Tracing ALS Degeneration: Insights from Spinal Cord and Cortex Transcriptomes
Abstract
:1. Introduction
2. Methods
2.1. Dataset and Pre-Processing
2.2. Variant Analysis
2.3. Differential Gene Expression Analysis
2.4. Variant Effect Prediction
2.5. Enrichment Analysis and Network Analysis of DEGs and VGs
2.6. Metabolic Activity Prediction of Differentially Expressed Genes in Frontal Cortex and Spinal Cord Samples
3. Results
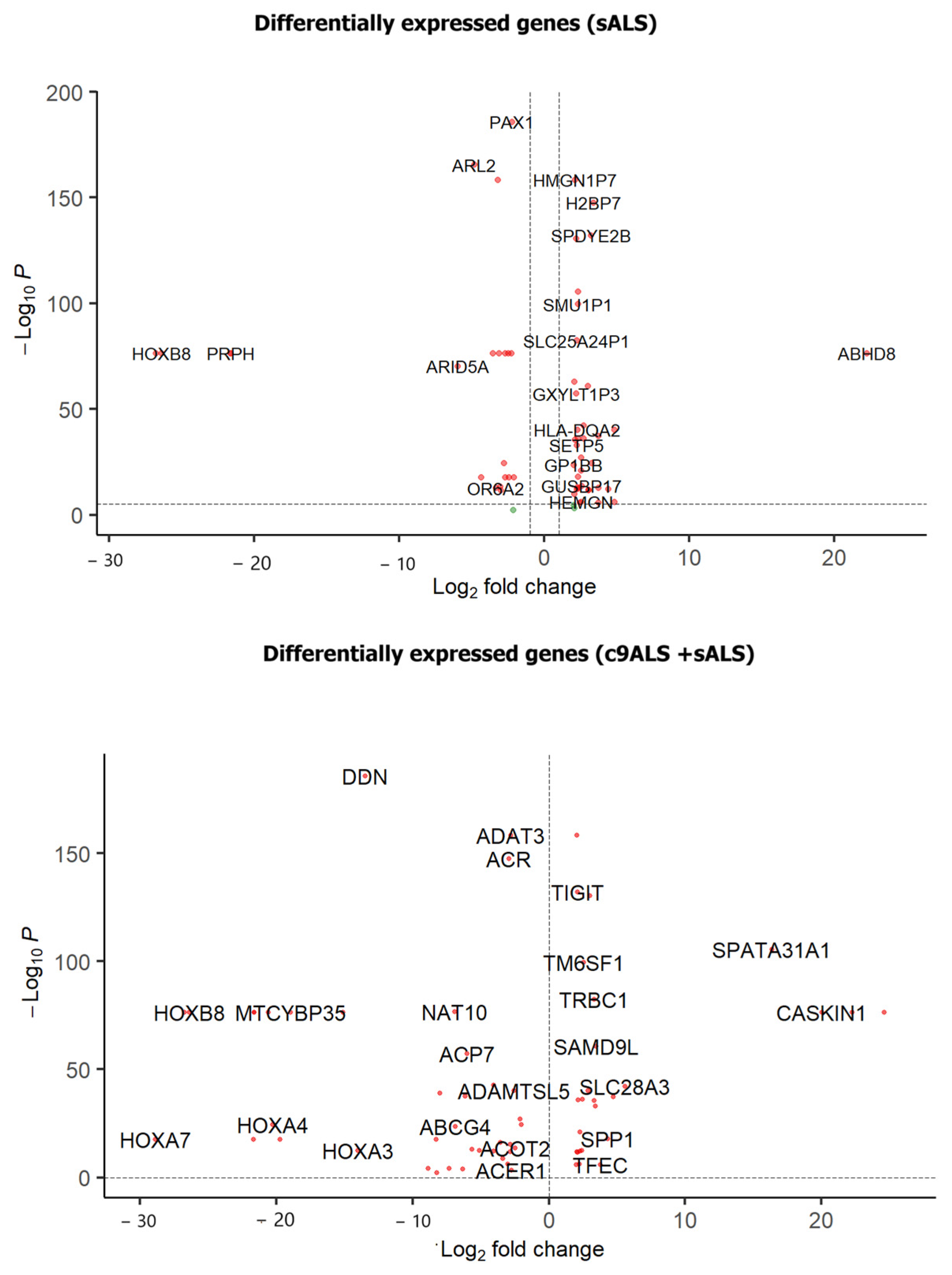
3.1. Differential Gene Expression Analysis
3.2. Novel Gene Expression Patterns Identified Through the DEG Analysis
3.2.1. Novel Genes of DGE1 (c9ALS+sALS) Study
3.2.2. Novel Expression Signature of Common Genes from DGE1 and DGE2
3.2.3. Novel Genes of DGE2 (sALS) Study
3.3. Differential Gene Expression Pattern Across Studies
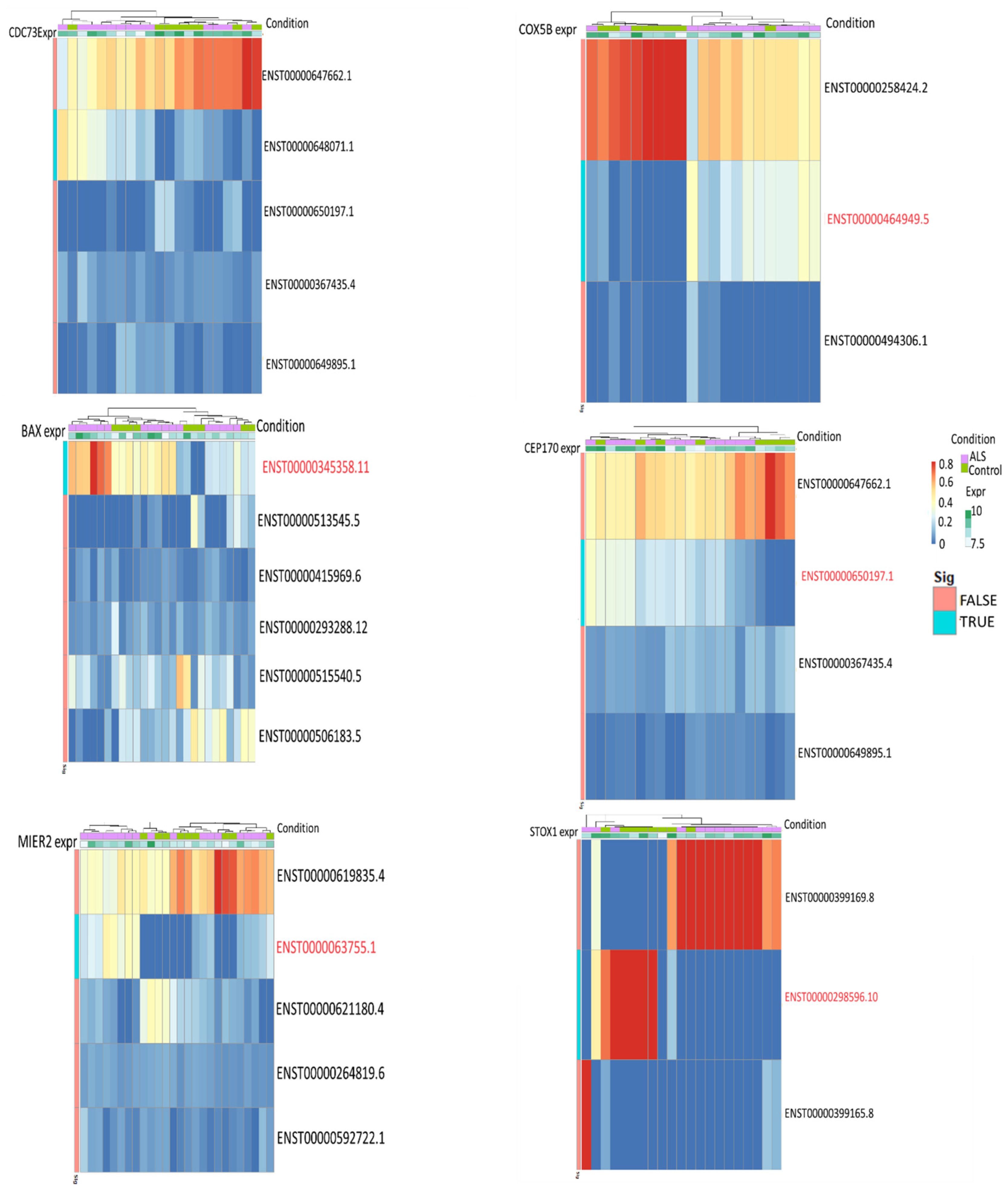
3.4. Differential Transcript Usage/Expression Analysis
3.5. Variant Analysis of Frontal Cortex and Spinal Cord ALS Samples
3.6. Variant Effect on Transcription Factor Binding
3.7. Variant Effect on miRNA Binding
3.8. Variant Effect on Epigenetic Regulation
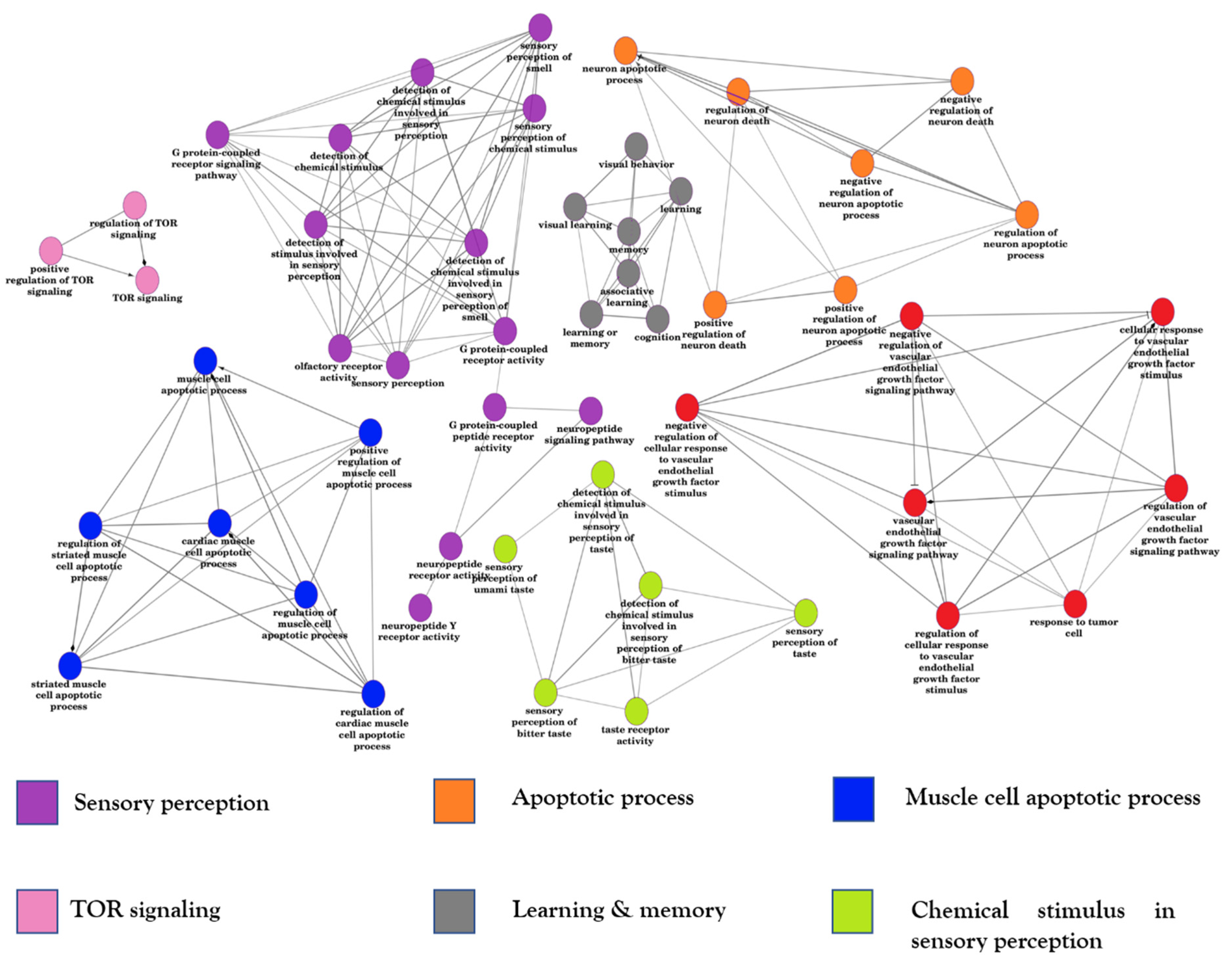
3.9. Function Enrichment on DEGs and Variant Genes
3.9.1. Function Enrichment Network of Spinal Cord and Frontal Cortex DEGs
3.9.2. Gene Set Enrichment Analysis on DEGs
3.9.3. Enrichment Study on Variant-Associated Genes
3.10. Metabolic Activity of DEGs
3.11. Limitations
4. Proposed Hypothesis and Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Miller, R.G.; Mitchell, J.D.; Moore, D.H. Riluzole for amyotrophic lateral sclerosis (ALS)/motor neuron disease (MND). Cochrane Database Syst. Rev. 2012, 2012, CD001447. [Google Scholar] [CrossRef] [PubMed]
- Rothstein, J.D. Edaravone: A new drug approved for ALS. Cell 2017, 171, 725. [Google Scholar] [CrossRef] [PubMed]
- Rizzuti, M.; Sali, L.; Melzi, V.; Scarcella, S.; Costamagna, G.; Ottoboni, L.; Quetti, L.; Brambilla, L.; Papadimitriou, D.; Verde, F.; et al. Genomic and transcriptomic advances in amyotrophic lateral sclerosis. Ageing Res. Rev. 2023, 92, 102126. [Google Scholar] [CrossRef] [PubMed]
- Varghese, A.M.; Ghosh, M.; Bhagat, S.K.; Vijayalakshmi, K.; Preethish-Kumar, V.; Vengalil, S.; Chevula, P.C.; Nashi, S.; Polavarapu, K.; Sharma, M.; et al. Chitotriosidase, a biomarker of amyotrophic lateral sclerosis, accentuates neurodegeneration in spinal motor neurons through neuroinflammation. J. Neuroinflamm. 2020, 17, 232. [Google Scholar] [CrossRef] [PubMed]
- Perrone, B.; La Cognata, V.; Sprovieri, T.; Ungaro, C.; Conforti, F.L.; Andò, S.; Cavallaro, S. Alternative Splicing of ALS Genes: Misregulation and Potential Therapies. Cell. Mol. Neurobiol. 2020, 40, 1–14. [Google Scholar] [CrossRef]
- La Cognata, V.; Gentile, G.; Aronica, E.; Cavallaro, S. Splicing players are differently expressed in sporadic amyotrophic lateral sclerosis molecular clusters and brain regions. Cells 2020, 9, 159. [Google Scholar] [CrossRef]
- Provenzano, F.; Torazza, C.; Bonifacino, T.; Bonanno, G.; Milanese, M. The Key Role of Astrocytes in Amyotrophic Lateral Sclerosis and Their Commitment to Glutamate Excitotoxicity. Int. J. Mol. Sci. 2023, 24, 15430. [Google Scholar] [CrossRef]
- Geloso, M.C.; Corvino, V.; Marchese, E.; Serrano, A.; Michetti, F.; D’ambrosi, N. The dual role of microglia in ALS: Mechanisms and therapeutic approaches. Front. Aging Neurosci. 2017, 9, 242. [Google Scholar] [CrossRef]
- D’Erchia, A.M.; Gallo, A.; Manzari, C.; Raho, S.; Horner, D.S.; Chiara, M.; Valletti, A.; Aiello, I.; Mastropasqua, F.; Ciaccia, L.; et al. Massive transcriptome sequencing of human spinal cord tissues provides new insights into motor neuron degeneration in ALS. Sci. Rep. 2017, 7, 10046. [Google Scholar] [CrossRef]
