Disentangling the Genetic Landscape of Peripartum Depression: A Multi-Polygenic Machine Learning Approach on an Italian Sample
Abstract
:1. Introduction
2. Materials and Methods
2.1. Participants
2.2. Polygenic Risk Score Calculation
2.3. Statistical Analysis
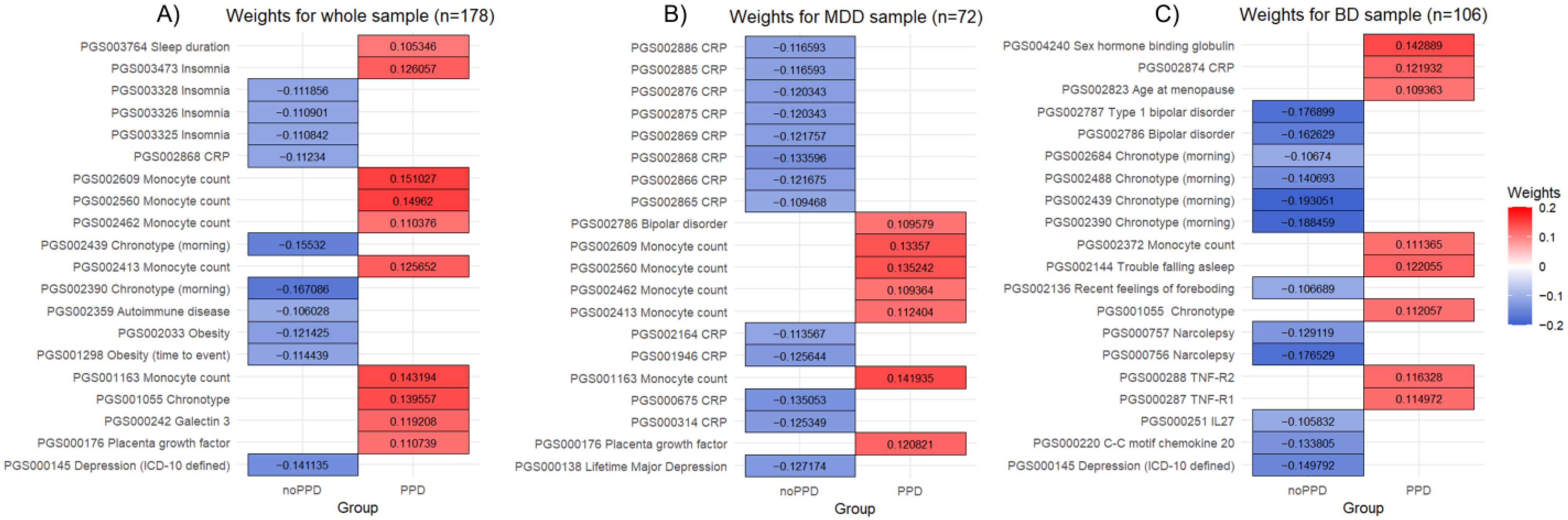
3. Results
3.1. Whole Sample
3.2. MDD Sample
3.3. BD Sample
3.4. Comparison Between MDD and BD Models
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Stewart, D.E.; Vigod, S. Postpartum Depression. N. Engl. J. Med. 2016, 375, 2177–2186. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Liu, J.; Shuai, H.; Cai, Z.; Fu, X.; Liu, Y.; Xiao, X.; Zhang, W.; Krabbendam, E.; Liu, S.; et al. Mapping Global Prevalence of Depression among Postpartum Women. Transl. Psychiatry 2021, 11, 543. [Google Scholar] [CrossRef]
- Payne, J.L.; Maguire, J. Pathophysiological Mechanisms Implicated in Postpartum Depression. Front. Neuroendocrinol. 2019, 52, 165–180. [Google Scholar] [CrossRef] [PubMed]
- American Psychiatric Association; DSM-5 Task Force. Diagnostic and Statistical Manual of Mental Disorders: DSM-5; American Psychiatric Association: Washington, DC, USA, 2013. [Google Scholar]
- Vesga-Lopez, O.; Blanco, C.; Keyes, K.; Olfson, M.; Grant, B.F.; Hasin, D.S. Psychiatric Disorders in Pregnant and Postpartum Women in the United States. Arch. Gen. Psychiatry 2008, 65, 805–815. [Google Scholar] [CrossRef] [PubMed]
- Batt, M.M.; Duffy, K.A.; Novick, A.M.; Metcalf, C.A.; Epperson, C.N. Is Postpartum Depression Different From Depression Occurring Outside of the Perinatal Period? A Review of the Evidence. Focus 2020, 18, 106–119. [Google Scholar] [CrossRef] [PubMed]
- Di Florio, A.; Meltzer-Brody, S. Is Postpartum Depression a Distinct Disorder? Curr. Psychiatry Rep. 2015, 17, 76. [Google Scholar] [CrossRef]
- Bloch, M.; Rotenberg, N.; Koren, D.; Klein, E. Risk Factors Associated with the Development of Postpartum Mood Disorders. J. Affect. Disord. 2005, 88, 9–18. [Google Scholar] [CrossRef]
- Pataky, E.A.; Ehlert, U. Longitudinal Assessment of Symptoms of Postpartum Mood Disorder in Women with and without a History of Depression. Arch. Womens Ment. Health 2020, 23, 391–399. [Google Scholar] [CrossRef]
- Sharma, V.; Doobay, M.; Baczynski, C. Bipolar Postpartum Depression: An Update and Recommendations. J. Affect. Disord. 2017, 219, 105–111. [Google Scholar] [CrossRef]
- Sharma, V.; Khan, M. Identification of Bipolar Disorder in Women with Postpartum Depression. Bipolar Disord. 2010, 12, 335–340. [Google Scholar] [CrossRef]
- Schiller, C.E.; Meltzer-Brody, S.; Rubinow, D.R. The Role of Reproductive Hormones in Postpartum Depression. CNS Spectr. 2015, 20, 48–59. [Google Scholar] [CrossRef] [PubMed]
- Bloch, M.; Schmidt, P.J.; Danaceau, M.; Murphy, J.; Nieman, L.; Rubinow, D.R. Effects of Gonadal Steroids in Women with a History of Postpartum Depression. Am. J. Psychiatry 2000, 157, 924–930. [Google Scholar] [CrossRef] [PubMed]
- Bränn, E.; Fransson, E.; White, R.A.; Papadopoulos, F.C.; Edvinsson, Å.; Kamali-Moghaddam, M.; Cunningham, J.L.; Sundström-Poromaa, I.; Skalkidou, A. Inflammatory Markers in Women with Postpartum Depressive Symptoms. J. Neurosci. Res. 2020, 98, 1309–1321. [Google Scholar] [CrossRef] [PubMed]
