Genome-Wide Association Study Reveals the Genetic Architecture of Growth and Meat Production Traits in a Chicken F2 Resource Population
Abstract
1. Introduction
2. Materials and Methods
2.1. Birds Involved in the Experiment
2.2. Phenotypic Characteristics
2.3. Sampling and DNA Extraction
2.4. SNP Genotyping and Quality Control
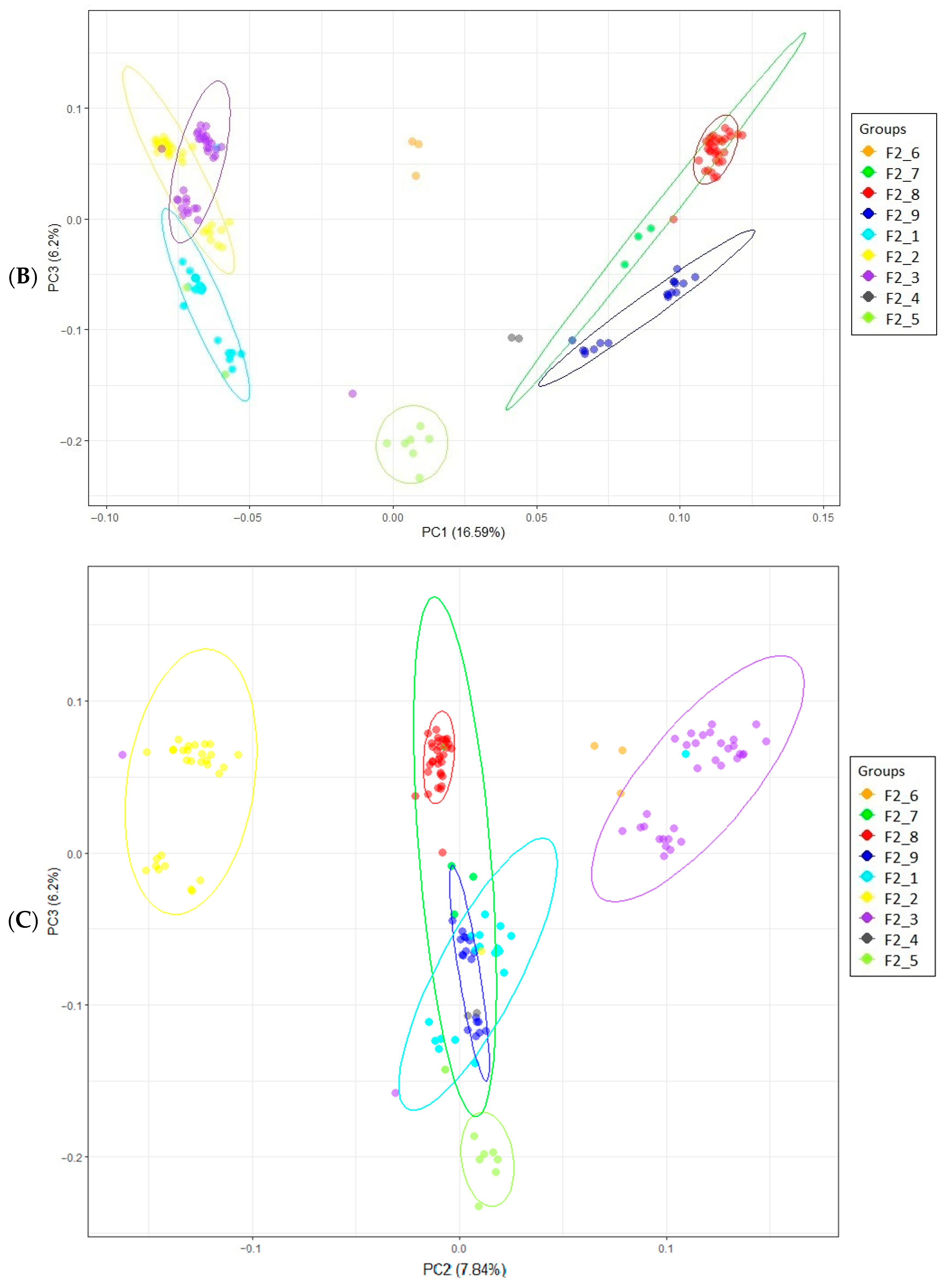
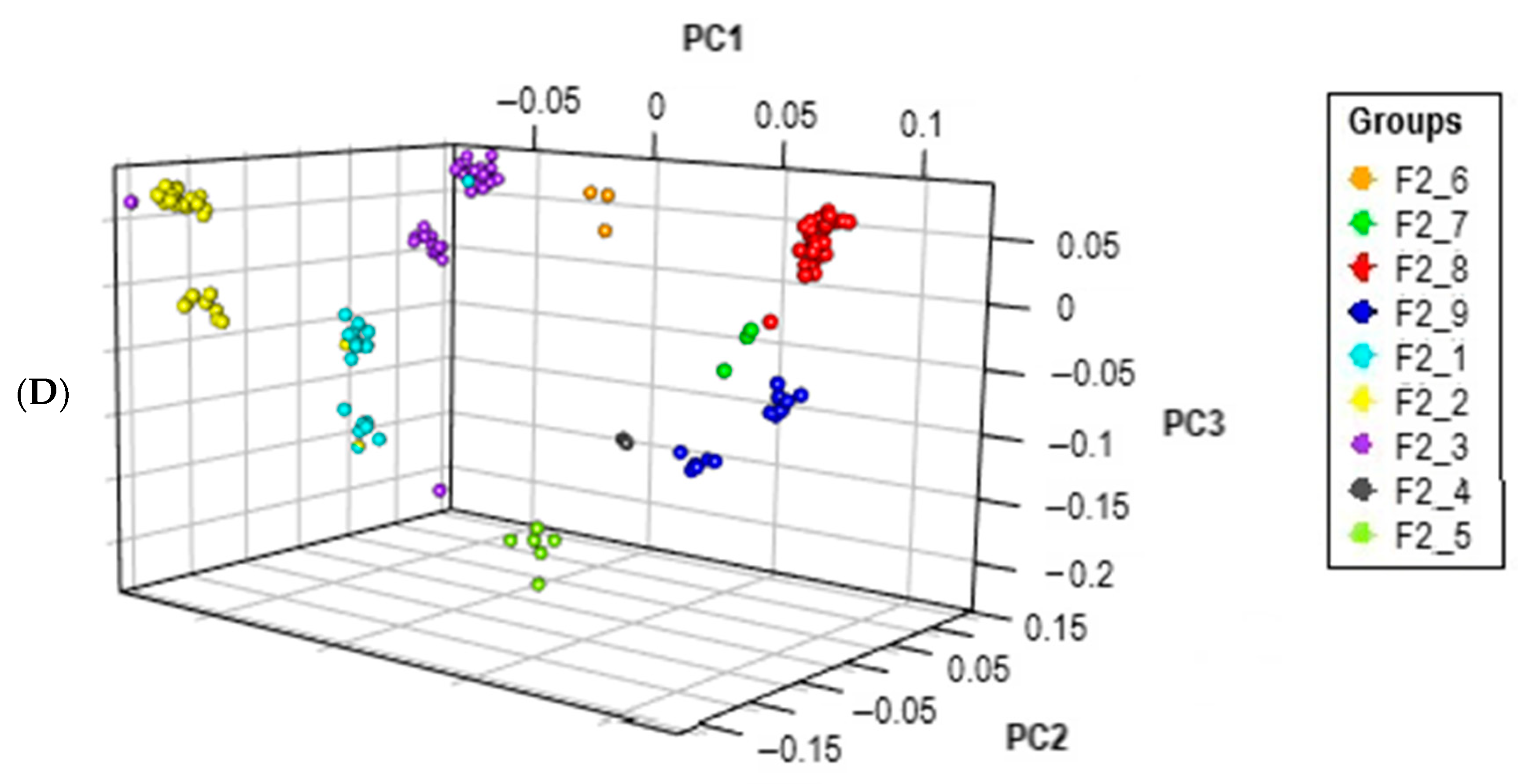
2.5. Principal Component Analysis