- MacLean, M.; López-Díez, R.; Vasquez, C.; Gugger, P.F.; Schmidt, A.M. Neuronal–glial communication perturbations in murine SOD1G93A spinal cord. Commun. Biol. 2022, 5, 177. [Google Scholar] [CrossRef]
- Yamashita, H.; Komine, O.; Fujimori-Tonou, N.; Yamanaka, K. Comprehensive expression analysis with cell-type-specific transcriptome in ALS-linked mutant SOD1 mice: Revisiting the active role of glial cells in disease. Front. Cell. Neurosci. 2023, 16, 1045647. [Google Scholar] [CrossRef] [PubMed]
- Scamps, F.; Aimond, F.; Hilaire, C.; Raoul, C. Synaptic Transmission and Motoneuron Excitability Defects in Amyotrophic Lateral Sclerosis; Exon Publications: Brisbane City, QLD, Australia, 2021; pp. 55–94. [Google Scholar]
- Cunha-Oliveira, T.; Montezinho, L.; Mendes, C.; Firuzi, O.; Saso, L.; Oliveira, P.J.; Silva, F.S.G. Oxidative stress in amyotrophic lateral sclerosis: Pathophysiology and opportunities for pharmacological intervention. Oxidative Med. Cell. Longev. 2020, 2020, 5021694. [Google Scholar] [CrossRef] [PubMed]
- McCombe, P.A.; DHenderson, R. The role of immune and inflammatory mechanisms in ALS. Curr. Mol. Med. 2011, 11, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Prudencio, M.; Belzil, V.V.; Batra, R.; Ross, C.A.; Gendron, T.F.; Pregent, L.J.; Murray, M.E.; Overstreet, K.K.; Piazza-Johnston, A.E.; Desaro, P.; et al. Distinct brain transcriptome profiles in C9orf72-associated and sporadic ALS. Nat. Neurosci. 2015, 18, 1175–1182. [Google Scholar] [CrossRef] [PubMed]
- Batra, R.; Hutt, K.; Vu, A.; Rabin, S.J.; Baughn, M.W.; Libby, R.T.; Hoon, S.; Ravits, J.; Yeo, G.W. Gene expression signatures of sporadic ALS motor neuron populations. bioRxiv 2016. [Google Scholar] [CrossRef]
- Patel, R.K.; Jain, M. NGS QC toolkit: A toolkit for quality control of next generation sequencing data. PLoS ONE 2012, 7, e30619. [Google Scholar] [CrossRef] [PubMed]
- Blankenberg, D.; Gordon, A.; Von Kuster, G.; Coraor, N.; Taylor, J.; Nekrutenko, A.; Team, G. Manipulation of FASTQ data with galaxy. Bioinformatics 2010, 26, 1783–1785. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Dobin, A.; Gingeras, T.R. Mapping RNA-seq Reads with STAR. Curr. Protoc. Bioinform. 2015, 51, 11–14. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 2011, 27, 2987–2993. [Google Scholar] [CrossRef] [PubMed]
- Koboldt, D.C.; Zhang, Q.; Larson, D.E.; Shen, D.; McLellan, M.D.; Lin, L.; Miller, C.A.; Mardis, E.R.; Ding, L.; Wilson, R.K. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012, 22, 568–576. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.L.; Marcora, E.; Pimenova, A.A.; Di Narzo, A.F.; Kapoor, M.; Jin, S.C.; Harari, O.; Bertelsen, S.; Fairfax, B.P.; Czajkowski, J.; et al. A common haplotype lowers PU.1 expression in myeloid cells and delays the onset of Alzheimer’s disease. Nat. Neurosci. 2017, 20, 1052–1061. [Google Scholar] [CrossRef]
- Deming, Y.; Li, Z.; Kapoor, M.; Harari, O.; Del Aguila, J.L.; Black, K.; Carrell, D.; Cai, Y.; Fernandez, M.V.; Budde, J.; et al. Genome-wide association study identifies four novel loci associated with Alzheimer’s endophenotypes and disease modifiers. Acta Neuropathol. 2017, 133, 839–856. [Google Scholar] [CrossRef]
- Fritsche, L.G.; Igl, W.; Bailey, J.N.C.; Grassmann, F.; Sengupta, S.; Bragg-Gresham, J.L.; Burdon, K.P.; Hebbring, S.J.; Wen, C.; Gorski, M.; et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat. Genet. 2016, 48, 134–143. [Google Scholar] [CrossRef]