- Osborne, L.M.; Yenokyan, G.; Fei, K.; Kraus, T.; Moran, T.; Monk, C.; Sperling, R. Innate Immune Activation and Depressive and Anxious Symptoms across the Peripartum: An Exploratory Study. Psychoneuroendocrinology 2019, 99, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Jin, J.; Tang, J. Inflammatory Pathophysiological Mechanisms Implicated in Postpartum Depression. Front. Pharmacol. 2022, 13, 955672. [Google Scholar] [CrossRef]
- Dørheim, S.K.; Bondevik, G.T.; Eberhard-Gran, M.; Bjorvatn, B. Sleep and Depression in Postpartum Women: A Population-Based Study. Sleep 2009, 32, 847–855. [Google Scholar] [CrossRef]
- Okun, M.L.; Mancuso, R.A.; Hobel, C.J.; Schetter, C.D.; Coussons-Read, M. Poor Sleep Quality Increases Symptoms of Depression and Anxiety in Postpartum Women. J. Behav. Med. 2018, 41, 703–710. [Google Scholar] [CrossRef]
- Sobol, M.; Błachnio, A.; Meisner, M.; Szyszkowska, J.; Jankowski, K.S. Sleep, Circadian Activity Patterns and Postpartum Depression: A Systematic Review and Meta-Analysis of Actigraphy Studies. J. Sleep Res. 2024, 33, e14116. [Google Scholar] [CrossRef]
- Obeysekare, J.L.; Cohen, Z.L.; Coles, M.E.; Pearlstein, T.B.; Monzon, C.; Flynn, E.E.; Sharkey, K.M. Delayed Sleep Timing and Circadian Rhythms in Pregnancy and Transdiagnostic Symptoms Associated with Postpartum Depression. Transl. Psychiatry 2020, 10, 14. [Google Scholar] [CrossRef]
- Zacher Kjeldsen, M.-M.; Bricca, A.; Liu, X.; Frokjaer, V.G.; Madsen, K.B.; Munk-Olsen, T. Family History of Psychiatric Disorders as a Risk Factor for Maternal Postpartum Depression: A Systematic Review and Meta-Analysis. JAMA Psychiatry 2022, 79, 1004–1013. [Google Scholar] [CrossRef]
- Lancaster, E.E.; Lapato, D.M.; Peterson, R.E. Understanding the Genetics of Peripartum Depression: Research Challenges, Strategies, and Opportunities. Front. Genet. 2022, 13, 1022188. [Google Scholar] [CrossRef] [PubMed]
- Viktorin, A.; Meltzer-Brody, S.; Kuja-Halkola, R.; Sullivan, P.F.; Landén, M.; Lichtenstein, P.; Magnusson, P.K.E. Heritability of Perinatal Depression and Genetic Overlap with Nonperinatal Depression. Am. J. Psychiatry 2016, 173, 158–165. [Google Scholar] [CrossRef] [PubMed]
- Luo, F.; Zhu, Z.; Du, Y.; Chen, L.; Cheng, Y. Risk Factors for Postpartum Depression Based on Genetic and Epigenetic Interactions. Mol. Neurobiol. 2023, 60, 3979–4003. [Google Scholar] [CrossRef]
- Border, R.; Johnson, E.C.; Evans, L.M.; Smolen, A.; Berley, N.; Sullivan, P.F.; Keller, M.C. No Support for Historical Candidate Gene or Candidate Gene-by-Interaction Hypotheses for Major Depression Across Multiple Large Samples. Am. J. Psychiatry 2019, 176, 376–387. [Google Scholar] [CrossRef]
- Wang, X.; Walker, A.; Revez, J.A.; Ni, G.; Adams, M.J.; McIntosh, A.M.; Wray, N.R.; Ripke, S.; Mattheisen, M.; Trzaskowski, M.; et al. Polygenic Risk Prediction: Why and When out-of-Sample Prediction R2 Can Exceed SNP-Based Heritability. Am. J. Human. Genet. 2023, 110, 1207–1215. [Google Scholar] [CrossRef] [PubMed]
- Murray, G.K.; Lin, T.; Austin, J.; McGrath, J.J.; Hickie, I.B.; Wray, N.R. Could Polygenic Risk Scores Be Useful in Psychiatry?: A Review. JAMA Psychiatry 2021, 78, 210–219. [Google Scholar] [CrossRef]
