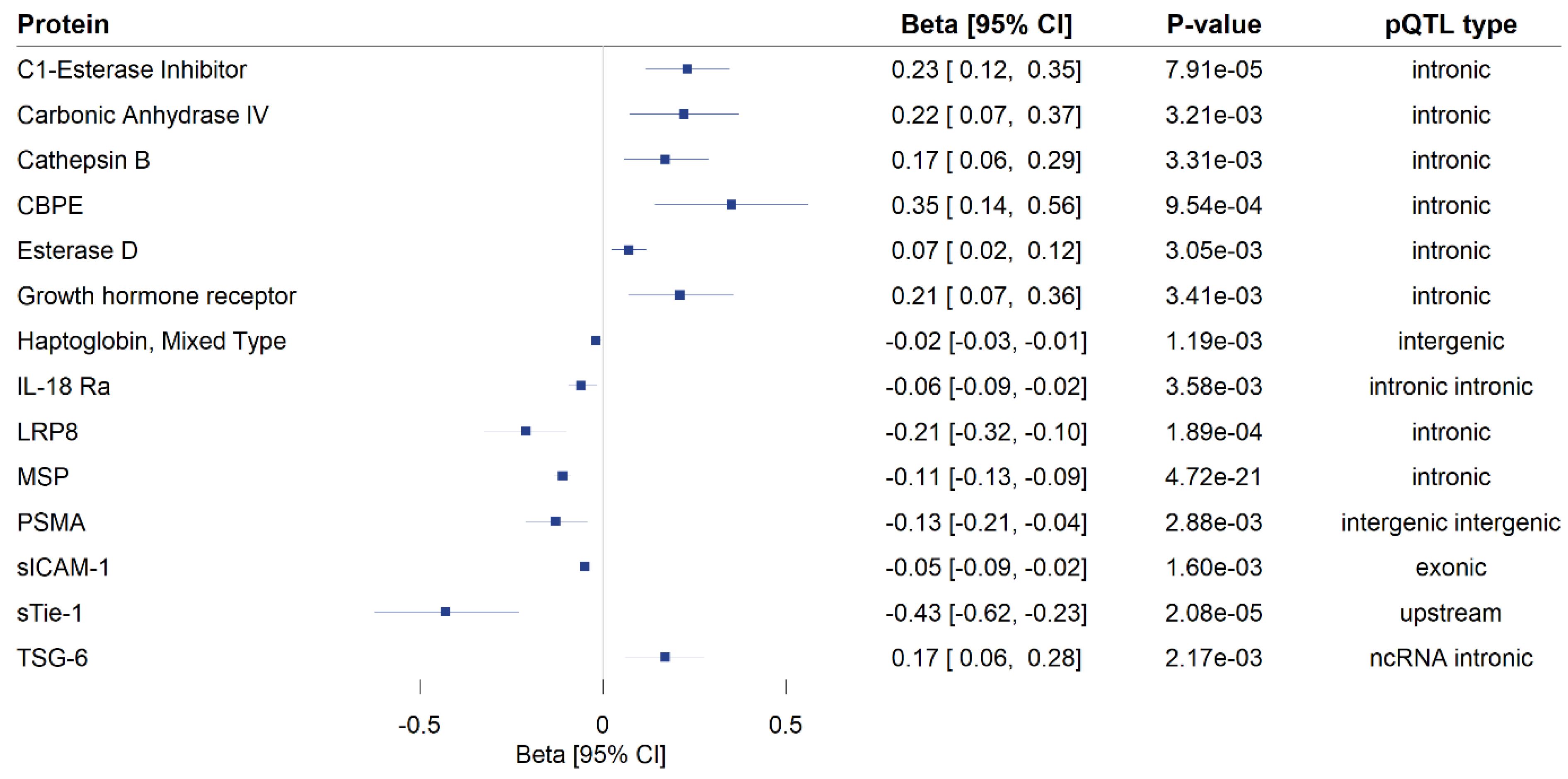
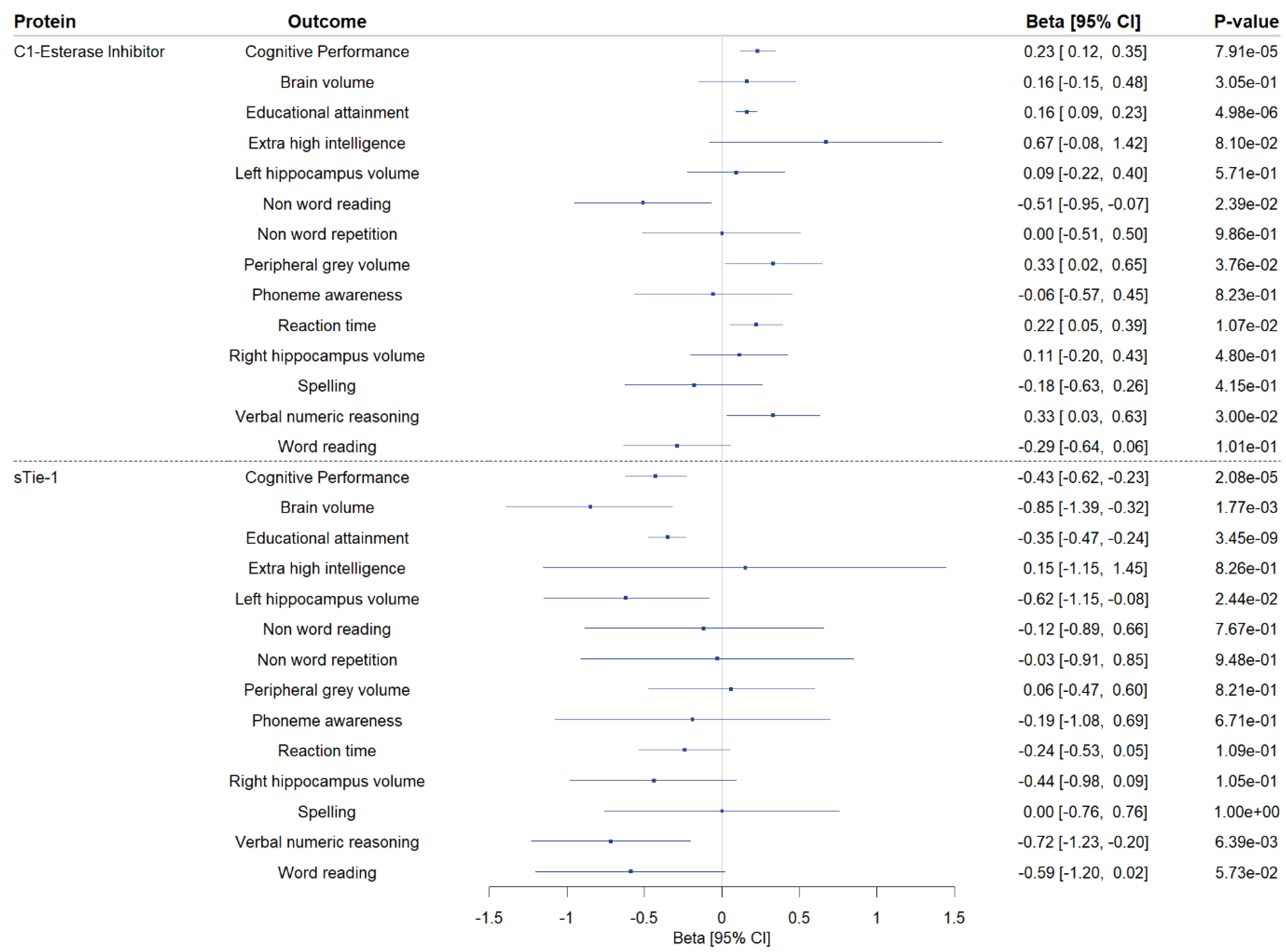
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sakka, L.; Coll, G.; Chazal, J. Anatomy and physiology of cerebrospinal fluid. Eur. Ann. Otorhinolaryngol. Head Neck Dis. 2011, 128, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Spector, R.; Robert Snodgrass, S.; Johanson, C.E. A balanced view of the cerebrospinal fluid composition and functions: Focus on adult humans. Exp. Neurol. 2015, 273, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Mollenhauer, B.; Parnetti, L.; Rektorova, I.; Kramberger, M.G.; Pikkarainen, M.; Schulz-Schaeffer, W.J.; Aarsland, D.; Svenningsson, P.; Farotti, L.; Verbeek, M.M.; et al. Biological confounders for the values of cerebrospinal fluid proteins in Parkinson’s disease and related disorders. J. Neurochem. 2016, 139 (Suppl. S1), 290–317. [Google Scholar] [CrossRef] [PubMed]
- Johar, I.; Mollenhauer, B.; Aarsland, D. Cerebrospinal Fluid Biomarkers of Cognitive Decline in Parkinson’s Disease. Int. Rev. Neurobiol. 2017, 132, 275–294. [Google Scholar] [PubMed]
- Blennow, K. Cerebrospinal fluid protein biomarkers for Alzheimer’s disease. NeuroRx 2004, 1, 213–225. [Google Scholar] [CrossRef] [PubMed]
- Blennow, K.; Dubois, B.; Fagan, A.M.; Lewczuk, P.; de Leon, M.J.; Hampel, H. Clinical utility of cerebrospinal fluid biomarkers in the diagnosis of early Alzheimer’s disease. Alzheimers Dement. 2015, 11, 58–69. [Google Scholar] [CrossRef]
- Daghlas, I. Mendelian randomization as a tool to inform drug development using human genetics. Camb. Prism. Precis. Med. 2023, 1, e16. [Google Scholar] [CrossRef]
- Schindler, U.; Rush, D.K.; Fielding, S. Nootropic Drugs-Animal-Models for Studying Effects on Cognition. Drug Dev. Res. 1984, 4, 567–576. [Google Scholar] [CrossRef]
- Olsson, B.; Portelius, E.; Cullen, N.C.; Sandelius, A.; Zetterberg, H.; Andreasson, U.; Höglund, K.; Irwin, D.J.; Grossman, M.; Weintraub, D.; et al. Association of Cerebrospinal Fluid Neurofilament Light Protein Levels with Cognition in Patients with Dementia, Motor Neuron Disease, and Movement Disorders. JAMA Neurol. 2019, 76, 318–325. [Google Scholar] [CrossRef]
