The Chicken HDAC4 Promoter and Its Regulation by MYC and HIF1A
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Bioinformatics Analysis of the HDAC4 Promoter
2.3. Amplification and Promoter Vectors Construction
2.4. Cell Culture and Treatments
2.5. Quantitative Real-Time PCR (qRT-PCR)
2.6. Dual-Luciferase Assay
2.7. CCK8 Analysis
2.8. Statistical Analysis
3. Results
3.1. Deletion Analysis of the Upstream Region of the HDAC4 Gene
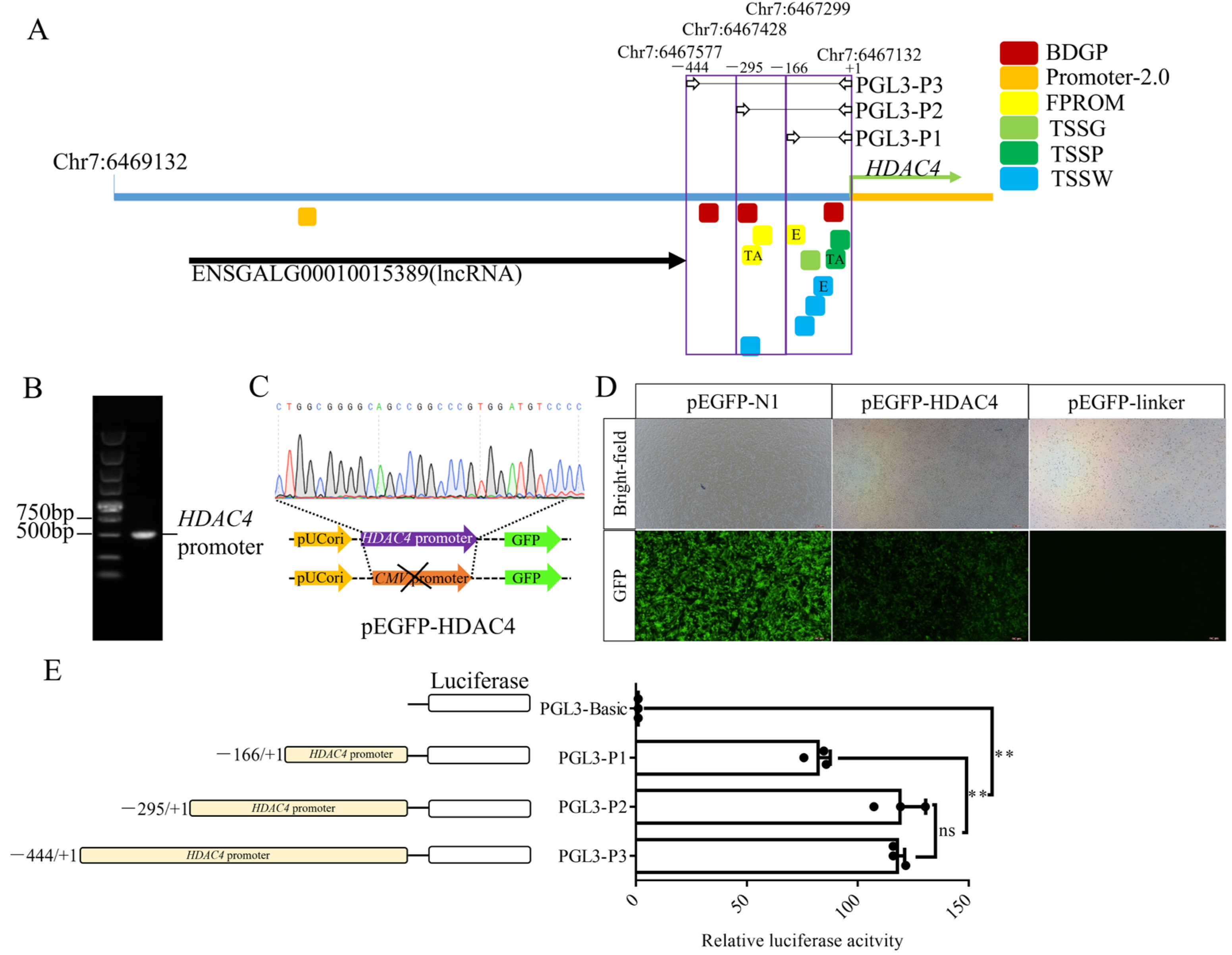
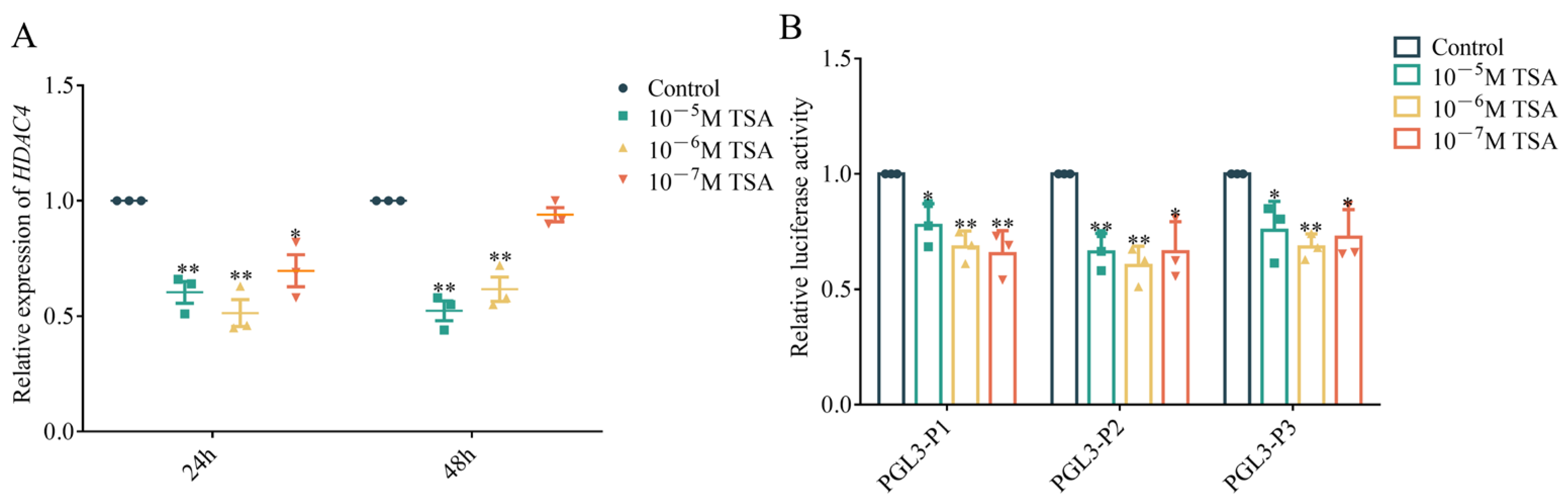
3.2. Effect of TSA on HDAC4 Transcription
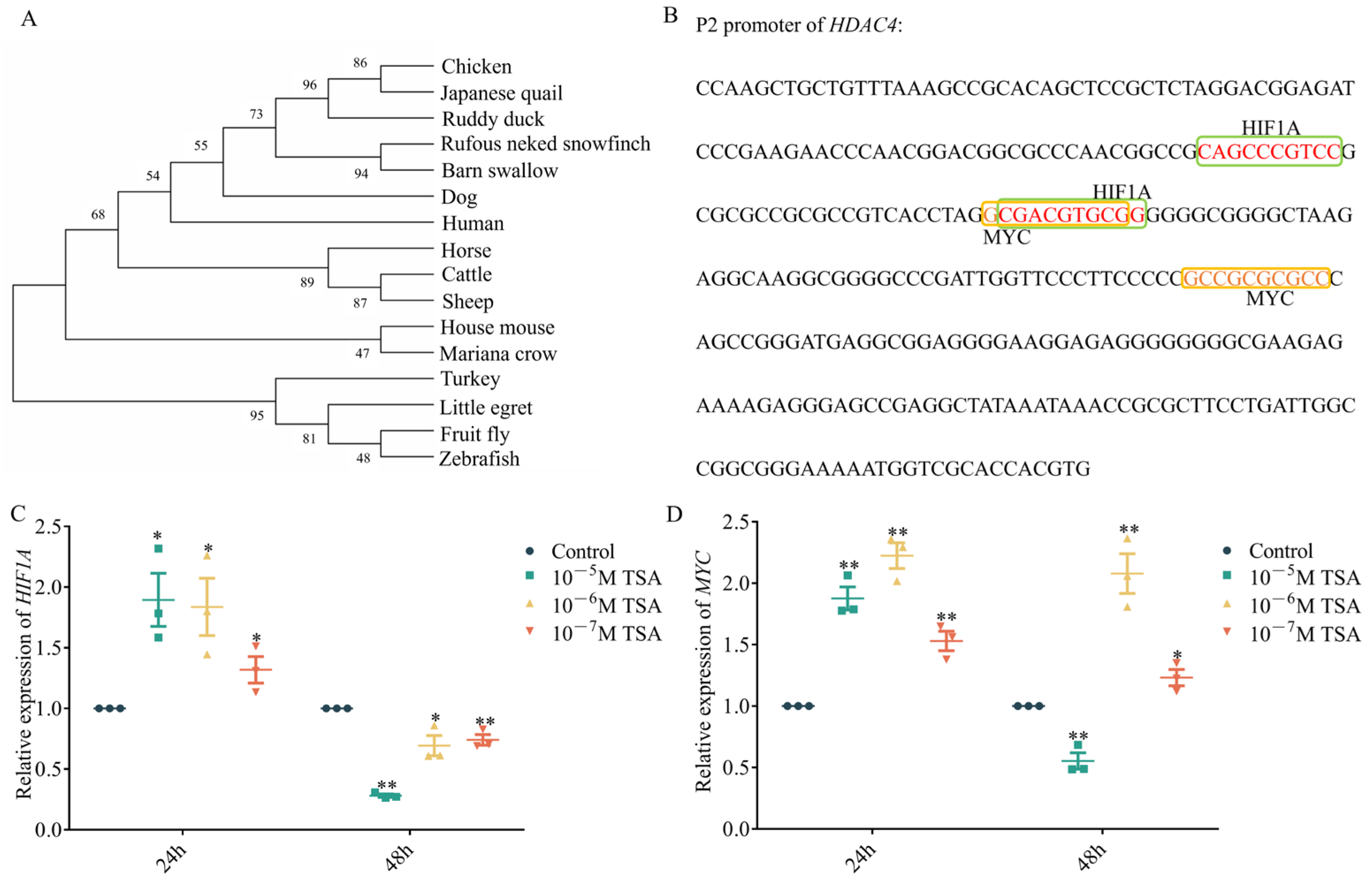
3.3. Bioinformatics of the HDAC4 Core Promoter Suggests Conserved Regulatory Sites
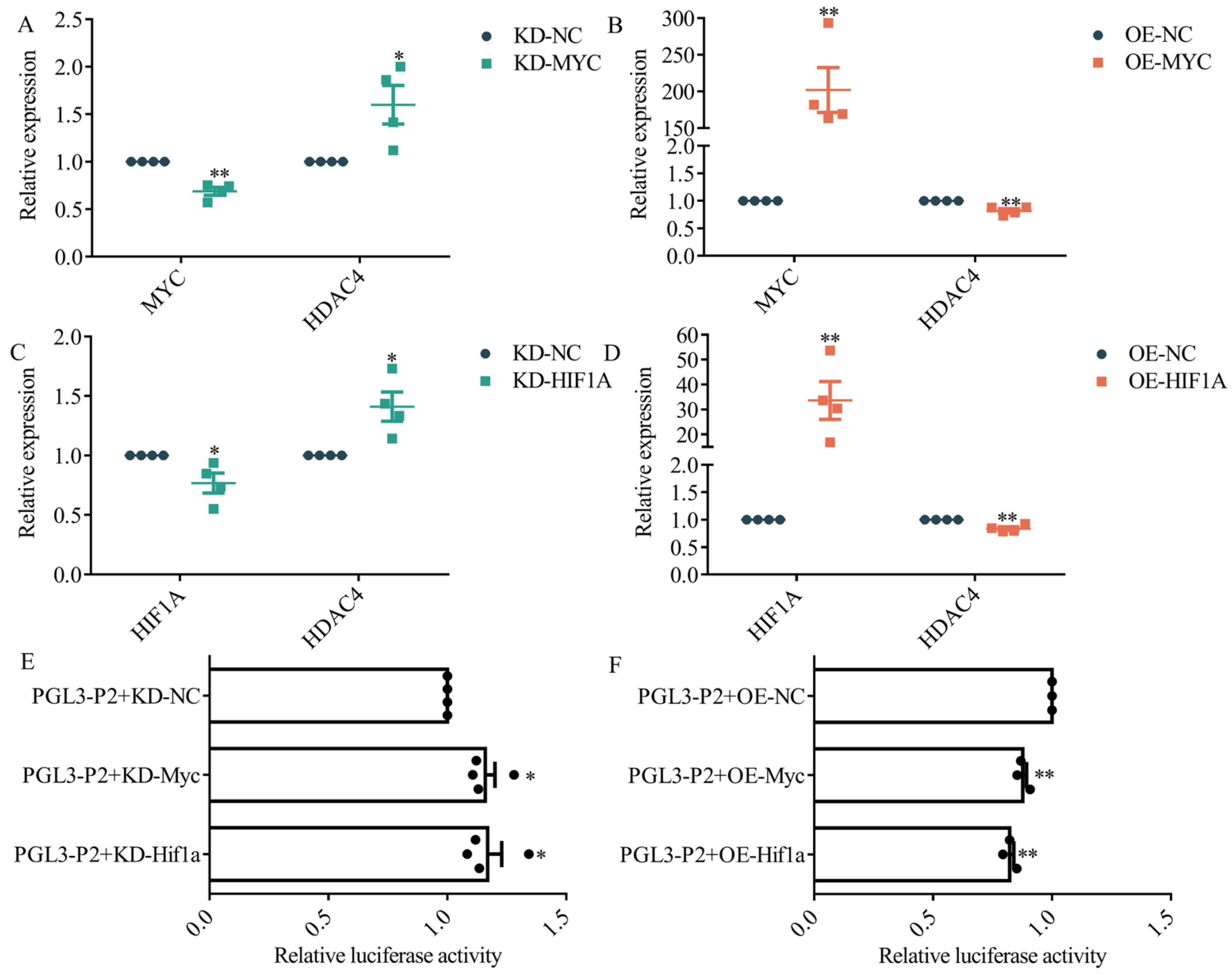
3.4. Effect of MYC and HIF1A Putative Binding Sites on HDAC4 Promoter Activity
3.5. MYC and HIF1A Inhibit HDAC4 Transcription
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kurdistani, S.K.; Tavazoie, S.; Grunstein, M. Mapping global histone acetylation patterns to gene expression. Cell 2004, 117, 721–733. [Google Scholar] [CrossRef] [PubMed]