- Byrne, E.M.; Carrillo-Roa, T.; Meltzer-Brody, S.; Penninx, B.W.; Sallis, H.M.; Viktorin, A.; Chapman, B.; Henders, A.K.; Pergadia, M.L.; Heath, A.C.; et al. Applying Polygenic Risk Scores to Postpartum Depression. Arch. Womens Ment. Health 2014, 17, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Rantalainen, V.; Binder, E.B.; Lahti-Pulkkinen, M.; Czamara, D.; Laivuori, H.; Villa, P.M.; Girchenko, P.; Kvist, T.; Hämäläinen, E.; Kajantie, E.; et al. Polygenic Prediction of the Risk of Perinatal Depressive Symptoms. Depress. Anxiety 2020, 37, 862–875. [Google Scholar] [CrossRef]
- Abdi, H. Partial Least Squares Regression and Projection on Latent Structure Regression (PLS Regression). WIREs Comput. Stat. 2010, 2, 97–106. [Google Scholar] [CrossRef]
- ACOG Committee Opinion No. 757: Screening for Perinatal Depression. Obstet. Gynecol. 2018, 132, e208–e212. [CrossRef]
- Centers for Disease Control and Prevention (CDC). Prevalence of Self-Reported Postpartum Depressive Symptoms—17 States, 2004–2005. MMWR Morb. Mortal. Wkly. Rep. 2008, 57, 361–366. [Google Scholar]
- Stewart, D.E.; Robertson, E.; Phil, M.; Dennis, C.-L.; Grace, S.L.; Wallington, T. Postpartum Depression: Literature Review of Risk Factors and Interventions; Toronto Public Health: Toronto, ON, Canada, 2003. [Google Scholar]
- Leckman, J.F.; Sholomskas, D.; Thompson, W.D.; Belanger, A.; Weissman, M.M. Best Estimate of Lifetime Psychiatric Diagnosis: A Methodological Study. Arch. Gen. Psychiatry 1982, 39, 879–883. [Google Scholar] [CrossRef] [PubMed]
- Hoekzema, E.; Barba-Müller, E.; Pozzobon, C.; Picado, M.; Lucco, F.; García-García, D.; Soliva, J.C.; Tobeña, A.; Desco, M.; Crone, E.A.; et al. Pregnancy Leads to Long-Lasting Changes in Human Brain Structure. Nat. Neurosci. 2017, 20, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Di Florio, A.; Forty, L.; Gordon-Smith, K.; Heron, J.; Jones, L.; Craddock, N.; Jones, I. Perinatal episodes across the mood disorder spectrum. JAMA Psychiatry 2013, 70, 168–175. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Anderson, C.A.; Pettersson, F.H.; Clarke, G.M.; Cardon, L.R.; Morris, A.P.; Zondervan, K.T. Data Quality Control in Genetic Case-Control Association Studies. Nat. Protoc. 2010, 5, 1564–1573. [Google Scholar] [CrossRef]
- Lambert, S.A.; Gil, L.; Jupp, S.; Ritchie, S.C.; Xu, Y.; Buniello, A.; McMahon, A.; Abraham, G.; Chapman, M.; Parkinson, H.; et al. The Polygenic Score Catalog as an Open Database for Reproducibility and Systematic Evaluation. Nat. Genet. 2021, 53, 420–425. [Google Scholar] [CrossRef]
- Boulesteix, A.-L.; Strimmer, K. Partial Least Squares: A Versatile Tool for the Analysis of High-Dimensional Genomic Data. Brief. Bioinform. 2007, 8, 32–44. [Google Scholar] [CrossRef]
- Cao, K.-A.L.; Rossouw, D.; Robert-Granié, C.; Besse, P. A Sparse PLS for Variable Selection When Integrating Omics Data. Stat. Appl. Genet. Mol. Biol. 2008, 7, 35. [Google Scholar] [CrossRef]
- Akarachantachote, N.; Chadcham, S.; Saithanu, K. Cutoff Threshold of Variable Importance in Projection for Variable Selection. Int. J. Pure Apllied Math. 2014, 94, 307–322. [Google Scholar] [CrossRef]