- Nicolas, A.; Kenna, K.P.; Renton, A.E.; Ticozzi, N.; Faghri, F.; Chia, R.; Dominov, J.A.; Kenna, B.J.; Nalls, M.A.; Keagle, P.; et al. Genome-wide analyses identify KIF5A as a novel ALS gene. Neuron 2018, 97, 1268–1283. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Soneson, C.; Love, M.I.; Robinson, M.D. Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000Research 2016, 4, 1521. [Google Scholar] [CrossRef]
- Robinson, M.D.; Nowicka, M. DRIMSeq: A Dirichlet-multinomial framework for multivariate count outcomes in genomics. F1000Research 2016, 5, 1356. [Google Scholar]
- Tekath, T.; Dugas, M. Differential transcript usage analysis of bulk and single-cell RNA-seq data with DTUrtle. Bioinformatics 2021, 37, 3781–3787. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.Y.; Lin YC, D.; Cui, S.; Huang, Y.; Tang, Y.; Xu, J.; Bao, J.; Li, Y.; Wen, J.; Zuo, H.; et al. MiRTarBase update 2022: An informative resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 2022, 50, D222–D230. [Google Scholar] [CrossRef] [PubMed]
- Kmetzsch, V.; Anquetil, V.; Saracino, D.; Rinaldi, D.; Camuzat, A.; Gareau, T.; Jornea, L.; Forlani, S.; Couratier, P.; Wallon, D.; et al. Plasma microRNA signature in presymptomatic and symptomatic subjects with C9orf72-associated frontotemporal dementia and amyotrophic lateral sclerosis. J. Neurol. Neurosurg. Psychiatry 2021, 92, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Ambrosini, G.; Bucher, P. SNP2TFBS—A database of regulatory SNPs affecting predicted transcription factor binding site affinity. Nucleic Acids Res. 2017, 45, D139–D144. [Google Scholar] [CrossRef]
- Wu, G.; Dawson, E.; Duong, A.; Haw, R.; Stein, L. ReactomeFIViz: The Reactome FI Cytoscape app for pathway and network-based data analysis. F1000Research 2014, 3, 146. [Google Scholar] [CrossRef]
- Zheng, Z.; Huang, D.; Wang, J.; Zhao, K.; Zhou, Y.; Guo, Z.; Zhai, S.; Xu, H.; Cui, H.; Yao, H.; et al. QTLbase: An integrative resource for quantitative trait loci across multiple human molecular phenotypes. Nucleic Acids Res. 2020, 48, D983–D991. [Google Scholar] [CrossRef]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’Ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef]
- Yu, G. clusterProfiler: An universal enrichment tool for functional and comparative study. bioRxiv 2018. [Google Scholar] [CrossRef]
- Greene, C.S.; Krishnan, A.; Wong, A.K.; Ricciotti, E.; Zelaya, R.A.; Himmelstein, D.S.; Zhang, R.; Hartmann, B.M.; Zaslavsky, E.; Sealfon, S.C.; et al. Understanding multicellular function and disease with human tissue-specific networks. Nat. Genet. 2015, 47, 569–576. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.-H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Mohanty, V.; Dede, M.; Tsai, K.; Daher, M.; Li, L.; Rezvani, K.; Chen, K. Characterizing cancer metabolism from bulk and single-cell RNA-seq data using METAFlux. Nat. Commun. 2023, 14, 4883. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Jaiswal, M.K.; Chien, J.-F.; Kozlenkov, A.; Jung, J.; Zhou, P.; Gardashli, M.; Pregent, L.J.; Engelberg-Cook, E.; Dickson, D.W.; et al. Divergent single cell transcriptome and epigenome alterations in ALS and FTD patients with C9orf72 mutation. Nat. Commun. 2023, 14, 5714. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Li, T.; Xu, Y.; Zhang, X.; Li, F.; Bai, J.; Chen, J.; Jiang, W.; Yang, K.; Ou, Q.; et al. CellMarker 2.0: An updated database of manually curated cell markers in human/mouse and web tools based on scRNA-seq data. Nucleic Acids Res. 2023, 51, D870–D876. [Google Scholar] [CrossRef] [PubMed]
- Basavarajappa, B.S.; Subbanna, S. Histone methylation regulation in neurodegenerative disorders. Int. J. Mol. Sci. 2021, 22, 4654. [Google Scholar] [CrossRef] [PubMed]