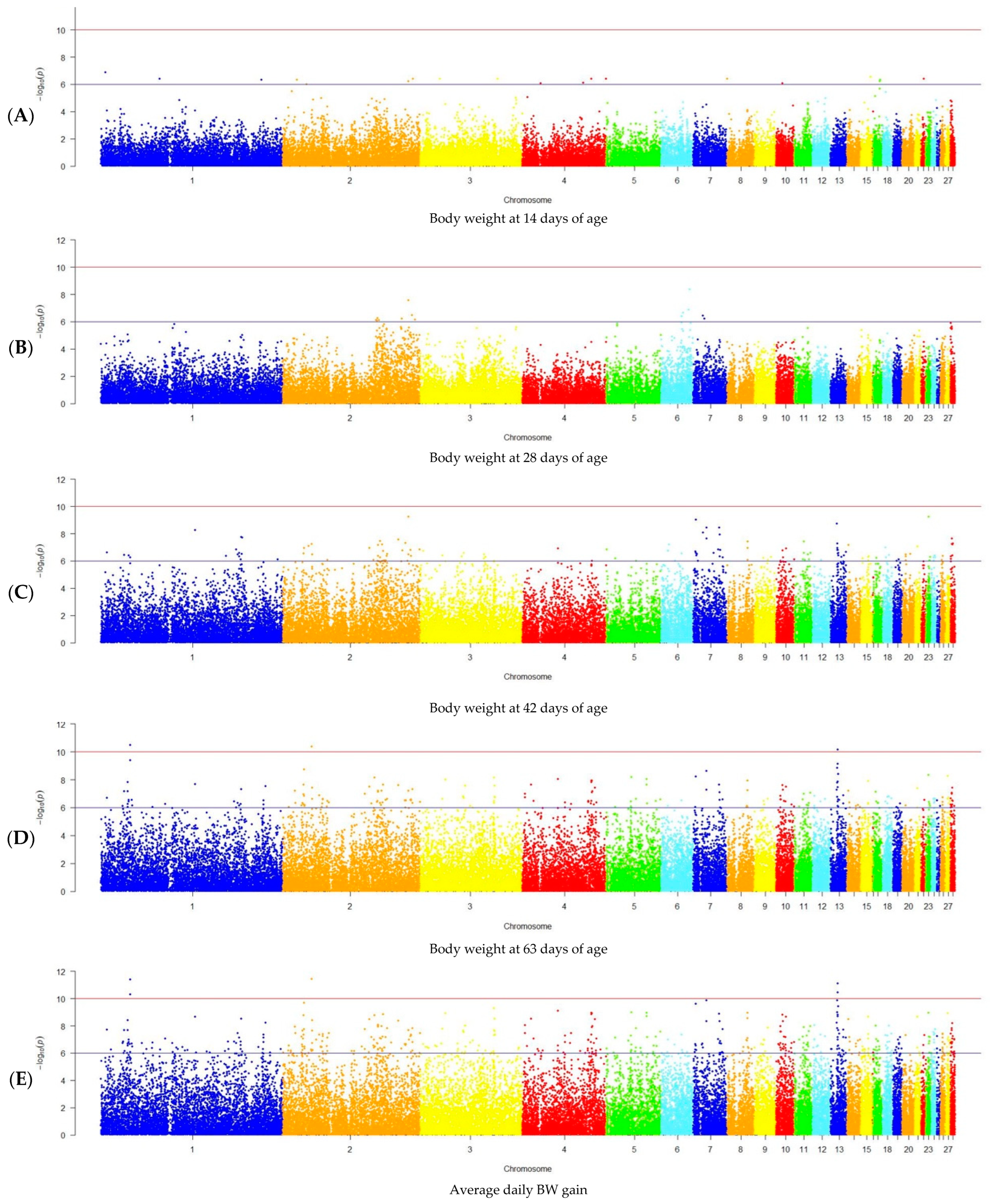
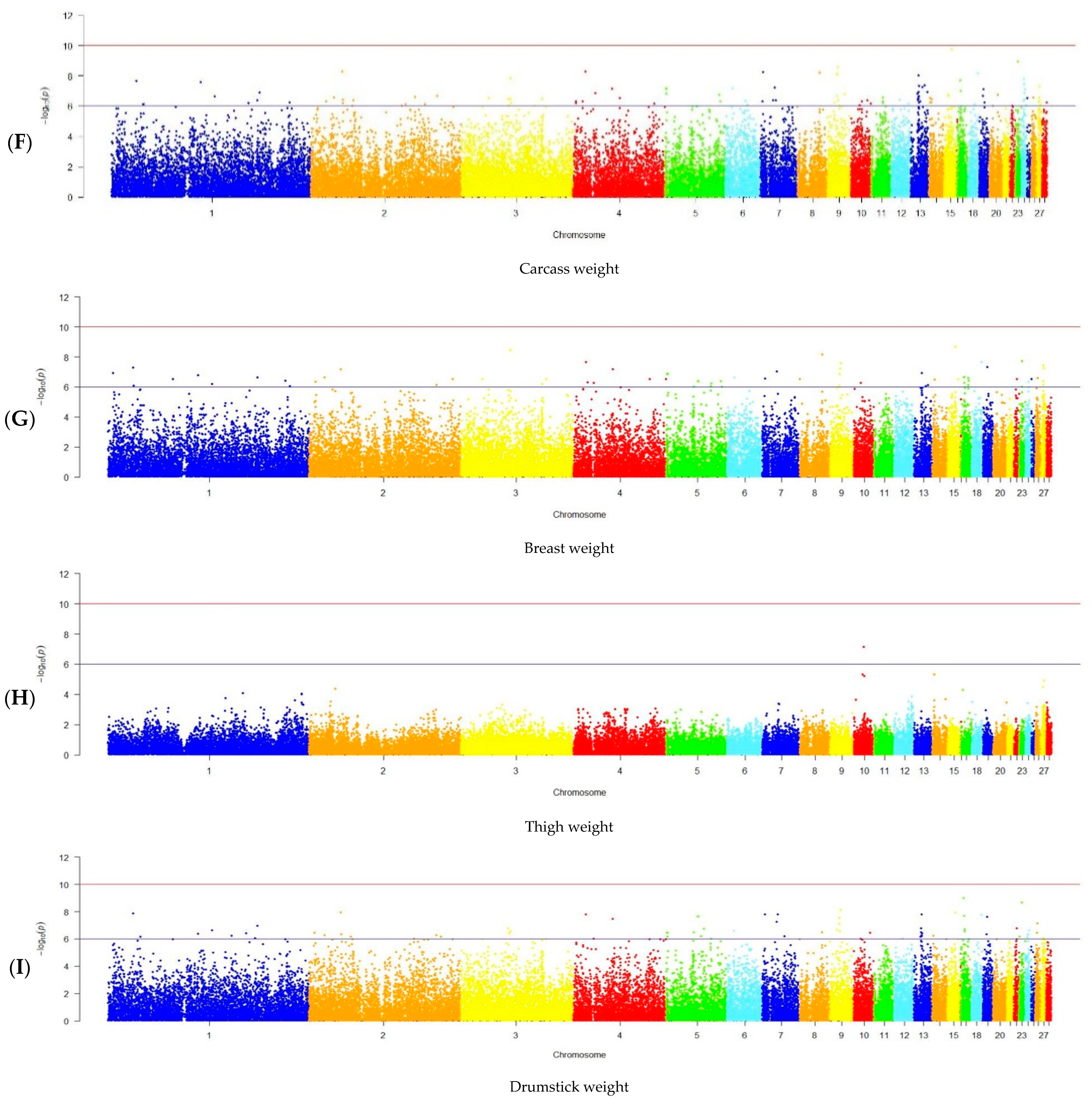
2.6. GWAS Analysis
3. Results
3.1. Population Stratification
3.2. GWAS Results
3.3. Candidate Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- OECD/FAO. OECD-FAO Agricultural Outlook 2019–2028; OECD Publishing: Paris, France; Food and Agriculture Organization of the United Nations: Rome, Italy, 2019; Available online: https://doi.org/10.1787/agr_outlook-2019-en (accessed on 30 August 2024).