- Chong, I.-G.; Jun, C.-H. Performance of Some Variable Selection Methods When Multicollinearity Is Present. Chemom. Intell. Lab. Syst. 2005, 78, 103–112. [Google Scholar] [CrossRef]
- Palermo, G.; Piraino, P.; Zucht, H.-D. Performance of PLS Regression Coefficients in Selecting Variables for Each Response of a Multivariate PLS for Omics-Type Data. Adv. Appl. Bioinform. Chem. 2009, 2, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Poletti, S.; Zanardi, R.; Mandelli, A.; Aggio, V.; Finardi, A.; Lorenzi, C.; Borsellino, G.; Carminati, M.; Manfredi, E.; Tomasi, E.; et al. Low-Dose Interleukin 2 Antidepressant Potentiation in Unipolar and Bipolar Depression: Safety, Efficacy, and Immunological Biomarkers. Brain Behav. Immun. 2024, 118, 52–68. [Google Scholar] [CrossRef]
- Martin, A.R.; Daly, M.J.; Robinson, E.B.; Hyman, S.E.; Neale, B.M. Predicting Polygenic Risk of Psychiatric Disorders. Biol. Psychiatry 2019, 86, 97–109. [Google Scholar] [CrossRef]
- Hosang, G.M.; Shakoor, S.; King, N.; Sanches, M.; Vincent, J.B.; Kennedy, J.L.; McGuffin, P.; Keers, R.; Zai, C.C. Interplay between Polygenic Risk for Mood Disorders and Stressful Life Events in Bipolar Disorder. J. Affect. Disord. 2024, 350, 565–572. [Google Scholar] [CrossRef] [PubMed]
- Lynall, M.-E.; Soskic, B.; Hayhurst, J.; Schwartzentruber, J.; Levey, D.F.; Pathak, G.A.; Polimanti, R.; Gelernter, J.; Stein, M.B.; Trynka, G.; et al. Genetic Variants Associated with Psychiatric Disorders Are Enriched at Epigenetically Active Sites in Lymphoid Cells. Nat. Commun. 2022, 13, 6102. [Google Scholar] [CrossRef]
- Sewell, M.D.E.; Jiménez-Sánchez, L.; Shen, X.; Edmondson-Stait, A.J.; Green, C.; Adams, M.J.; Rifai, O.M.; McIntosh, A.M.; Lyall, D.M.; Whalley, H.C.; et al. Associations between Major Psychiatric Disorder Polygenic Risk Scores and Blood-Based Markers in UK Biobank. Brain Behav. Immun. 2021, 97, 32–41. [Google Scholar] [CrossRef]
- Beumer, W.; Gibney, S.M.; Durophage, R.C.; Pont-Lezica, L.; Doorduin, J.; Klein, H.C.; Steiner, J.; Connor, T.J.; Harkin, A.; Versnel, M.A.; et al. The Immune Theory of Psychiatric Diseases: A Key Role for Activated Microglia and Circulating Monocytes. J. Leukoc. Biol. 2012, 92, 959–975. [Google Scholar] [CrossRef]
- Hasselmann, H.; Gamradt, S.; Taenzer, A.; Nowacki, J.; Zain, R.; Patas, K.; Ramien, C.; Paul, F.; Wingenfeld, K.; Piber, D.; et al. Pro-Inflammatory Monocyte Phenotype and Cell-Specific Steroid Signaling Alterations in Unmedicated Patients with Major Depressive Disorder. Front. Immunol. 2018, 9, 2693. [Google Scholar] [CrossRef]
- Simon, M.S.; Schiweck, C.; Arteaga-Henríquez, G.; Poletti, S.; Haarman, B.C.M.; Dik, W.A.; Schwarz, M.; Vrieze, E.; Mikova, O.; Joergens, S.; et al. Monocyte Mitochondrial Dysfunction, Inflammaging, and Inflammatory Pyroptosis in Major Depression. Prog. Neuropsychopharmacol. Biol. Psychiatry 2021, 111, 110391. [Google Scholar] [CrossRef]
- Bergink, V.; Burgerhout, K.M.; Weigelt, K.; Pop, V.J.; de Wit, H.; Drexhage, R.C.; Kushner, S.A.; Drexhage, H.A. Immune System Dysregulation in First-Onset Postpartum Psychosis. Biol. Psychiatry 2013, 73, 1000–1007. [Google Scholar] [CrossRef] [PubMed]