- Kvartsberg, H.; Duits, F.H.; Ingelsson, M.; Andreasen, N.; Ohrfelt, A.; Andersson, K.; Brinkmalm, G.; Lannfelt, L.; Minthon, L.; Hansson, O.; et al. Cerebrospinal fluid levels of the synaptic protein neurogranin correlates with cognitive decline in prodromal Alzheimer’s disease. Alzheimers Dement. 2015, 11, 1180–1190. [Google Scholar] [CrossRef]
- Kern, S.; Syrjanen, J.A.; Blennow, K.; Zetterberg, H.; Skoog, I.; Waern, M.; Hagen, C.E.; Van Harten, A.C.; Knopman, D.S.; Jack, C.R.; et al. Association of Cerebrospinal Fluid Neurofilament Light Protein with Risk of Mild Cognitive Impairment Among Individuals Without Cognitive Impairment. JAMA Neurol. 2019, 76, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Davies, N.M.; Holmes, M.V.; Davey Smith, G. Reading Mendelian randomisation studies: A guide, glossary, and checklist for clinicians. BMJ 2018, 362, k601. [Google Scholar] [CrossRef] [PubMed]
- Burgess, S.; Butterworth, A.; Thompson, S.G. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 2013, 37, 658–665. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Farias, F.H.G.; Ibanez, L.; Suhy, A.; Sadler, B.; Fernandez, M.V.; Wang, F.; Bradley, J.L.; Eiffert, B.; Bahena, J.A.; et al. Genomic atlas of the proteome from brain, CSF and plasma prioritizes proteins implicated in neurological disorders. Nat. Neurosci. 2021, 24, 1302–1312. [Google Scholar] [CrossRef] [PubMed]
- Zuber, V.; Grinberg, N.F.; Gill, D.; Manipur, I.; Slob, E.A.W.; Patel, A.; Wallace, C.; Burgess, S. Combining evidence from Mendelian randomization and colocalization: Review and comparison of approaches. Am. J. Hum. Genet. 2022, 109, 767–782. [Google Scholar] [CrossRef] [PubMed]
- Gold, L.; Ayers, D.; Bertino, J.; Bock, C.; Bock, A.; Brody, E.N.; Carter, J.; Cunningham, V.; Dalby, A.; Eaton, B.; et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS ONE 2010, 5, e15004. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.J.; Wedow, R.; Okbay, A.; Kong, E.; Maghzian, O.; Zacher, M.; Nguyen-Viet, T.A.; Bowers, P.; Sidorenko, J.; Karlsson Linnér, R.; et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat. Genet. 2018, 50, 1112–1121. [Google Scholar] [CrossRef] [PubMed]