- Getun, I.V.; Wu, Z.; Fallahi, M.; Ouizem, S.; Liu, Q.; Li, W.; Costi, R.; Roush, W.R.; Cleveland, J.L.; Bois, P. Functional Roles of Acetylated Histone Marks at Mouse Meiotic Recombination Hot Spots. Mol. Cell. Biol. 2017, 37, e00942-15. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Cui, J.; Li, D.; Ran, S.; Huang, J.; Chen, G. Morin exhibits a neuroprotective effect in MPTP-induced Parkinson’s disease model via TFEB/AMPK-mediated mitophagy. Phytomedicine 2023, 116, 154866. [Google Scholar] [CrossRef]
- Yuan, J.H.; Yang, F.; Chen, B.F.; Lu, Z.; Huo, X.S.; Zhou, W.P.; Wang, F.; Sun, S.H. The histone deacetylase 4/SP1/microrna-200a regulatory network contributes to aberrant histone acetylation in hepatocellular carcinoma. Hepatology 2011, 54, 2025–2035. [Google Scholar] [CrossRef]
- Sujit, K.M.; Sarkar, S.; Singh, V.; Pandey, R.; Agrawal, N.K.; Trivedi, S.; Singh, K.; Gupta, G.; Rajender, S. Genome-wide differential methylation analyses identifies methylation signatures of male infertility. Hum. Reprod. 2018, 33, 2256–2267. [Google Scholar] [CrossRef]
- Song, W.; Zhang, D.; Mi, J.; Du, W.; Yang, Y.; Chen, R.; Tian, C.; Zhao, X.; Zou, K. E-cadherin maintains the undifferentiated state of mouse spermatogonial progenitor cells via beta-catenin. Cell Biosci. 2022, 12, 141. [Google Scholar] [CrossRef]