- Krause, D.; Jobst, A.; Kirchberg, F.; Kieper, S.; Härtl, K.; Kästner, R.; Myint, A.-M.; Müller, N.; Schwarz, M.J. Prenatal Immunologic Predictors of Postpartum Depressive Symptoms: A Prospective Study for Potential Diagnostic Markers. Eur. Arch. Psychiatry Clin. Neurosci. 2014, 264, 615–624. [Google Scholar] [CrossRef] [PubMed]
- Manusama, O.; Singh, S.; Brooimans, R.A.; Wijkhuijs, A.; van der Ent, M.; Drexhage, H.A.; Dalm, V.A. Reduced Numbers of Naïve CD4 + T Cells and an Altered CD4/CD8 Balance in Depressed Common Variable Immune Deficiency (CVID) Patients. Is Thymosin-A1 a Possible Treatment? Int. Immunopharmacol. 2023, 119, 110168. [Google Scholar] [CrossRef] [PubMed]
- Felger, J.C.; Haroon, E.; Patel, T.A.; Goldsmith, D.R.; Wommack, E.C.; Woolwine, B.J.; Le, N.-A.; Feinberg, R.; Tansey, M.G.; Miller, A.H. What Does Plasma CRP Tell Us about Peripheral and Central Inflammation in Depression? Mol. Psychiatry 2020, 25, 1301–1311. [Google Scholar] [CrossRef]
- Fernandes, B.S.; Steiner, J.; Molendijk, M.L.; Dodd, S.; Nardin, P.; Gonçalves, C.-A.; Jacka, F.; Köhler, C.A.; Karmakar, C.; Carvalho, A.F.; et al. C-Reactive Protein Concentrations across the Mood Spectrum in Bipolar Disorder: A Systematic Review and Meta-Analysis. Lancet Psychiatry 2016, 3, 1147–1156. [Google Scholar] [CrossRef]
- Silva-Fernandes, A.; Conde, A.; Marques, M.; Caparros-Gonzalez, R.A.; Fransson, E.; Mesquita, A.R.; Figueiredo, B.; Skalkidou, A. Inflammatory Biomarkers and Perinatal Depression: A Systematic Review. PLoS ONE 2024, 19, e0280612. [Google Scholar] [CrossRef]
- Miller, E.S.; Hoxha, D.; Pinheiro, E.; Grobman, W.A.; Wisner, K.L. The Association of Serum C-Reactive Protein with the Occurrence and Course of Postpartum Depression. Arch. Womens Ment. Health 2019, 22, 129–132. [Google Scholar] [CrossRef]
- Ancelin, M.-L.; Farré, A.; Carrière, I.; Ritchie, K.; Chaudieu, I.; Ryan, J. C-Reactive Protein Gene Variants: Independent Association with Late-Life Depression and Circulating Protein Levels. Transl. Psychiatry 2015, 5, e499. [Google Scholar] [CrossRef]
- Zwicker, A.; Fabbri, C.; Rietschel, M.; Hauser, J.; Mors, O.; Maier, W.; Zobel, A.; Farmer, A.; Aitchison, K.J.; McGuffin, P.; et al. Genetic Disposition to Inflammation and Response to Antidepressants in Major Depressive Disorder. J. Psychiatr. Res. 2018, 105, 17–22. [Google Scholar] [CrossRef]
- Poletti, S.; Mazza, M.G.; Benedetti, F. Inflammatory Mediators in Major Depression and Bipolar Disorder. Transl. Psychiatry 2024, 14, 247. [Google Scholar] [CrossRef]
- Poletti, S.; Vai, B.; Mazza, M.G.; Zanardi, R.; Lorenzi, C.; Calesella, F.; Cazzetta, S.; Branchi, I.; Colombo, C.; Furlan, R.; et al. A Peripheral Inflammatory Signature Discriminates Bipolar from Unipolar Depression: A Machine Learning Approach. Prog. Neuropsychopharmacol. Biol. Psychiatry 2021, 105, 110136. [Google Scholar] [CrossRef]
- Taylor, B.J.; Hasler, B.P. Chronotype and Mental Health: Recent Advances. Curr. Psychiatry Rep. 2018, 20, 59. [Google Scholar] [CrossRef]
- Mokros, L.; Nowakowska-Domagała, K.; Witusik, A.; Pietras, T. Evening Chronotype as a Bipolar Feature among Patients with Major Depressive Disorder: The Results of a Pilot Factor Analysis. Braz. J. Psychiatry 2020, 44, 35–40. [Google Scholar] [CrossRef]