- Pereira, P.M.D.C.C.; Vicente, A.F.D.R.B. Meat nutritional composition and nutritive role in the human diet. Meat Sci. 2013, 93, 586–592. [Google Scholar] [CrossRef] [PubMed]
- Mir, N.A.; Rafiq, A.; Kumar, F.; Singh, V.; Shukla, V. Determinants of broiler chicken meat quality and factors affecting them: A review. J. Food Sci. Technol. 2017, 54, 2997–3009. [Google Scholar] [CrossRef] [PubMed]
- Marchewka, J.; Sztandarski, P.; Solka, M.; Louton, H.; Rath, K.; Vogt, L.; Rauch, E.; Ruijter, D.; de Jong, I.C.; Horbańczuk, J.O. Linking key husbandry factors to the intrinsic quality of broiler meat. Poult. Sci. 2023, 102, 102384. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.; Liu, R.; Li, C.; Xu, X.; Zhou, G. Meat quality and flavor compounds of soft-boiled chickens: Effect of Chinese yellow-feathered chicken breed and slaughter age. Poult. Sci. 2022, 101, 102168. [Google Scholar] [CrossRef]
- Dalle Zotte, A.; Gleeson, E.; Franco, D.; Cullere, M.; Lorenzo, J.M. Proximate composition, amino acid profile, and oxidative stability of slow-growing indigenous chickens compared with commercial broiler chickens. Foods 2020, 9, 546. [Google Scholar] [CrossRef]
- Jung, Y.-K.; Jeon, H.-J.; Jung, S.; Choe, J.-H.; Lee, J.-H.; Heo, K.-N.; Kang, B.-S.; Jo, C.-R. Comparison of quality traits of thigh meat from Korean native chickens and broilers. Food Sci. Anim. 2011, 31, 684–692. [Google Scholar] [CrossRef]
- Berri, C.; Wacrenier, N.; Millet, N.; Le Bihan-Duval, E. Effect of selection for improved body composition on muscle and meat characteristics of broilers from experimental and commercial lines. Poult. Sci. 2001, 80, 833–838. [Google Scholar] [CrossRef]
- Tůmová, E.; Chodová, D.; Skřivanová, E.; Laloučková, K.; Šubrtová-Salmonová, H.; Ketta, M.; Machander, V.; Cotozzolo, E. Research Note: The effects of genotype, sex, and feeding regime on performance, carcasses characteristic, and microbiota in chickens. Poult. Sci. 2021, 100, 760–764. [Google Scholar] [CrossRef]
- Suzuki, S.; Kobayashi, M.; Murai, A.; Tsudzuki, M.; Ishikawa, A. Characterization of growth, fat deposition, and lipid metabolism-related gene expression in lean and obese meat-type chickens. J. Poult. Sci. 2019, 56, 101–111. [Google Scholar] [CrossRef]
- Petracci, M.; Cavani, C. Muscle growth and poultry meat quality issues. Nutrients 2012, 4, 1–12. [Google Scholar] [CrossRef]
- Mueller, S.; Taddei, L.; Albiker, D.; Kreuzer, M.; Siegrist, M.; Messikommer, R.E.; Gangnat, I.D.M. Growth, carcass, and meat quality of 2 dual-purpose chickens and a layer hybrid grown for 67 or 84 D compared with slow-growing broilers. J. Appl. Poult. Res. 2020, 29, 185–196. [Google Scholar] [CrossRef]
- Xiong, X.; Liu, X.; Zhu, X.; Tan, Y.; Wang, Z.; Xu, J.; Tu, X.; Rao, Y.; Duan, J.; Zhao, W.; et al. A mutation in PHKG1 causes high drip loss and low meat quality in Chinese Ningdu yellow chickens. Poult. Sci. 2022, 101, 101556. [Google Scholar] [CrossRef]
- Elkhazen, A.; Larbi, M.; M’hamdi, N.; Haddad, B. Comparison of meat quality of local poultry and Arbors acres reared in two farming systems in Tunisia. J. New Sci. 2016, 34, 1922–1929. [Google Scholar]
- Jin, S.; Yang, L.; Zang, H.; Xu, Y.; Chen, X.; Chen, X.; Liu, P.; Geng, Z. Influence of free-range days on growth performance, carcass traits, meat quality, lymphoid organ indices, and blood biochemistry of Wannan Yellow chickens. Poult. Sci. 2019, 98, 6602–6610. [Google Scholar] [CrossRef]
- Yang, L.; Wang, X.; He, T.; Xiong, F.; Chen, X.; Chen, X.; Jin, S.; Geng, Z. Association of residual feed intake with growth performance, carcass traits, meat quality, and blood variables in native chickens. J. Anim. Sci. 2020, 98, skaa121. [Google Scholar] [CrossRef] [PubMed]
- Bughio, E.; Hussain, J.; Mahmud, A.; Khalique, A. Effects of production system and feeding regimen on carcass and meat quality traits of Naked Neck chicken. S. Afr. J. Anim. Sci. 2021, 51, 250–261. [Google Scholar] [CrossRef]
- Arthur, J.A.; Albers, G.A. Industrial perspective on problems and issues associated with poultry breeding. In Poultry Genetics, Breeding and Biotechnology; Muir, W.M., Aggrey, S.E., Eds.; CAB International: Wallingford, UK; Cambridge, UK, 2003; pp. 1–12. [Google Scholar] [CrossRef]
- da Silva, D.C.F.; de Arruda, A.M.V.; Gonçalves, A.A. Quality characteristics of broiler chicken meat from free-range and industrial poultry system for the consumers. J. Food Sci. Technol. 2017, 54, 1818–1826. [Google Scholar] [CrossRef]
- Santhi, D.; Kalaikannan, A. Enrichment of chicken meat with dietary fibre sources as functional ingredients. Worlds Poult. Sci. J. 2023, 79, 783–806. [Google Scholar] [CrossRef]
- Bessei, W. Welfare of broilers: A review. Worlds Poult. Sci. J. 2006, 62, 455–466. [Google Scholar] [CrossRef]
- Petracci, M.; Mudalal, S.; Soglia, F.; Cavani, C. Meat quality in fast-growing broiler chickens. Worlds Poult. Sci. J. 2015, 71, 363–374. [Google Scholar] [CrossRef]
- Scheuermann, G.N.; Bilgili, S.F.; Hess, J.B.; Mulvaney, D.R. Breast muscle development in commercial broiler chickens. Poult. Sci. 2003, 82, 1648–1658. [Google Scholar] [CrossRef] [PubMed]