- Cong, T. Analysis of the Expression Characteristics of HDAC4 in Mouse and Chicken Spermatogonia Stem Cells. Ph.D. Thesis, Nanjing Agricultural University, Nanjing, China, 2021. [Google Scholar]
- Zhang, Z.; Elsayed, A.K.; Shi, Q.; Zhang, Y.; Zuo, Q.; Li, D.; Lian, C.; Tang, B.; Xiao, T.; Xu, Q.; et al. Crucial genes and pathways in chicken germ stem cell differentiation. J. Biol. Chem. 2015, 290, 13605–13621. [Google Scholar] [CrossRef]
- Wu, H.; Liu, C.; Yang, Q.; Xin, C.; Du, J.; Sun, F.; Zhou, L. MIR145-3p promotes autophagy and enhances bortezomib sensitivity in multiple myeloma by targeting HDAC4. Autophagy 2020, 16, 683–697. [Google Scholar] [CrossRef]
- Yao, W.; Kulyar, M.F.; Ding, Y.; Du, H.; Hong, J.; Loon, K.S.; Nawaz, S.; Li, J. The Effect of miR-140-5p with HDAC4 towards Growth and Differentiation Signaling of Chondrocytes in Thiram-Induced Tibial Dyschondroplasia. Int. J. Mol. Sci. 2023, 24, 10975. [Google Scholar] [CrossRef]
- Wang, B.; Li, D.; Filkowski, J.; Rodriguez-Juarez, R.; Storozynsky, Q.; Malach, M.; Carpenter, E.; Kovalchuk, O. A dual role of miR-22 modulated by RelA/p65 in resensitizing fulvestrant-resistant breast cancer cells to fulvestrant by targeting FOXP1 and HDAC4 and constitutive acetylation of p53 at Lys382. Oncogenesis 2018, 7, 54. [Google Scholar] [CrossRef]