- Melo, M.C.A.; Abreu, R.L.C.; Linhares Neto, V.B.; de Bruin, P.F.C.; de Bruin, V.M.S. Chronotype and Circadian Rhythm in Bipolar Disorder: A Systematic Review. Sleep Med. Rev. 2017, 34, 46–58. [Google Scholar] [CrossRef]
- Vidafar, P.; Yocum, A.K.; Han, P.; McInnis, M.G.; Burgess, H.J. Late Chronotype Predicts More Depressive Symptoms in Bipolar Disorder over a 5 Year Follow-up Period. Int. J. Bipolar Disord. 2021, 9, 28. [Google Scholar] [CrossRef]
- Jeong Jeong, H.; Moon, E.; Min Park, J.; Dae Lee, B.; Min Lee, Y.; Choi, Y.; In Chung, Y. The Relationship between Chronotype and Mood Fluctuation in the General Population. Psychiatry Res. 2015, 229, 867–871. [Google Scholar] [CrossRef]
- Duarte Faria, A.; Cardoso, T.d.A.; Campos Mondin, T.; Souza, L.D.d.M.; Magalhaes, P.V.d.S.; Patrick Zeni, C.; Silva, R.A.d.; Kapczinski, F.; Jansen, K. Biological Rhythms in Bipolar and Depressive Disorders: A Community Study with Drug-Naïve Young Adults. J. Affect. Disord. 2015, 186, 145–148. [Google Scholar] [CrossRef]
- Garbazza, C.; Hackethal, S.; Migliore, E.; D’Agostino, A.; Serrati, C.; Fanti, V.; Riccardi, S.; Baiardi, S.; Cicolin, A.; Borgwardt, S.; et al. Influence of Chronotype on the Incidence and Severity of Perinatal Depression in the “Life-ON” Study. J. Affect. Disord. 2022, 317, 245–255. [Google Scholar] [CrossRef]
- Yeom, J.W.; Lee, H.-J. Exploring the Relationship Between Circadian Rhythm Shifts and Postpartum Depression. Chronobiol. Med. 2023, 5, 53–57. [Google Scholar] [CrossRef]
- Melloni, E.M.T.; Paolini, M.; Dallaspezia, S.; Lorenzi, C.; Poletti, S.; d’Orsi, G.; Yoshiike, T.; Zanardi, R.; Colombo, C.; Benedetti, F. Melatonin Secretion Patterns Are Associated with Cognitive Vulnerability and Brain Structure in Bipolar Depression. Chronobiol. Int. 2023, 40, 1279–1290. [Google Scholar] [CrossRef]
- Parry, B.L.; Meliska, C.J.; Sorenson, D.L.; Lopez, A.M.; Martinez, L.F.; Nowakowski, S.; Elliott, J.A.; Hauger, R.L.; Kripke, D.F. Plasma Melatonin Circadian Rhythm Disturbances During Pregnancy and Postpartum in Depressed Women and Women with Personal or Family Histories of Depression. Am. J. Psychiatry 2008, 165, 1551–1558. [Google Scholar] [CrossRef]
- Sharkey, K.M.; Pearlstein, T.B.; Carskadon, M.A. Circadian Phase Shifts and Mood across the Perinatal Period in Women with a History of Major Depressive Disorder: A Preliminary Communication. J. Affect. Disord. 2013, 150, 1103–1108. [Google Scholar] [CrossRef]
- Emamian, F.; Khazaie, H.; Okun, M.L.; Tahmasian, M.; Sepehry, A.A. Link between Insomnia and Perinatal Depressive Symptoms: A Meta-Analysis. J. Sleep Res. 2019, 28, e12858. [Google Scholar] [CrossRef]
- Li, H.; Li, H.; Zhong, J.; Wu, Q.; Shen, L.; Tao, Z.; Zhang, H.; Song, S. Association between Sleep Disorders during Pregnancy and Risk of Postpartum Depression: A Systematic Review and Meta-Analysis. Arch. Womens Ment. Health 2023, 26, 259–267. [Google Scholar] [CrossRef]
- Fields, J.C.; Graham, H.L.; Brandt, J.S.; Bodenlos, K.; Ananth, C.V. Risk of Postpartum Readmission for Depression in Relation to Ischaemic Placental Disease: A Population-Based Study. EClinicalMedicine 2023, 60, 102011. [Google Scholar] [CrossRef]
- Selby, C. Sex Hormone Binding Globulin: Origin, Function and Clinical Significance. Ann. Clin. Biochem. 1990, 27, 532–541. [Google Scholar] [CrossRef]