- Dutta, K.; Thammisetty, S.S.; Boutej, H.; Bareil, C.; Julien, J.-P. Mitigation of ALS pathology by neuron-specific inhibition of nuclear factor kappa B signaling. J. Neurosci. 2020, 40, 5137–5154. [Google Scholar] [CrossRef]
- Jiang, X.; Guan, Y.; Zhao, Z.; Meng, F.; Wang, X.; Gao, X.; Liu, J.; Chen, Y.; Zhou, F.; Zhou, S.; et al. Potential roles of the WNT signaling pathway in amyotrophic lateral sclerosis. Cells 2021, 10, 839. [Google Scholar] [CrossRef] [PubMed]
- Yerbury, J.; Chisholm, C.; Lum, J.; Farrawell, N. Ubiquitin homeostasis disruption, a common cause of proteostasis collapse in amyotrophic lateral sclerosis? Neural Regen. Res. 2022, 17, 2218–2220. [Google Scholar] [CrossRef]
- Shen, D.; Ji, Y.; Qiu, C.; Wang, K.; Gao, Z.; Liu, B.; Shen, Y.; Gong, L.; Yang, X.; Chen, X.; et al. Single-cell RNA sequencing analysis of microglia dissected the energy metabolism and revealed potential biomarkers in amyotrophic lateral sclerosis. Mol. Neurobiol. 2024, 61, 4473–4487. [Google Scholar] [CrossRef]
- Ekegren, T.; Grandström, E.; Lindholm, D.; Aquilonius, S.M. Upregulation of Bax protein and increased DNA degradation in ALS spinal cord motor neurons. Acta Neurol. Scand. 1999, 100, 317–321. [Google Scholar] [CrossRef]
- Choudhury, S.; Ganguly, A.; Chakrabarti, K.; Sharma, R.K.; Gupta, S.K. DNA vaccine encoding chimeric protein encompassing epitopes of human ZP3 and ZP4: Immunogenicity and characterization of antibodies. J. Reprod. Immunol. 2009, 79, 137–147. [Google Scholar] [CrossRef] [PubMed]
- Montibeller, L.; de Belleroche, J. Amyotrophic lateral sclerosis (ALS) and Alzheimer’s disease (AD) are characterised by differential activation of ER stress pathways: Focus on UPR target genes. Cell Stress Chaperones 2018, 23, 897–912. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Zheng, Y.; Pham, A.D.; Mandal, S.S.; Erdjument-Bromage, H.; Tempst, P.; Reinberg, D. Monoubiquitination of human histone H2B: The factors involved and their roles in HOX gene regulation. Mol. Cell 2005, 20, 601–611. [Google Scholar] [CrossRef]
- Schmidt, M.F.; Gan, Z.Y.; Komander, D.; Dewson, G. Ubiquitin signalling in neurodegeneration: Mechanisms and therapeutic opportunities. Cell Death Differ. 2021, 28, 570–590. [Google Scholar] [CrossRef] [PubMed]
- Derwish, R.; Paterno, G.D.; Gillespie, L.L. Differential HDAC1 and 2 recruitment by members of the MIER family. PLoS ONE 2017, 12, e0169338. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.K.; Thombre, R.; Wang, J. Autophagy as a common pathway in amyotrophic lateral sclerosis. Neurosci. Lett. 2019, 697, 34–48. [Google Scholar] [CrossRef]
- Doridot, L.; Châtre, L.; Ducat, A.; Vilotte, J.L.; Lombes, A.; Méhats, C.; Barbaux, S.; Calicchio, R.; Ricchetti, M.; Vaiman, D. Nitroso-redox balance and mitochondrial homeostasis are regulated by STOX1, a pre-eclampsia-associated gene. Antioxid. Redox Signal. 2014, 21, 819–834. [Google Scholar] [CrossRef]
- Barber, S.C.; Mead, R.J.; Shaw, P.J. Oxidative stress in ALS: A mechanism of neurodegeneration and a therapeutic target. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2006, 1762, 1051–1067. [Google Scholar] [CrossRef]
- Chen, W.; Yu, J.; Xie, R.; Zhang, S.; Zhou, T.; Xiong, C.; Huang, D.; Zhong, M. Effect of silencing CITED1 gene to regulate PI3K/AKT pathway on the biological function of PTC cells and its mechanism. Cell. Mol. Biol. 2023, 69, 113–117. [Google Scholar] [CrossRef]
- Pias, E.K.; Ekshyyan, O.Y.; Rhoads, C.A.; Fuseler, J.; Harrison, L.; Aw, T.Y. Differential effects of superoxide dismutase isoform expression on hydroperoxide-induced apoptosis in PC-12 cells. J. Biol. Chem. 2003, 278, 13294–13301. [Google Scholar] [CrossRef]