- Sudlow, C.; Gallacher, J.; Allen, N.; Beral, V.; Burton, P.; Danesh, J.; Downey, P.; Elliott, P.; Green, J.; Landray, M.; et al. UK biobank: An open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015, 12, e1001779. [Google Scholar] [CrossRef]
- Lencz, T.; Knowles, E.; Davies, G.; Guha, S.; Liewald, D.C.; Starr, J.M.; Djurovic, S.; Melle, I.; Sundet, K.; Christoforou, A.; et al. Molecular genetic evidence for overlap between general cognitive ability and risk for schizophrenia: A report from the Cognitive Genomics consorTium (COGENT). Mol. Psychiatry 2014, 19, 168–174. [Google Scholar] [CrossRef]
- Smith, S.M.; Douaud, G.; Chen, W.; Hanayik, T.; Alfaro-Almagro, F.; Sharp, K.; Elliott, L.T. An expanded set of genome-wide association studies of brain imaging phenotypes in UK Biobank. Nat. Neurosci. 2021, 24, 737–745. [Google Scholar] [CrossRef]
- Eising, E.; Mirza-Schreiber, N.; de Zeeuw, E.L.; Wang, C.A.; Truong, D.T.; Allegrini, A.G.; Shapland, C.Y.; Zhu, G.; Wigg, K.G.; Gerritse, M.L.; et al. Genome-wide analyses of individual differences in quantitatively assessed reading- and language-related skills in up to 34,000 people. Proc. Natl. Acad. Sci. USA 2022, 119, e2202764119. [Google Scholar] [CrossRef] [PubMed]
- Zabaneh, D.; Krapohl, E.; Gaspar, H.A.; Curtis, C.; Lee, S.H.; Patel, H.; Newhouse, S.; Wu, H.M.; Simpson, M.A.; Putallaz, M.; et al. A genome-wide association study for extremely high intelligence. Mol. Psychiatry 2018, 23, 1226–1232. [Google Scholar] [CrossRef] [PubMed]
- Davies, G.; Lam, M.; Harris, S.E.; Trampush, J.W.; Luciano, M.; Hill, W.D.; Hagenaars, S.P.; Ritchie, S.J.; Marioni, R.E.; Fawns-Ritchie, C.; et al. Study of 300,486 individuals identifies 148 independent genetic loci influencing general cognitive function. Nat. Commun. 2018, 9, 2098. [Google Scholar] [CrossRef] [PubMed]
- The 1000 Genomes Project Consortium. An integrated map of genetic variation from 1092 human genomes. Nature 2012, 491, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Hemani, G.; Zheng, J.; Elsworth, B.; Wade, K.H.; Haberland, V.; Baird, D.; Laurin, C.; Burgess, S.; Bowden, J.; Langdon, R.; et al. The MR-Base platform supports systematic causal inference across the human phenome. eLife 2018, 7, e34408. [Google Scholar] [CrossRef] [PubMed]