- Kerlan, V.; Nahoul, K.; Le Martelot, M.-T.; Bercovici, J.-P. Longitudinal Study of Maternal Plasma Bioavailable Testosterone and Androstanediol Glucuronide Levels during Pregnancy. Clin. Endocrinol. 1994, 40, 263–267. [Google Scholar] [CrossRef]
- Zhu, H.; Sun, Y.; Guo, S.; Zhou, Q.; Jiang, Y.; Shen, Y.; Zhou, Z.; Du, Z.; Zhou, H. Causal Relationship between Sex Hormone-Binding Globulin and Major Depression: A Mendelian Randomization Study. Acta Psychiatr. Scand. 2023, 148, 426–436. [Google Scholar] [CrossRef]
- Meijsen, J.J.; Shen, H.; Vemuri, M.; Rasgon, N.L.; Koenen, K.C.; Duncan, L.E. Shared Genetic Influences on Depression and Menopause Symptoms. Psychol. Med. 2023, 53, 2241–2251. [Google Scholar] [CrossRef]
- Deecher, D.; Andree, T.H.; Sloan, D.; Schechter, L.E. From Menarche to Menopause: Exploring the Underlying Biology of Depression in Women Experiencing Hormonal Changes. Psychoneuroendocrinology 2008, 33, 3–17. [Google Scholar] [CrossRef]
- Bauer, A.E.; Liu, X.; Byrne, E.M.; Sullivan, P.F.; Wray, N.R.; Agerbo, E.; Nyegaard, M.; Grove, J.; Musliner, K.L.; Ingstrup, K.G.; et al. Genetic Risk Scores for Major Psychiatric Disorders and the Risk of Postpartum Psychiatric Disorders. Transl. Psychiatry 2019, 9, 288. [Google Scholar] [CrossRef]
- Kiewa, J.; Meltzer-Brody, S.; Milgrom, J.; Guintivano, J.; Hickie, I.B.; Whiteman, D.C.; Olsen, C.M.; Colodro-Conde, L.; Medland, S.E.; Martin, N.G.; et al. Perinatal Depression Is Associated with a Higher Polygenic Risk for Major Depressive Disorder than Non-Perinatal Depression. Depress. Anxiety 2022, 39, 182–191. [Google Scholar] [CrossRef]
- Munk-Olsen, T.; Di Florio, A.; Madsen, K.B.; Albiñana, C.; Mægbæk, M.L.; Bergink, V.; Frøkjær, V.G.; Agerbo, E.; Vilhjálmsson, B.J.; Werge, T.; et al. Postpartum and Non-Postpartum Depression: A Population-Based Matched Case-Control Study Comparing Polygenic Risk Scores for Severe Mental Disorders. Transl. Psychiatry 2023, 13, 346. [Google Scholar] [CrossRef]
- Liu, X.; Agerbo, E.; Li, J.; Meltzer-Brody, S.; Bergink, V.; Munk-Olsen, T. Depression and Anxiety in the Postpartum Period and Risk of Bipolar Disorder: A Danish Nationwide Register-Based Cohort Study. J. Clin. Psychiatry 2017, 78, e469–e476. [Google Scholar] [CrossRef]
- Luppino, F.S.; de Wit, L.M.; Bouvy, P.F.; Stijnen, T.; Cuijpers, P.; Penninx, B.W.J.H.; Zitman, F.G. Overweight, Obesity, and Depression: A Systematic Review and Meta-Analysis of Longitudinal Studies. Arch. Gen. Psychiatry 2010, 67, 220–229. [Google Scholar] [CrossRef]
- Scammell, B.H.; Tchio, C.; Song, Y.; Nishiyama, T.; Louie, T.L.; Dashti, H.S.; Nakatochi, M.; Zee, P.C.; Daghlas, I.; Momozawa, Y.; et al. Multi-Ancestry Genome-Wide Analysis Identifies Shared Genetic Effects and Common Genetic Variants for Self-Reported Sleep Duration. Hum. Mol. Genet. 2023, 32, 2797–2807. [Google Scholar] [CrossRef]
- Dashti, H.S.; Jones, S.E.; Wood, A.R.; Lane, J.M.; van Hees, V.T.; Wang, H.; Rhodes, J.A.; Song, Y.; Patel, K.; Anderson, S.G.; et al. Genome-Wide Association Study Identifies Genetic Loci for Self-Reported Habitual Sleep Duration Supported by Accelerometer-Derived Estimates. Nat. Commun. 2019, 10, 1100. [Google Scholar] [CrossRef]