- Devatkal, S.K.; Naveena, B.M.; Kotaiah, T. Quality, composition, and consumer evaluation of meat from slow-growing broilers relative to commercial broilers. Poult. Sci. 2019, 98, 6177–6186. [Google Scholar] [CrossRef]
- Reyer, H.; Hawken, R.; Murani, E.; Ponsuksili, S.; Wimmers, K. The genetics of feed conversion efficiency traits in a commercial broiler line. Sci. Rep. 2015, 5, 16387. [Google Scholar] [CrossRef] [PubMed]
- Patreva, L.S.; Kovalenko, V.P.; Tereshchenko, O.V.; Katerynych, O.O. Miasne Ptakhivnytstvo [Poultry Meat Production]; Mykolaivskyi DAU: Mykolaiv, Ukraine, 2010; ISBN 978-966-8205-60-6. (In Ukrainian) [Google Scholar]
- Vakhrameev, A.B.; Narushin, V.G.; Larkina, T.A.; Barkova, O.Y.; Peglivanyan, G.K.; Dysin, A.P.; Dementieva, N.V.; Shcherbakov, Y.S.; Pozovnikova, M.V.; Griffin, D.K.; et al. Pectoral angle: A glance at a traditional phenotypic trait in chickens from a new perspective. J. Agric. Sci. 2023, 161, 606–615. [Google Scholar] [CrossRef]
- Dou, D.; Shen, L.; Zhou, J.; Cao, Z.; Luan, P.; Li, Y.; Xiao, F.; Guo, H.; Li, H.; Zhang, H. Genome-wide association studies for growth traits in broilers. BMC Genom. Data 2022, 23, 1. [Google Scholar] [CrossRef] [PubMed]
- Mebratie, W.; Madsen, P.; Hawken, R.; Romé, H.; Marois, D.; Henshall, J.; Bovenhuis, H.; Jensen, J. Genetic parameters for body weight and different definitions of residual feed intake in broiler chickens. Genet. Sel. Evol. 2019, 51, 53. [Google Scholar] [CrossRef]
- Chu, T.T.; Madsen, P.; Norberg, E.; Wang, L.; Marois, D.; Henshall, J.; Jensen, J. Genetic analysis on body weight at different ages in broiler chicken raised in commercial environment. J. Anim. Breed. Genet. 2020, 137, 245–259. [Google Scholar] [CrossRef]
- Dou, T.; Li, Z.; Wang, K.; Liu, L.; Rong, H.; Xu, Z.; Huang, Y.; Gu, D.; Chen, X.; Hu, W.; et al. Regulation of myostatin expression is associated with growth and muscle development in commercial broiler and DMC muscle. Mol. Biol. Rep. 2018, 45, 511–522. [Google Scholar] [CrossRef]
- Guo, X.; Zhang, H.; Wang, H.; He, X.-X.; Wang, J.-X.; Wei, W.; Liu, M.; Xu, J.-M.; Liu, Y.-N.; Jiang, R.-S. Identification of key modules and hub genes involved in regulating the color of chicken breast meat using WGCNA. Animals 2023, 13, 2356. [Google Scholar] [CrossRef]
- Romanov, M.N.; Miao, Y.W.; Wilson, P.W.; Morris, A.; Sharp, P.J.; Dunn, I.C. Detection and Assay of Polymorphism in Reproductive Gene Loci in a Commercial Broiler Breeder Population for Use in Association Studies. In Proceedings of the Conference “From Jay Lush to Genomics: Visions for Animal Breeding and Genetics”, Ames, IA, USA, 16–18 May 1999; Dekkers, J.C.M., Lamont, S.J., Rothschild, M.F., Eds.; Department of Animal Science, Iowa State University: Ames, IA, USA, 1999; p. 155. Available online: https://web.archive.org/web/20050314091227/http:/www.agbiotechnet.com/proceedings/jaylush.asp#15 (accessed on 14 March 2005).
- Dunn, I.C.; Miao, Y.-W.; Morris, A.; Romanov, M.N.; Wilson, P.W.; Waddington, D.; Sharp, P.J. Candidate genes and reproductive traits in a commercial broiler breeder population, an association study. J. Anim. Sci. 2001, 79 (Suppl. S1), 43. [Google Scholar]
- Dunn, I.C.; Miao, Y.-W.; Morris, A.; Romanov, M.N.; Waddington, D.W.; Wilson, P.W.; Sharp, P.J. Association between candidate genes and reproductive traits in a commercial broiler breeder population. Br. Poult. Sci. 2001, 42 (Suppl. S1), S113–S114. [Google Scholar] [CrossRef]
- Dementieva, N.V.; Fedorova, E.S.; Krutikova, A.A.; Mitrofanova, O.V.; Stanishevskaya, O.I.; Pleshanov, N.V.; Smaragdov, M.G.; Kudinov, A.A.; Terletsky, V.P.; Romanov, M.N. Genetic variability of indels in the prolactin and dopamine receptor D2 genes and their association with the yield of allanto-amniotic fluid in Russian White laying hens. Tarım Bilim. Derg. J. Agric. Sci. 2020, 26, 373–379. [Google Scholar] [CrossRef]
- Pickering, N.K.; Auvray, B.; Dodds, K.G.; McEwan, J.C. Genomic prediction and genome-wide association study for dagginess and host internal parasite resistance in New Zealand sheep. BMC Genom. 2015, 16, 958. [Google Scholar] [CrossRef] [PubMed]
- Nikitkina, E.V.; Dementieva, N.V.; Shcherbakov, Y.S.; Atroshchenko, M.M.; Kudinov, A.A.; Samoylov, O.I.; Pozovnikova, M.V.; Dysin, A.P.; Krutikova, A.A.; Musidray, A.A.; et al. Genome-wide association study for frozen-thawed sperm motility in stallions across various horse breeds. Anim. Biosci. 2022, 35, 1827. [Google Scholar] [CrossRef]
- Dementieva, N.V.; Dysin, A.P.; Shcherbakov, Y.S.; Nikitkina, E.V.; Musidray, A.A.; Petrova, A.V.; Mitrofanova, O.V.; Plemyashov, K.V.; Azovtseva, A.I.; Griffin, D.K.; et al. Risk of sperm disorders and impaired fertility in frozen–thawed bull semen: A genome-wide association study. Animals 2024, 14, 251. [Google Scholar] [CrossRef] [PubMed]
- Dementeva, N.V.; Kudinov, A.A.; Mitrofanova, O.V.; Stanishevskaya, O.I.; Fedorova, E.S.; Romanov, M.N. Genome-wide association study of reproductive traits in a gene pool breed of the Russian White chickens. Reprod. Domest. Anim. 2018, 53 (Suppl. S2), 123–124. [Google Scholar] [CrossRef]