- Linker, K.; Pautz, A.; Fechir, M.; Hubrich, T.; Greeve, J.; Kleinert, H. Involvement of KSRP in the post-transcriptional regulation of human iNOS expression-complex interplay of KSRP with TTP and HuR. Nucleic Acids Res. 2005, 33, 4813–4827. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-Y.; Patterson, M.; Mikkola, H.K.A.; Lowry, W.E.; Kurdistani, S.K. Dynamic Distribution of Linker Histone H1.5 in Cellular Differentiation. PLOS Genet. 2012, 8, e1002879. [Google Scholar] [CrossRef] [PubMed]
- Busse, M.; Feta, A.; Presto, J.; Wilén, M.; Grønning, M.; Kjellén, L.; Kusche-Gullberg, M. Contribution of EXT1, EXT2, and EXTL3 to heparan sulfate chain elongation. J. Biol. Chem. 2007, 282, 32802–32810. [Google Scholar] [CrossRef] [PubMed]
- Patella, F.; Leucci, E.; Evangelista, M.; Parker, B.; Wen, J.; Mercatanti, A.; Rizzo, M.; Chiavacci, E.; Lund, A.H.; Rainaldi, G. MiR-492 impairs the angiogenic potential of endothelial cells. J. Cell. Mol. Med. 2013, 17, 1006–1015. [Google Scholar] [CrossRef]
- Calamini, B.; Santarsiero, B.D.; Boutin, J.A.; Mesecar, A.D. Kinetic, thermodynamic and X-ray structural insights into the interaction of melatonin and analogues with quinone reductase 2. Biochem. J. 2008, 413, 81–91. [Google Scholar] [CrossRef]
- Lan, L.; Nakajima, S.; Kapetanaki, M.G.; Hsieh, C.L.; Fagerburg, M.; Thickman, K.; Rodriguez-Collazo, P.; Leuba, S.H.; Levine, A.S.; Rapić-Otrin, V. Monoubiquitinated histone H2A destabilizes photolesioncontaining nucleosomes with concomitant release of UV-damaged DNA-binding protein E3 ligase. J. Biol. Chem. 2012, 287, 12036–12049. [Google Scholar] [CrossRef]
- Knox, R.J.; Jenkins, T.C.; Hobbs, S.M.; Chen, S.; Melton, R.G.; Burke, P.J. Bioactivation of 5-(Aziridin-1-yl)-2,4-dinitrobenzamide (CB 1954) by human NAD(P)H quinone oxidoreductase 2: A novel co-substrate-mediated antitumor prodrug therapy. Cancer Res. 2000, 60, 4179–4186. [Google Scholar]
- Sato, Y.; Terawaki, S.; Oikawa, D.; Shimizu, K.; Okina, Y.; Ito, H.; Tokunaga, F. Involvement of heterologous ubiquitination including linear ubiquitination in Alzheimer’s disease and amyotrophic lateral sclerosis. Front. Mol. Biosci. 2023, 10, 1089213. [Google Scholar] [CrossRef]
- Dhahri, M.; Alghrably, M.; Mohammed, H.A.; Badshah, S.L.; Noreen, N.; Mouffouk, F.; Rayyan, S.; Qureshi, K.A.; Mahmood, D.; Lachowicz, J.I.; et al. Natural polysaccharides as preventive and therapeutic horizon for neurodegenerative diseases. Pharmaceutics 2021, 14, 1. [Google Scholar] [CrossRef]
- Tang, X.; Jang, S.W.; Okada, M.; Chan, C.B.; Feng, Y.; Liu, Y.; Luo, S.W.; Hong, Y.; Rama, N.; Xiong, W.C.; et al. Netrin-1 mediates neuronal survival through PIKE-L interaction with the dependence receptor UNC5B. Nat. Cell Biol. 2008, 10, 698–706. [Google Scholar] [CrossRef]
- Vantaggiato, C.; Bondioni, S.; Airoldi, G.; Bozzato, A.; Borsani, G.; Rugarli, E.I.; Bresolin, N.; Clementi, E.; Bassi, M.T. Senataxin modulates neurite growth through fibroblast growth factor 8 signalling. Brain 2011, 134, 1808–1828. [Google Scholar] [CrossRef] [PubMed]
- Bonzo, J.R.; Norris, A.A.; Esham, M.; Moncman, C.L. The nebulette repeat domain is necessary for the proper maintenance of tropomyosin with the cardiac sarcomere. Exp. Cell Res. 2008, 314, 3519–3530. [Google Scholar] [CrossRef] [PubMed]
- Araujo, B.G.; Souza e Silva, L.F.; de Barros Torresi, J.L.; Siena, A.; Valerio BC, O.; Brito, M.D.; Rosenstock, T.R. Decreased Mitochondrial Function, Biogenesis, and Degradation in Peripheral Blood Mononuclear Cells from Amyotrophic Lateral Sclerosis Patients as a Potential Tool for Biomarker Research. Mol. Neurobiol. 2020, 57, 5084–5102. [Google Scholar] [CrossRef] [PubMed]