| Variables | PPD (n = 62) | No PPD (n = 116) | t/x2 | Cohen’s D/Cramer’s V | p-Value |
|---|---|---|---|---|---|
| Age (years) | 48.18 ± 9.40 | 53.85 ± 8.89 | −3.976 | 0.620 | <0.001 |
| Education (years) | 12.05 ± 4.19 | 10.91 ± 3.79 | 1.846 | 0.285 | 0.067 |
| BMI | 26.38 ± 5.51 | 25.80 ± 4.80 | 0.683 | 0.112 | 0.496 |
| Smoking Status | 37/79 | 20/42 | 1.938 | 0.104 | 0.585 |
| Alcohol Status | 4/58 | 3/113 | 4.410 | 0.157 | 0.353 |
| Number of pregnancies | 2.07 ± 1.05 | 1.95 ± 0.99 | 0.732 | 0.118 | 0.465 |
| Sex of children (male/female) | 50/52 | 100/70 | 2.477 | 0.118 | 0.116 |
| Menopause (yes/no) | 21/41 | 67/49 | 9.223 | −0.228 | 0.002 |
| Current hormone replacement therapy (yes/no) | 3/59 | 9/107 | 0.548 | −0.055 | 0.459 |
| Number of PPD episodes | 1.21 ± 0.52 | NA | NA | NA | NA |
| Duration of illness (years) | 19.31 ± 11.65 | 18.45 ± 11.65 | 0.468 | 0.074 | 0.640 |
| Number of mood episodes | 9.86 ± 12.02 | 9.12 ± 12.14 | 0.377 | 0.061 | 0.707 |
| Number of depressive episodes | 7.97 ± 9.81 | 6.50 ± 7.21 | 1.137 | 0.171 | 0.257 |
| Number of manic episodes | 3.14 ± 4.45 | 4.50 ± 7.18 | −1.051 | 0.228 | 0.296 |
| MDD Sample | Psychiatric PRSs (n = 67) | Hormone and Pregnancy PRSs (n = 89) | Immune and Inflammation PRSs (n = 131) | Sleep and Circadian PRSs (n = 54) |
|---|---|---|---|---|
| PPD | 12 | 10 | 19 | 7 |
| No PPD | 8 | 14 | 28 | 9 |
| Total | 20 | 24 | 47 | 16 |
| BD Sample | Psychiatric PRSs (n = 67) | Hormone and Pregnancy PRSs (n = 89) | Immune and Inflammation PRSs (n = 131) | Sleep and Circadian PRSs (n = 54) |
|---|---|---|---|---|
| PPD | 3 | 17 | 21 | 9 |
| No PPD | 10 | 15 | 6 | 15 |
| Total | 13 | 33 | 28 | 24 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Harrington, Y.A.; Fortaner-Uyà, L.; Paolini, M.; Poletti, S.; Lorenzi, C.; Spadini, S.; Melloni, E.M.T.; Agnoletto, E.; Zanardi, R.; Colombo, C.; et al. Disentangling the Genetic Landscape of Peripartum Depression: A Multi-Polygenic Machine Learning Approach on an Italian Sample. Genes 2024, 15, 1517. https://doi.org/10.3390/genes15121517
Harrington YA, Fortaner-Uyà L, Paolini M, Poletti S, Lorenzi C, Spadini S, Melloni EMT, Agnoletto E, Zanardi R, Colombo C, et al. Disentangling the Genetic Landscape of Peripartum Depression: A Multi-Polygenic Machine Learning Approach on an Italian Sample. Genes. 2024; 15(12):1517. https://doi.org/10.3390/genes15121517
Chicago/Turabian StyleHarrington, Yasmin A., Lidia Fortaner-Uyà, Marco Paolini, Sara Poletti, Cristina Lorenzi, Sara Spadini, Elisa M. T. Melloni, Elena Agnoletto, Raffaella Zanardi, Cristina Colombo, and et al. 2024. "Disentangling the Genetic Landscape of Peripartum Depression: A Multi-Polygenic Machine Learning Approach on an Italian Sample" Genes 15, no. 12: 1517. https://doi.org/10.3390/genes15121517
APA StyleHarrington, Y. A., Fortaner-Uyà, L., Paolini, M., Poletti, S., Lorenzi, C., Spadini, S., Melloni, E. M. T., Agnoletto, E., Zanardi, R., Colombo, C., & Benedetti, F. (2024). Disentangling the Genetic Landscape of Peripartum Depression: A Multi-Polygenic Machine Learning Approach on an Italian Sample. Genes, 15(12), 1517. https://doi.org/10.3390/genes15121517