- Guo, J.; Qu, L.; Dou, T.C.; Shen, M.M.; Hu, Y.P.; Ma, M.; Wang, K.H. Genome-wide association study provides insights into the genetic architecture of bone size and mass in chickens. Genome 2020, 63, 133–143. [Google Scholar] [CrossRef]
- Zhou, G.; Liu, T.; Wang, Y.; Qu, H.; Shu, D.; Jia, X.; Luo, C. Genome-wide association studies provide insight into the genetic determination for hyperpigmentation of the visceral peritoneum in broilers. Front. Genet. 2022, 13, 820297. [Google Scholar] [CrossRef]
- Romanov, M.N.; Shakhin, A.V.; Abdelmanova, A.S.; Volkova, N.A.; Efimov, D.N.; Fisinin, V.I.; Korshunova, L.G.; Anshakov, D.V.; Dotsev, A.V.; Griffin, D.K.; et al. Dissecting selective signatures and candidate genes in grandparent lines subject to high selection pressure for broiler production and in a local Russian chicken breed of Ushanka. Genes 2024, 15, 524. [Google Scholar] [CrossRef]
- Abdelmanova, A.S.; Dotsev, A.V.; Romanov, M.N.; Stanishevskaya, O.I.; Gladyr, E.A.; Rodionov, A.N.; Vetokh, A.N.; Volkova, N.A.; Fedorova, E.S.; Gusev, I.V.; et al. Unveiling comparative genomic trajectories of selection and key candidate genes in egg-type Russian White and meat-type White Cornish chickens. Biology 2021, 10, 876. [Google Scholar] [CrossRef]
- Moiseeva, I.G. Variability and heritability of some features of egg quality in Russkaya Belaya chickens. Tr. Akad. Nauk SSSR Inst. Genet. 1964, 31, 302–308. (In Russian) [Google Scholar]
- Moiseeva, I.G. Soderzhanie lipidov i kholesterina v iatsakh kur russkoĭ beloĭ porody v sviazi s produktivnost’iu [The lipid and cholesterin contents of hen’s eggs of the Russian white breed in relation to productiveness]. Tr. Akad. Nauk SSSR Inst. Genet. 1965, 33, 119–128. Available online: https://www.cabidigitallibrary.org/doi/full/10.5555/19660103382 (accessed on 24 April 2024). (In Russian). [PubMed]
- Dementeva, N.V.; Romanov, M.N.; Kudinov, A.A.; Mitrofanova, O.V.; Stanishevskaya, O.I.; Terletsky, V.P.; Fedorova, E.S.; Nikitkina, E.V.; Plemyashov, K.V. Studying the structure of a gene pool population of the Russian White chicken breed by genome-wide SNP scan. Sel’skokhozyaistvennaya Biol. Agric. Biol. 2017, 52, 1166–1174. [Google Scholar] [CrossRef]
- Kudinov, A.A.; Dementieva, N.V.; Mitrofanova, O.V.; Stanishevskaya, O.I.; Fedorova, E.S.; Larkina, T.A.; Mishina, A.I.; Plemyashov, K.V.; Griffin, D.K.; Romanov, M.N. Genome-wide association studies targeting the yield of extraembryonic fluid and production traits in Russian White chickens. BMC Genom. 2019, 20, 270. [Google Scholar] [CrossRef] [PubMed]
- Moiseeva, I.G. Content of lipids and cholesterol in eggs of Russian White chickens. Collect. Works Young Sci. All-Union Res. Tech. Poult. Inst. 1966, 8, 225–235. (In Russian) [Google Scholar]
- Bondarenko, Y.V.; Sergheyeva, V.D.; Kuranova, E.N.; Krasnozhon, S.A.; Romanov, M.N. Autosexing maternal form of meat-type chickens. Ptitsevodstvo Poult. Farm. 1987, 40, 6–11. (In Russian) [Google Scholar]
- Romanov, M.N.; Yakubovskaya, S.N.; Nachalnaya, Z.P. Problems of Cage Keeping of Meat-Type Chickens and Broilers; Information Bulletin; Ukrainian Poultry Research Institute: Kharkov, Russia, 1990; Issue 4. (In Russian) [Google Scholar]
- Tereshchenko, A.V.; Artemenko, A.B.; Pudov, V.Y. A hidden source of increasing the production of broiler chickens. Eksklyuziv Agro [Exclus. Agro] 2007, 4, 64–65. (In Russian) [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018; Available online: https://www.r-project.org/ (accessed on 30 August 2024).
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Purcell, S. PLINK 1.9; Center for Human Genetic Research: Boston, MA, USA; Massachusetts General Hospital: Boston, MA, USA; Broad Institute of Harvard & MIT: Cambridge, MA, USA, 2017; Available online: https://zzz.bwh.harvard.edu/plink/index.shtml (accessed on 30 August 2024).
- DataCamp. Principal Component Analysis in R Tutorial; Tutorials, R Programming; DataCamp, Inc.: New York, NY, USA, 2023; Available online: https://www.datacamp.com/tutorial/pca-analysis-r (accessed on 30 August 2024).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009; ISBN 978-0-387-98141-3. [Google Scholar] [CrossRef]
- GitHub. ggplot2. Tidyverse; GitHub, Inc.: San Francisco, CA, USA, 2023; Available online: https://github.com/tidyverse/ggplot2 (accessed on 30 August 2024).
- R Core Team. R-4; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://cran.r-project.org/src/base/R-4/ (accessed on 30 August 2024).
- GitHub. qqman; GitHub, Inc.: San Francisco, CA, USA, 2024; Available online: https://github.com/qqman (accessed on 30 August 2024).
- Turner, S.D. qqman: An R package for visualizing GWAS results using Q-Q and Manhattan plots. J. Open Source Softw. 2018, 3, 731. [Google Scholar] [CrossRef]
- NCBI. Gallus gallus Genome Assembly GRCg6a; Genome; National Library of Medicine: Bethesda, MD, USA, 2018. Available online: https://ncbi.nlm.nih.gov/datasets/genome/GCF_000002315.6/ (accessed on 30 August 2024).
- NCBI. Gallus gallus (Chicken); Genome; National Library of Medicine: Bethesda, MD, USA, 2024. Available online: https://www.ncbi.nlm.nih.gov/datasets/genome/?taxon=9031 (accessed on 30 August 2024).