- Greco, M.F.; Minelli, C.; Sheehan, N.A.; Thompson, J.R. Detecting pleiotropy in Mendelian randomisation studies with summary data and a continuous outcome. Stat. Med. 2015, 34, 2926–2940. [Google Scholar] [CrossRef] [PubMed]
- Burgess, S.; Bowden, J.; Fall, T.; Ingelsson, E.; Thompson, S.G. Sensitivity Analyses for Robust Causal Inference from Mendelian Randomization Analyses with Multiple Genetic Variants. Epidemiology 2017, 28, 30–42. [Google Scholar] [CrossRef]
- Bowden, J.; Davey Smith, G.; Haycock, P.C.; Burgess, S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet. Epidemiol. 2016, 40, 304–314. [Google Scholar] [CrossRef]
- Giambartolomei, C.; Vukcevic, D.; Schadt, E.E.; Franke, L.; Hingorani, A.D.; Wallace, C.; Plagnol, V. Bayesian Test for Colocalisation between Pairs of Genetic Association Studies Using Summary Statistics. PLoS Genet. 2014, 10, e1004383. [Google Scholar] [CrossRef]
- Wallace, C. Eliciting priors and relaxing the single causal variant assumption in colocalisation analyses. PLoS Genet. 2020, 16, e1008720. [Google Scholar] [CrossRef]
- Thom, C.S.; Voight, B.F. Genetic colocalization atlas points to common regulatory sites and genes for hematopoietic traits and hematopoietic contributions to disease phenotypes. BMC Med. Genom. 2020, 13, 89. [Google Scholar] [CrossRef] [PubMed]
- Kunkle, B.W.; Grenier-Boley, B.; Sims, R.; Bis, J.C.; Damotte, V.; Naj, A.C.; Boland, A.; Vronskaya, M.; Van Der Lee, S.J.; Amlie-Wolf, A.; et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Abeta, tau, immunity and lipid processing. Nat. Genet. 2019, 51, 414–430. [Google Scholar] [CrossRef] [PubMed]
- Soderholm, M.; Pedersen, A.; Lorentzen, E.; Stanne, T.M.; Bevan, S.; Olsson, M.; Cole, J.W.; Fernandez-Cadenas, I.; Hankey, G.J.; Jimenez-Conde, J.; et al. Genome-wide association meta-analysis of functional outcome after ischemic stroke. Neurology 2019, 92, e1271–e1283. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. International Statistical Classification of Diseases and Related Health Problems, 10th Revision; World Health Organization: Geneva, Switzerland, 1992. [Google Scholar]