- Mochida, G.H.; Mahajnah, M.; Hill, A.D.; Basel-Vanagaite, L.; Gleason, D.; Hill, R.S.; Bodell, A.; Crosier, M.; Straussberg, R.; Walsh, C.A. A truncating mutation of TRAPPC9 is associated with autosomal-recessive intellectual disability and postnatal microcephaly. Am. J. Hum. Genet. 2009, 85, 897–902. [Google Scholar] [CrossRef]
- Blankman, J.L.; Long, J.Z.; Trauger, S.A.; Siuzdak, G.; Cravatt, B.F. ABHD12 controls brain lysophosphatidylserine pathways that are deregulated in a murine model of the neurodegenerative disease PHARC. Proc. Natl. Acad. Sci. USA 2013, 110, 1500–1505. [Google Scholar] [CrossRef]
- Marcadier, J.L.; Smith, A.M.; Pohl, D.; Schwartzentruber, J.; Al-Dirbashi, O.Y.; FORGE Canada Consortium; Majewski, J.; Ferdinandusse, S.; Wanders, R.J.; Bulman, D.E.; et al. Mutations in ALDH6A1 encoding methylmalonate semialdehyde dehydrogenase are associated with dysmyelination and transient methylmalonic aciduria. Orphanet J. Rare Dis. 2013, 8, 98. [Google Scholar] [CrossRef]
- Senda, M.; Ito, A.; Tsuchida, A.; Hagiwara, T.; Kaneda, T.; Nakamura, Y.; Kasama, K.; Kiso, M.; Yoshikawa, K.; Katagiri, Y.; et al. Identification and expression of a sialyltransferase responsible for the synthesis of disialylgalactosylgloboside in normal and malignant kidney cells: Downregulation of ST6GalNAc VI in renal cancers. Biochem. J. 2007, 402, 459–470. [Google Scholar] [CrossRef]
- Schartz, N.D.; Tenner, A.J. The good, the bad, and the opportunities of the complement system in neurodegenerative disease. J. Neuroinflamm. 2020, 17, 354. [Google Scholar] [CrossRef]
- Hu, D.; Mayeda, A.; Trembley, J.H.; Lahti, J.M.; Kidd, V.J. CDK11 complexes promote pre-mRNA splicing. J. Biol. Chem. 2003, 278, 8623–8629. [Google Scholar] [CrossRef]
- Rader, D.J. A new feature on the cholesterol-lowering landscape. Nat. Med. 2001, 7, 1282–1284. [Google Scholar] [CrossRef]
- Zhao, W.; Beers, D.R.; Hooten, K.G.; Sieglaff, D.H.; Zhang, A.; Kalyana-Sundaram, S.; Traini, C.M.; Halsey, W.S.; Hughes, A.M.; Sathe, G.M.; et al. Characterization of gene expression phenotype in amyotrophic lateral sclerosis monocytes. JAMA Neurol. 2017, 74, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Taghibiglou, C.; Lu, J.; Mackenzie, I.R.; Wang, Y.T.; Cashman, N.R. Sterol regulatory element binding protein-1 (SREBP1) activation in motor neurons in excitotoxicity and amyotrophic lateral sclerosis (ALS): Indip, a potential therapeutic peptide. Biochem. Biophys. Res. Commun. 2011, 413, 159–163. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.-Y.; Ren, M.; Wang, J.; Jiang, H.-Q.; Yin, X.; Qi, Y.; Wang, X.-D.; Dong, G.-T.; Wang, T.-H.; Yang, Y.-Q.; et al. Notch pathway is activated in cell culture and mouse models of mutant SOD1-related familial amyotrophic lateral sclerosis, with suppression of its activation as an additional mechanism of neuroprotection for lithium and valproate. Neuroscience 2015, 301, 276–288. [Google Scholar] [CrossRef] [PubMed]
- Deivasigamani, S.; Verma, H.K.; Ueda, R.; Ratnaparkhi, A.; Ratnaparkhi, G.S. A genetic screen identifies Tor as an interactor of VAPB in a Drosophila model of amyotrophic lateral sclerosis. Biol. Open 2014, 3, 1127–1138. [Google Scholar] [CrossRef] [PubMed]
- Tibshirani, M.; Tradewell, M.L.; Mattina, K.R.; Minotti, S.; Yang, W.; Zhou, H.; Strong, M.J.; Hayward, L.J.; Durham, H.D. Cytoplasmic sequestration of FUS/TLS associated with ALS alters histone marks through loss of nuclear protein arginine methyltransferase 1. Hum. Mol. Genet. 2015, 24, 773–786. [Google Scholar] [CrossRef]
- Lenzi, J.; Pagani, F.; De Santis, R.; Limatola, C.; Bozzoni, I.; Di Angelantonio, S.; Rosa, A. Differentiation of control and ALS mutant human iPSCs into functional skeletal muscle cells, a tool for the study of neuromuscolar diseases. Stem Cell Res. 2016, 17, 140–147. [Google Scholar] [CrossRef]