- Kinsella, R.J.; Kähäri, A.; Haider, S.; Zamora, J.; Proctor, G.; Spudich, G.; Almeida-King, J.; Staines, D.; Derwent, P.; Kerhornou, A.; et al. Ensembl BioMarts: A hub for data retrieval across taxonomic space. Database 2011, 2011, bar030. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Gong, Y.; Wu, C.; Jiang, J.; Wang, Y.; Li, K. Variation of meat quality traits among five genotypes of chicken. Poult. Sci. 2009, 88, 2212–2218. [Google Scholar] [CrossRef]
- Sun, Y.; Zhao, G.; Liu, R.; Zheng, M.; Hu, Y.; Wu, D.; Zhang, L.; Li, P.; Wen, J. The identification of 14 new genes for meat quality traits in chicken using a genome-wide association study. BMC Genom. 2013, 14, 458. [Google Scholar] [CrossRef] [PubMed]
- Devatkal, S.K.; Vishnuraj, M.R.; Kulkarni, V.V.; Kotaiah, T. Carcass and meat quality characterization of indigenous and improved variety of chicken genotypes. Poult. Sci. 2018, 97, 2947–2956. [Google Scholar] [CrossRef]
- Tarsani, E.; Kranis, A.; Maniatis, G.; Avendano, S.; Hager-Theodorides, A.L.; Kominakis, A. Discovery and characterization of functional modules associated with body weight in broilers. Sci. Rep. 2019, 9, 9125. [Google Scholar] [CrossRef]
- Habimana, R.; Ngeno, K.; Okeno, T.O.; Hirwa, C.D.A.; Keambou Tiambo, C.; Yao, N.K. Genome-wide association study of growth performance and immune response to Newcastle disease virus of indigenous chicken in Rwanda. Front. Genet. 2021, 12, 723980. [Google Scholar] [CrossRef]
- Li, Y.D.; Bai, X.; Liu, X.; Wang, W.J.; Li, Z.W.; Wang, N.; Xiao, F.; Gao, H.H.; Guo, H.S.; Li, H.; et al. Integration of genome-wide association study and selection signatures reveals genetic determinants for skeletal muscle production traits in an F2 chicken population. J. Integr. Agric. 2022, 21, 2065–2075. [Google Scholar] [CrossRef]
- Wang, S.; Wang, Y.; Li, Y.; Xiao, F.; Guo, H.; Gao, H.; Wang, N.; Zhang, H.; Li, H. Genome-wide association study and selective sweep analysis reveal the genetic architecture of body weights in a chicken F2 resource population. Front. Vet. Sci. 2022, 9, 875454. [Google Scholar] [CrossRef]
- Yang, R.; Xu, Z.; Wang, Q.; Zhu, D.; Bian, C.; Ren, J.; Huang, Z.; Zhu, X.; Tian, Z.; Wang, Y.; et al. Genome-wide association study and genomic prediction for growth traits in yellow-plumage chicken using genotyping-by-sequencing. Genet. Sel. Evol. 2021, 53, 82. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Y.; Li, Y.; Wu, J.; Wang, X.; Bian, C.; Tian, Y.; Sun, G.; Han, R.; Liu, X.; et al. Genome-wide association study reveals the genetic determinism of growth traits in a Gushi-Anka F2 chicken population. Heredity 2021, 126, 293–307. [Google Scholar] [CrossRef]
- Pampouille, E.; Berri, C.; Boitard, S.; Hennequet-Antier, C.; Beauclercq, S.A.; Godet, E.; Praud, C.; Jégo, Y.; Le Bihan-Duval, E. Mapping QTL for white striping in relation to breast muscle yield and meat quality traits in broiler chickens. BMC Genom. 2018, 19, 202. [Google Scholar] [CrossRef]
- Paredes, M.; Vásquez, B. Growth, carcass characteristics, weight of internal organs and meat proximate composition of six genotypes in chickens reared in Andean region of northern Peruvian. Sci. Agropecu. 2020, 11, 365–374. [Google Scholar] [CrossRef]
- Sun, J.; Tan, X.; Yang, X.; Bai, L.; Kong, F.; Zhao, G.; Wen, J.; Liu, R. Identification of candidate genes for meat color of chicken by combing selection signature analyses and differentially expressed genes. Genes 2022, 13, 307. [Google Scholar] [CrossRef] [PubMed]
- Dunn, I.C.; Miao, Y.-W.; Morris, A.; Romanov, M.; Wilson, P.W.; Sharp, P.J. The Detection and Assay of Polymorphism in Candidate Reproductive Gene Loci in a Commercial Broiler Breeder Population for Association Studies. In Proceedings of the Poultry Genetics Symposium, Mariensee, Germany, 6–8 October 1999; Preisinger, R., Ed.; Working Group 3 of WPSA, Lohmann Tierzucht: Cuxhaven, Germany, 1999; p. 113. [Google Scholar]
- Zhang, G.X.; Fan, Q.C.; Zhang, T.; Wang, J.Y.; Wang, W.H.; Xue, Q.; Wang, Y.J. Genome-wide association study of growth traits in the Jinghai Yellow chicken. Genet. Mol. Res. 2015, 14, 15331–15338. [Google Scholar] [CrossRef] [PubMed]
- Walugembe, M.; Amuzu-Aweh, E.N.; Botchway, P.K.; Naazie, A.; Aning, G.; Wang, Y.; Saelao, P.; Kelly, T.; Gallardo, R.A.; Zhou, H.; et al. Genetic basis of response of Ghanaian local chickens to infection with a lentogenic new castle disease virus. Front. Genet. 2020, 11, 739. [Google Scholar] [CrossRef]
- Kang, H.; Zhao, D.; Xiang, H.; Li, J.; Zhao, G.; Li, H. Large-scale transcriptome sequencing in broiler chickens to identify candidate genes for breast muscle weight and intramuscular fat content. Genet. Sel. Evol. 2021, 53, 66. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Duan, X.; An, B.; Chang, T.; Liang, M.; Xu, L.; Zhang, L.; Li, J.; E, G.; Gao, H. Genome-wide association study based on random regression model reveals candidate genes associated with longitudinal data in Chinese Simmental beef cattle. Animals 2021, 11, 2524. [Google Scholar] [CrossRef]
- Wang, W.; Zhang, Y.; Zhang, X.; Li, C.; Yuan, L.; Zhang, D.; Zhao, Y.; Li, X.; Cheng, J.; Lin, C.; et al. Heritability and recursive influence of host genetics on the rumen microbiota drive body weight variance in male Hu sheep lambs. Microbiome 2023, 11, 197. [Google Scholar] [CrossRef]
- Mohammadi, H.; Khalababdi Farahani, A.H.; Moradi, M.H. The genome-wide study in Japanese quail for traits related to feed efficiency using a single step approach. Anim. Prod. 2022, 24, 117–126. [Google Scholar] [CrossRef]
- Tiezzi, F.; Brito, L.F.; Howard, J.; Huang, Y.J.; Gray, K.; Schwab, C.; Fix, J.; Maltecca, C. Genomics of heat tolerance in reproductive performance investigated in four independent maternal lines of pigs. Front. Genet. 2020, 11, 629. [Google Scholar] [CrossRef] [PubMed]