- Carroll, R.J.; Bastarache, L.; Denny, J.C. R PheWAS: Data analysis and plotting tools for phenome-wide association studies in the R environment. Bioinformatics 2014, 30, 2375–2376. [Google Scholar] [CrossRef]
- Pendergrass, S.A.; Ritchie, M.D. Phenome-Wide Association Studies: Leveraging Comprehensive Phenotypic and Genotypic Data for Discovery. Curr. Genet. Med. Rep. 2015, 3, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Carithers, L.J.; Ardlie, K.; Barcus, M.; Branton, P.A.; Britton, A.; Buia, S.A.; Compton, C.C.; DeLuca, D.S.; Peter-Demchok, J.; Gelfand, E.T.; et al. A Novel Approach to High-Quality Postmortem Tissue Procurement: The GTEx Project. Biopreserv. Biobank. 2015, 13, 311–319. [Google Scholar] [CrossRef]
- Caliezi, C.; Wuillemin, W.A.; Zeerleder, S.; Redondo, M.; Eisele, B.; Hack, C.E. C1-esterase inhibitor: An anti-inflammatory agent and its potential use in the treatment of diseases other than hereditary angioedema. Pharmacol. Rev. 2000, 52, 91–112. [Google Scholar]
- Farfara, D.; Feierman, E.; Richards, A.; Revenko, A.S.; MacLeod, R.A.; Norris, E.H.; Strickland, S. Knockdown of circulating C1 inhibitor induces neurovascular impairment, glial cell activation, neuroinflammation, and behavioral deficits. Glia 2019, 67, 1359–1373. [Google Scholar] [CrossRef]
- Gesuete, R.; Storini, C.; Fantin, A.; Stravalaci, M.; Zanier, E.R.; Orsini, F.; Vietsch, H.; Mannesse, M.M.; Ziere, B.; Gobbi, M.; et al. Recombinant C1 Inhibitor in Brain Ischemic Injury. Ann. Neurol. 2009, 66, 332–342. [Google Scholar] [CrossRef]
- Longhi, L.; Perego, C.; Ortolano, F.; Zanier, E.R.; Bianchi, P.; Stocchetti, N.; McIntosh, T.K.; De Simoni, M.G. C1-inhibitor attenuates neurobehavioral deficits and reduces contusion volume after controlled cortical impact brain injury in mice. Crit. Care Med. 2009, 37, 659–665. [Google Scholar] [CrossRef]