- Lindblad, C.; Neumann, S.; Kolbeinsdóttir, S.; Zachariadis, V.; Thelin, E.P.; Enge, M.; Thams, S.; Brundin, L.; Svensson, M. Stem cell-derived brainstem mouse astrocytes obtain a neurotoxic phenotype in vitro upon neuroinflammation. J. Inflamm. 2023, 20, 22. [Google Scholar] [CrossRef]
- Badu-Mensah, A.; Guo, X.; Nimbalkar, S.; Cai, Y.; Hickman, J.J. ALS mutations in both human skeletal muscle and motoneurons differentially affects neuromuscular junction integrity and function. Biomaterials 2022, 289, 121752. [Google Scholar] [CrossRef]
- Vaughan, S.K.; Sutherland, N.M.; Zhang, S.; Hatzipetros, T.; Vieira, F.; Valdez, G. The ALS-inducing factors, TDP43A315T and SOD1G93A, directly affect and sensitize sensory neurons to stress. Sci. Rep. 2018, 8, 16582. [Google Scholar] [CrossRef]
- Violatto, M.B.; Pasetto, L.; Casarin, E.; Tondello, C.; Schiavon, E.; Talamini, L.; Marchini, G.; Cagnotto, A.; Morelli, A.; Lanno, A.; et al. Development of a Nanoparticle-Based Approach for the Blood–Brain Barrier Passage in a Murine Model of Amyotrophic Lateral Sclerosis. Cells 2022, 11, 4003. [Google Scholar] [CrossRef]
- Chiu, I.M.; Chen, A.; Zheng, Y.; Kosaras, B.; Tsiftsoglou, S.A.; Vartanian, T.K.; Brown, R.H.; Carroll, M.C. T lymphocytes potentiate endogenous neuroprotective inflammation in a mouse model of ALS. Proc. Natl. Acad. Sci. USA 2008, 105, 17913–17918. [Google Scholar] [CrossRef] [PubMed]
- Frakes, A.E.; Ferraiuolo, L.; Haidet-Phillips, A.M.; Schmelzer, L.; Braun, L.; Miranda, C.J.; Ladner, K.J.; Bevan, A.K.; Foust, K.D.; Godbout, J.P.; et al. Microglia induce motor neuron death via the classical NF-κB pathway in amyotrophic lateral sclerosis. Neuron 2014, 81, 1009–1023. [Google Scholar] [CrossRef] [PubMed]
- Marlin, E.; Valencia, M.; Peregrín, N.; Ferrero, R.; Nicolás, M.J.; Vinueza-Gavilanes, R.; Pineda-Lucena, A.; Artieda, J.; Arrasate, M.; Aragón, T. Pharmacological inhibition of the integrated stress response accelerates disease progression in an amyotrophic lateral sclerosis mouse model. Br. J. Pharmacol. 2024, 181, 495–508. [Google Scholar] [CrossRef] [PubMed]
- Tsioras, K.; Smith, K.C.; Edassery, S.L.; Garjani, M.; Li, Y.; Williams, C.; McKenna, E.D.; Guo, W.; Wilen, A.P.; Hark, T.J.; et al. Analysis of proteome-wide degradation dynamics in ALS SOD1 iPSC-derived patient neurons reveals disrupted VCP homeostasis. Cell Rep. 2023, 42, 113160. [Google Scholar] [CrossRef] [PubMed]
- Masala, A.; Sanna, S.; Esposito, S.; Rassu, M.; Galioto, M.; Zinellu, A.; Carru, C.; Carrì, M.T.; Iaccarino, C.; Crosio, C. Epigenetic changes associated with the expression of amyotrophic lateral sclerosis (ALS) causing genes. Neuroscience 2018, 390, 1–11. [Google Scholar] [CrossRef]
- Rubio, M.A.; Herrando-Grabulosa, M.; Navarro, X. Sensory involvement in amyotrophic lateral sclerosis. Int. J. Mol. Sci. 2022, 23, 15521. [Google Scholar] [CrossRef]
- Ricciardi, D.; Todisco, V.; Tedeschi, G.; Trojsi, F.; Cirillo, G. Altered sensory-motor plasticity in amyotrophic lateral sclerosis and complex regional pain type I syndrome: A shared mechanism? Neurol. Sci. 2020, 41, 1919–1921. [Google Scholar] [CrossRef]
- Loeffler, J.P.; Picchiarelli, G.; Dupuis, L.; Gonzalez De Aguilar, J.L. The role of skeletal muscle in amyotrophic lateral sclerosis. Brain Pathol. 2016, 26, 227–236. [Google Scholar] [CrossRef]
- Castets, P.; Ham, D.J.; Rüegg, M.A. The TOR pathway at the neuromuscular junction: More than a metabolic player? Front. Mol. Neurosci. 2020, 13, 162. [Google Scholar] [CrossRef]