- Do, D.N.; Strathe, A.B.; Ostersen, T.; Jensen, J.; Mark, T.; Kadarmideen, H.N. Genome-wide association study reveals genetic architecture of eating behavior in pigs and its implications for humans obesity by comparative mapping. PLoS ONE 2013, 8, e71509. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Chu, Q.; Guo, G.; Dong, G.; Li, X.; Zhang, Q.; Zhang, S.; Zhang, Z.; Wang, Y. Genome-wide association studies identified multiple genetic loci for body size at four growth stages in Chinese Holstein cattle. PLoS ONE 2017, 12, e0175971. [Google Scholar] [CrossRef] [PubMed]
- Fan, B.; Onteru, S.K.; Du, Z.Q.; Garrick, D.J.; Stalder, K.J.; Rothschild, M.F. Genome-wide association study identifies Loci for body composition and structural soundness traits in pigs. PLoS ONE 2011, 6, e14726. [Google Scholar] [CrossRef]
- Liu, R.; Sun, Y.; Zhao, G.; Wang, F.; Wu, D.; Zheng, M.; Chen, J.; Zhang, L.; Hu, Y.; Wen, J. Genome-wide association study identifies loci and candidate genes for body composition and meat quality traits in Beijing-You chickens. PLoS ONE 2013, 8, e61172. [Google Scholar] [CrossRef]
- Wang, W.; Zhang, T.; Wang, J.; Zhang, G.; Wang, Y.; Zhang, Y.; Zhang, J.; Li, G.; Xue, Q.; Han, K.; et al. Genome-wide association study of 8 carcass traits in Jinghai Yellow chickens using specific-locus amplified fragment sequencing technology. Poult. Sci. 2016, 95, 500–506. [Google Scholar] [CrossRef]
- Volkova, N.A.; Romanov, M.N.; Abdelmanova, A.S.; Larionova, P.V.; German, N.Y.; Vetokh, A.N.; Shakhin, A.V.; Volkova, L.A.; Sermyagin, A.A.; Anshakov, D.V.; et al. Genome-wide association study revealed putative SNPs and candidate genes associated with growth and meat traits in Japanese quail. Genes 2024, 15, 294. [Google Scholar] [CrossRef]
- Abdalla, B.A.; Chen, J.; Nie, Q.; Zhang, X. Genomic insights into the multiple factors controlling abdominal fat deposition in a chicken model. Front. Genet. 2018, 9, 262. [Google Scholar] [CrossRef] [PubMed]
- Moreira, G.C.M.; Boschiero, C.; Cesar, A.S.M.; Reecy, J.M.; Godoy, T.F.; Pértille, F.; Ledur, M.C.; Moura, A.S.A.M.T.; Garrick, D.J.; Coutinho, L.L. Integration of genome wide association studies and whole genome sequencing provides novel insights into fat deposition in chicken. Sci. Rep. 2018, 8, 16222. [Google Scholar] [CrossRef]
- Volkova, N.A.; German, N.Y.; Larionova, P.V.; Vetokh, A.N.; Romanov, M.N.; Zinovieva, N.A. Identification of SNPs and candidate genes associated with abdominal fat deposition in quails (Coturnix japonica). Sel’skokhozyaistvennaya Biol. Agric. Biol. 2023, 58, 1079–1087. [Google Scholar] [CrossRef]
- Li, F.; Hu, G.; Zhang, H.; Wang, S.; Wang, Z.; Li, H. Epistatic effects on abdominal fat content in chickens: Results from a genome-wide SNP-SNP interaction analysis. PLoS ONE 2013, 8, e81520. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Cui, H.; Jin, Y.; Zhao, W.; Liu, X.; Wang, Y.; Ding, J.; Liu, L.; Wen, J.; Zhao, G. Fatty acid metabolism-related genes are associated with flavor-presenting aldehydes in Chinese local chicken. Front. Genet. 2022, 13, 902180. [Google Scholar] [CrossRef] [PubMed]
- Valdés-Hernández, J.; Folch, J.M.; Crespo-Piazuelo, D.; Passols, M.; Sebastià, C.; Criado-Mesas, L.; Castelló, A.; Sánchez, A.; Ramayo-Caldas, Y. Identification of candidate regulatory genes for intramuscular fatty acid composition in pigs by transcriptome analysis. Genet. Sel. Evol. 2024, 56, 12. [Google Scholar] [CrossRef]
- Lawal, R.A.; Hanotte, O. Domestic chicken diversity: Origin, distribution, and adaptation. Anim. Genet. 2021, 52, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xue, X.; Liu, Y.; Abied, A.; Ding, Y.; Zhao, S.; Wang, W.; Ma, L.; Guo, J.; Guan, W.; et al. Genome-wide comparative analyses reveal selection signatures underlying adaptation and production in Tibetan and Poll Dorset sheep. Sci. Rep. 2021, 11, 2466. [Google Scholar] [CrossRef]
- Pauciullo, A.; Küpper, J.; Brandt, H.; Donat, K.; Iannuzzi, L.; Erhardt, G. Wingless-type MMTV integration site family member 2 (WNT2) gene is associated with resistance to MAP in faecal culture and antibody response in Holstein cattle. Anim. Genet. 2015, 46, 122–132. [Google Scholar] [CrossRef]
- Drobik-Czwarno, W.; Wolc, A.; Petal, C.R.; Miedzinska, K.; Dekkers, J.; Fulton, J.E.; Smith, J. Candidate genes associated with survival following highly pathogenic avian influenza infection in chickens. Int. J. Mol. Sci. 2024, 25, 10056. [Google Scholar] [CrossRef]

| Trait | Mean | SD | Min–Max | CV, % |
|---|---|---|---|---|
| BW at 14-day age, g | 215.7 | 45.7 | 92.8–396.1 | 21.2 |
| BW at 28-day age, g | 611.6 | 111.7 | 341.6–902.4 | 18.3 |
| BW at 42-day age, g | 1132.9 | 207.4 | 644.2–1690.1 | 18.3 |
| BW at 63-day age, g | 1829.1 | 377.9 | 963.9–2747.7 | 20.7 |
| Average daily BW gain, g | 28.7 | 6.1 | 14.6–43.0 | 21.4 |
| Slaughter weight, % | 71.1 | 4.1 | 55.4–80.1 | 5.8 |
| Dressed carcass weight, g | 1346.4 | 309.1 | 665.3–2032.1 | 23.0 |
| Breast weight, g | 385.3 | 100.5 | 144.8–632.6 | 26.1 |
| Thigh weight, g | 104.1 | 25.8 | 49.4–163.2 | 24.8 |
| Drumstick weight, g | 88.6 | 18.3 | 42.7–133.3 | 20.6 |
| Wing weight, g | 77.8 | 15.8 | 33.4–119.1 | 20.3 |
| Trait | No. of SNPs | Chromosomes |
|---|---|---|
| BW at 14-day age | - | - |
| BW at 28-day age | 2 | 2, 6 |
| BW at 42-day age | 34 | 1, 2, 6–8, 11, 13–14, 21, 23, 28 |
| BW at 63-day age | 69 | 1–5, 7, 8, 10, 11, 13–15, 17, 21, 23, 27, 28 |
| Average daily BW gain | 148 | 1–15, 17, 18, 20–28 |
| Slaughter weight | - | - |
| Dressed carcass weight | 30 | 1–9, 13, 15, 17–19, 23, 24, 27 |
| Breast weight | 16 | 1-4, 7-9, 15, 18, 19, 23, 27 |
| Thigh weight | 1 | 10 |
| Drumstick weight | 21 | 1–2, 4–5, 7, 9, 13, 15, 17–19, 23, 26 |
| Wing weight | - | - |
| GGA 1 | SNP | Position, bp | Traits 2 | Genes |