- Jiang, H.; Jia, D.; Zhang, B.; Yang, W.; Dong, Z.; Sun, X.; Cui, X.; Ma, L.; Wu, J.; Hu, K.; et al. Exercise improves cardiac function and glucose metabolism in mice with experimental myocardial infarction through inhibiting HDAC4 and upregulating GLUT1 expression. Basic Res. Cardiol. 2020, 115, 28. [Google Scholar] [CrossRef] [PubMed]
- De Sousa, G.R.; Salomao, K.B.; Nagano, L.; Riemondy, K.A.; Chagas, P.S.; Veronez, L.C.; Saggioro, F.P.; Marie, S.; Yunes, J.A.; Cardinalli, I.A.; et al. Identification of HDAC4 as a potential therapeutic target and prognostic biomarker for ZFTA-fused ependymomas. Cancer Gene Ther. 2023, 30, 1105–1113. [Google Scholar] [CrossRef]
- Liu, X.; Feng, M.; Hao, X.; Gao, Z.; Wu, Z.; Wang, Y.; Du, L.; Wang, C. m6A methylation regulates hypoxia-induced pancreatic cancer glycolytic metabolism through ALKBH5-HDAC4-HIF1alpha positive feedback loop. Oncogene 2023, 42, 2047–2060. [Google Scholar] [CrossRef] [PubMed]