- Rickman, O.J.; Baple, E.L.; Crosby, A.H. Lipid metabolic pathways converge in motor neuron degenerative diseases. Brain 2020, 143, 1073–1087. [Google Scholar] [CrossRef]
- Kumar, P.; Sharoyko, V.V.; Spégel, P.; Gullberg, U.; Mulder, H.; Olsson, I.; Ajore, R. The transcriptional co-repressor myeloid translocation gene 16 inhibits glycolysis and stimulates mitochondrial respiration. PLoS ONE 2013, 8, e68502. [Google Scholar] [CrossRef] [PubMed]
- Velazquez-Caldelas, T.E.; Zamora-Fuentes, J.M.; Hernandez-Lemus, E. Coordinated inflammation and immune response transcriptional regulation in breast cancer molecular subtypes. Front. Immunol. 2024, 15, 1357726. [Google Scholar] [CrossRef] [PubMed]
- Rivers-Auty, J.; Hoyle, C.; Pointer, A.; Lawrence, C.; Pickering-Brown, S.; Brough, D.; Ryan, S. C9orf72 dipeptides activate the NLRP3 inflammasome. Brain Commun. 2024, 6, fcae282. [Google Scholar] [CrossRef]
- Yang, T.; Wei, Q.; Li, C.; Ou, R.; Lin, J.; Cheng, Y.; Xiao, Y.; Shang, H. Peripheral immunity involvement in the cognitive impairment of sporadic amyotrophic lateral sclerosis. Front. Neurol. 2024, 15, 1405275. [Google Scholar] [CrossRef] [PubMed]
- Vieira de Sá, R.; Sudria-Lopez, E.; Cañizares Luna, M.; Harschnitz, O.; van den Heuvel, D.M.; Kling, S.; Vonk, D.; Westeneng, H.J.; Karst, H.; Bloemenkamp, L.; et al. ATAXIN-2 intermediate-length polyglutamine expansions elicit ALS-associated metabolic and immune phenotypes. Nat. Commun. 2024, 15, 7484. [Google Scholar] [CrossRef] [PubMed]
- Liguori, F.; Alberti, F.; Amadio, S.; Angelini, D.F.; Pilesi, E.; Vitale, G.; Tesoriere, G.; Borsellino, G.; Vernì, F.; Volonté, C. Pan-neuronal expression of human mutant SOD1 in Drosophila impairs survival and motor performance, induces early neuroinflammation and chromosome aberrations. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2024, 1870, 167192. [Google Scholar] [CrossRef]
- Chiarotto, G.B.; Nardo, G.; Trolese, M.C.; França, M.C.; Bendotti, C.; De Oliveira, A.L.R. The emerging role of the major histocompatibility complex class I in amyotrophic lateral sclerosis. Int. J. Mol. Sci. 2017, 18, 2298. [Google Scholar] [CrossRef]


| Chr | Ref/Alt | SNP | Novel/Reported Variant | Affected miRNA (Expression) | miRNA Targets (Expression) |
|---|---|---|---|---|---|
| Chr10 | C/T | rs17091403 | Novel | miR-2110 (UP) | UNCB5 (DOWN) |
| Chr 9 | G/A | rs1844035 | Novel | miR-4477b (UP) | SETX, NEBL (DOWN) |
| Chr17 | A/G | rs771797645 | Novel | mir-548aa (DOWN) | CITED2, SOD2 (UP) |
| Chr17 | G/C | rs745666 | Novel | miR-3615 (DOWN) | KHSRP, HIST1H1B (UP) |
| Chr17 | A/G | rs5432522 | Novel | miR-548d-5p (DOWN) | EXT1, SP1 (UP) |
| Chr20 | G/A | rs2427556 | Novel | miR-941 (DOWN) | DDB1, NQO2 (UP) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sneha, N.P.; Dharshini, S.A.P.; Taguchi, Y.-h.; Gromiha, M.M. Tracing ALS Degeneration: Insights from Spinal Cord and Cortex Transcriptomes. Genes 2024, 15, 1431. https://doi.org/10.3390/genes15111431
Sneha NP, Dharshini SAP, Taguchi Y-h, Gromiha MM. Tracing ALS Degeneration: Insights from Spinal Cord and Cortex Transcriptomes. Genes. 2024; 15(11):1431. https://doi.org/10.3390/genes15111431
Chicago/Turabian StyleSneha, Nela Pragathi, S. Akila Parvathy Dharshini, Y.-h. Taguchi, and M. Michael Gromiha. 2024. "Tracing ALS Degeneration: Insights from Spinal Cord and Cortex Transcriptomes" Genes 15, no. 11: 1431. https://doi.org/10.3390/genes15111431
APA StyleSneha, N. P., Dharshini, S. A. P., Taguchi, Y.-h., & Gromiha, M. M. (2024). Tracing ALS Degeneration: Insights from Spinal Cord and Cortex Transcriptomes. Genes, 15(11), 1431. https://doi.org/10.3390/genes15111431