|---|---|---|---|---|
| 1 | Gga_rs14800862 | 24,842,665 | DCW, BrW, DW | CTTNBP2, CFTR, ASZ1, WNT2, ST7, CAPZA2 |
| 1 | Gga_rs14902811 | 152,430,990 | BW42, BW63, ADBWG | - |
| 1 | Gga_rs14902833 | 152,488,231 | BW42, BW63, ADBWG | SLC2A13 |
| 1 | GGaluGA050529 | 152,453,938 | BW42, BW63, ADBWG | SLC2A13 |
| 1 | GGaluGA034658 | 102,412,092 | BW42, BW63, ADBWG | - |
| 2 | Gga_rs14160005 | 31,441,781 | BW42, BW63, ADBWG, DCW, BrW, DW | IGF2BP3, TRA2A, CCDC126, FAM221A, STK31, NPY, PALS2, DFNA5 |
| 2 | Gga_rs14248546 | 125,490,179 | BW42, BW63, ADBWG | TRIQK |
| 2 | Gga_rs15168561 | 136,710,388 | BW28, BW42, BW63, ADBWG | ENPP2, TAF2, DSCC1, DEPTOR, COL14A1 |
| 2 | Gga_rs16088599 | 103,517,528 | BW42, BW63, ADBWG | OSBPL1A, IMPACT, ZNF521 |
| 3 | Gga_rs14356736 | 48,921,434 | BW63, ADBWG, DCW, BrW | PLEKHG1, MTHFD1L, AKAP12, ZBTB2, RMND1, ARMT1, CCDC170, ESR1 |
| 4 | Gga_rs13516467 | 38,746,248 | BW63, ADBWG, DCW, BrW, DW | NPNT, GSTCD, INTS12, ARHGEF38, PPA2, TET2 |
| 4 | GGaluGA246480 | 12,518,793 | DCW, BrW, DW | SLC16A2, RLIM, NEXMIF, gga-mir-1573, ABCB7, UPRT, ZDHHC15 |
| 7 | Gga_rs13737657 | 14,269,161 | BW42, BW63, ADBWG, DCW, BrW, DW | U4, PDE1A, PPP1R1C, ITPRID2, NEUROD1, ITGA4 |
| 7 | Gga_rs14622272 | 28,057,143 | BW42, BW63, ADBWG | KALRN, ACADL, UMPS, ITGB5, HEG1, MYL1, ZNF148, SNX4, OSBPL11, LMLN, DTX3L |
| 7 | Gga_rs14622611 | 28,327,789 | BW42, BW63, ADBWG | MYL1, OSBPL11, LMLN, DTX3L, PARP9, LANCL1, FAIM, CEP70, ESYT3, CFAP221, SCTR, TMEM37, DBI, C7H2ORF76, STEAP3, CPS1, C1QL2, MARCO, EN1 |
| 7 | Gga_rs15848860 | 14,393,379 | BW42, BW63, ADBWG, DCW, BrW, DW | U4, PDE1A, PPP1R1C, ITPRID2, NEUROD1, ITGA4 |
| 7 | GGaluGA308586 | 2,639,082 | BW42, BW63, ADBWG, DCW, DW | CNTNAP5, MAP2, MRAS, gga-mir-3530, TMEM177, PTPN4, EPB41L5, RALB, INHBB, GLI2, UNC80, TFCP2L1, CLASP1, NIFK, TSN, IQCB1, EAF2, SLC15A2, HSPBAP1, SLC49A4, SEMA5B, PDIA5, SEC22A, ADCY6, KANSL1L, HACD2, MYLK, CCDC14, KALRN, ACADL, UMPS, ITGB5, HEG1, MYL1, ZNF148, SNX4, OSBPL11, LMLN, DTX3L, PARP9, LANCL1, FAIM, CEP70, ESYT3, CFAP221, SCTR, TMEM37, DBI, C7H2ORF76 |
| 8 | Gga_rs16640785 | 22,847,287 | BW63, ADBWG, DCW, BrW | TRABD2B, SLC5A9, SPATA6, gga-mir-1809 |
| 8 | GGaluGA330152 | 22,760,396 | BW42, BW63, ADBWG | TRABD2B |
| 9 | Gga_rs15947559 | 11,450,206 | DCW, BrW, DW | PLOD2 |
| 13 | Gga_rs15677377 | 8,879,549 | BW63, ADBWG, DCW, DW | TTC1, ADRA1B, IL12B, FBXO38, HTR4, gga-mir-458a, SLC26A2 |
| 13 | Gga_rs15679261 | 8,271,910 | BW42, BW63, ADBWG | GABRB2, ATP10B |
| 13 | Gga_rs15680269 | 7,909,523 | BW42, BW63, ADBWG | - |
| 13 | GGaluGA093626 | 9,139,110 | BW63, ADBWG, DCW | gga-mir-458a, HTR4, SLC26A2, CSNK1A1, gga-mir-145, gga-mir-143, IL17B, PCYOX1L, GRPEL2, AFAP1L1, ABLIM3 |
| 14 | Gga_rs15003767 | 2,062,529 | BW42, BW63, ADBWG | FAM20C, FOXL3 |
| 15 | GGaluGA109523 | 8,381,798 | BW63, ADBWG, DCW, BrW, DW | DGCR2, VPS29L, VPREB3, CHCHD10, MMP11, SMARCB1, DERL3, SLC2A11, SLC2A11L1, MIF, DDX51, GSTT1, DDTL, CABIN1, TBX6, CRKL |
| 17 | Gga_rs14102454 | 3,408,140 | BW63, ADBWG, DCW, DW | PAPPA, ASTN2 |
| 18 | Gga_rs16347495 | 9,967,210 | DCW, BrW, DW | TIMP2, USP36, CYTH1, PGS1, SOCS3, AFMID, TK1, SYNGR2, TMC6, ARL16, HGS, MRPL12, GCGR, MCRIP1, PPP1R27, P4HB, ARHGDIA, ALYREF, NPB, PCYT2, SIRT7, MAFG, PYCR1, NME1, SPAG9, PITPNM3, FBXO39, TEKT1, SMTNL2 |
| 19 | GGaluGA126188 | 4,370,123 | DCW, BrW, DW | CUX1, PRKRIP1, ORAI3, ALKBH4, LRWD1, RASA4B, UPK3B, DTX2, SSC4D, YWHAG, HSPB1, SRRM3, MDH2, TMEM120A, POR, TAF15, MMP28, RASL10B, AP2B1 |
| 21 | Gga_rs15182225 | 2,760,476 | BW42, BW63, ADBWG | TNFRSF18, gga-mir-429, gga-mir-200a, gga-mir-200b, gga-mir-6680, C1orf159 |
| 23 | GGaluGA188509 | 2,994,311 | BW42, BW63, ADBWG, DCW, BrW, DW | EPB41, TMEM200B, SRSF4, MECR, PTPRU, gga-mir-1724, PTPRU |
| 27 | Gga_rs13620324 | 4,812,782 | BW63, ADBWG, BrW | CRHR1, ITGB3, METTL2B, TLK2, MRC2, TANC2 |
| 28 | Gga_rs14306444 | 1,714,462 | BW42, BW63, ADBWG | ZBTB7A, PIAS4, EEF2, gga-mir-1434, NMRK2, ATCAY, NRTN, DUS3L, LARP6L, RFX2, ACSBG2, MLLT1, ACER1, ANP32B, ZNF414, MYO1F, ADAMTS10, gga-mir-6615, ZAP70 |
| 28 | Gga_rs14306581 | 1,592,968 | BW42, BW63, ADBWG | NCLN, CELF5, HSD11B1L, MICOS13, gga-mir-1774, FSD1, YJU2, gga-mir-6593, ZBTB7A, PIAS4, EEF2, gga-mir-1434, NMRK2, ATCAY, NRTN, DUS3L, LARP6L, RFX2, ACSBG2, MLLT1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Volkova, N.A.; Romanov, M.N.; Vetokh, A.N.; Larionova, P.V.; Volkova, L.A.; Abdelmanova, A.S.; Sermyagin, A.A.; Griffin, D.K.; Zinovieva, N.A. Genome-Wide Association Study Reveals the Genetic Architecture of Growth and Meat Production Traits in a Chicken F2 Resource Population. Genes 2024, 15, 1246. https://doi.org/10.3390/genes15101246
Volkova NA, Romanov MN, Vetokh AN, Larionova PV, Volkova LA, Abdelmanova AS, Sermyagin AA, Griffin DK, Zinovieva NA. Genome-Wide Association Study Reveals the Genetic Architecture of Growth and Meat Production Traits in a Chicken F2 Resource Population. Genes. 2024; 15(10):1246. https://doi.org/10.3390/genes15101246
Chicago/Turabian StyleVolkova, Natalia A., Michael N. Romanov, Anastasia N. Vetokh, Polina V. Larionova, Ludmila A. Volkova, Alexandra S. Abdelmanova, Alexander A. Sermyagin, Darren K. Griffin, and Natalia A. Zinovieva. 2024. "Genome-Wide Association Study Reveals the Genetic Architecture of Growth and Meat Production Traits in a Chicken F2 Resource Population" Genes 15, no. 10: 1246. https://doi.org/10.3390/genes15101246
APA StyleVolkova, N. A., Romanov, M. N., Vetokh, A. N., Larionova, P. V., Volkova, L. A., Abdelmanova, A. S., Sermyagin, A. A., Griffin, D. K., & Zinovieva, N. A. (2024). Genome-Wide Association Study Reveals the Genetic Architecture of Growth and Meat Production Traits in a Chicken F2 Resource Population. Genes, 15(10), 1246. https://doi.org/10.3390/genes15101246