- Korhonen, E.A.; Lampinen, A.; Giri, H.; Anisimov, A.; Kim, M.; Allen, B.; Fang, S.; D’Amico, G.; Sipilä, T.J.; Lohela, M.; et al. Tie1 controls angiopoietin function in vascular remodeling and inflammation. J. Clin. Investig. 2016, 126, 3495–3510. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yao, T.; Cai, J.; Zhang, Q.; Li, S.; Li, H.; Fu, X.; Wu, J. A novel cis-regulatory variant modulating TIE1 expression associated with attention deficit hyperactivity disorder in Han Chinese children. J. Affect. Disord. 2022, 300, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Honda, T.; Kobayashi, K.; Mikoshiba, K.; Nakajima, K. Regulation of Cortical Neuron Migration by the Reelin Signaling Pathway. Neurochem. Res. 2011, 36, 1270–1279. [Google Scholar] [CrossRef] [PubMed]
- van der Meer, D.; Kaufmann, T.; Shadrin, A.A.; Makowski, C.; Frei, O.; Roelfs, D.; Monereo-Sánchez, J.; Linden, D.E.; Rokicki, J.; Alnæs, D.; et al. The genetic architecture of human cortical folding. Sci. Adv. 2021, 7, eabj9446. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zagkos, L.; Dib, M.-J.; Cronjé, H.T.; Elliott, P.; Dehghan, A.; Tzoulaki, I.; Gill, D.; Daghlas, I. Cerebrospinal Fluid C1-Esterase Inhibitor and Tie-1 Levels Affect Cognitive Performance: Evidence from Proteome-Wide Mendelian Randomization. Genes 2024, 15, 71. https://doi.org/10.3390/genes15010071
Zagkos L, Dib M-J, Cronjé HT, Elliott P, Dehghan A, Tzoulaki I, Gill D, Daghlas I. Cerebrospinal Fluid C1-Esterase Inhibitor and Tie-1 Levels Affect Cognitive Performance: Evidence from Proteome-Wide Mendelian Randomization. Genes. 2024; 15(1):71. https://doi.org/10.3390/genes15010071
Chicago/Turabian StyleZagkos, Loukas, Marie-Joe Dib, Héléne T. Cronjé, Paul Elliott, Abbas Dehghan, Ioanna Tzoulaki, Dipender Gill, and Iyas Daghlas. 2024. "Cerebrospinal Fluid C1-Esterase Inhibitor and Tie-1 Levels Affect Cognitive Performance: Evidence from Proteome-Wide Mendelian Randomization" Genes 15, no. 1: 71. https://doi.org/10.3390/genes15010071
APA StyleZagkos, L., Dib, M.-J., Cronjé, H. T., Elliott, P., Dehghan, A., Tzoulaki, I., Gill, D., & Daghlas, I. (2024). Cerebrospinal Fluid C1-Esterase Inhibitor and Tie-1 Levels Affect Cognitive Performance: Evidence from Proteome-Wide Mendelian Randomization. Genes, 15(1), 71. https://doi.org/10.3390/genes15010071