- Zang, W.J.; Hu, Y.L.; Qian, C.Y.; Feng, Y.; Liu, J.Z.; Yang, J.L.; Huang, H.; Zhu, Y.Z.; Xue, W.J. HDAC4 promotes the growth and metastasis of gastric cancer via autophagic degradation of MEKK3. Br. J. Cancer 2022, 127, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Li, J.; Zheng, T.; Chen, T.; Yuan, Z. KLF7 Alleviates Atherosclerotic Lesions and Inhibits Glucose Metabolic Reprogramming in Macrophages by Regulating HDAC4/miR-148b-3p/NCOR1. Gerontology 2022, 68, 1291–1310. [Google Scholar] [CrossRef]
- Liu, Z.M.; Wang, X.; Li, C.X.; Liu, X.Y.; Guo, X.J.; Li, Y.; Chen, Y.L.; Ye, H.X.; Chen, H.S. SP1 Promotes HDAC4 Expression and Inhibits HMGB1 Expression to Reduce Intestinal Barrier Dysfunction, Oxidative Stress, and Inflammatory Response after Sepsis. J. Innate Immun. 2022, 14, 366–379. [Google Scholar] [CrossRef]
- Taufique, S.; Prabhat, A.; Kumar, V. Illuminated night alters hippocampal gene expressions and induces depressive-like responses in diurnal corvids. Eur. J. Neurosci. 2018, 48, 3005–3018. [Google Scholar] [CrossRef]
- Wu, N.; Gu, T.; Lu, L.; Cao, Z.; Song, Q.; Wang, Z.; Zhang, Y.; Chang, G.; Xu, Q.; Chen, G. Roles of miRNA-1 and miRNA-133 in the proliferation and differentiation of myoblasts in duck skeletal muscle. J. Cell. Physiol. 2019, 234, 3490–3499. [Google Scholar] [CrossRef]
- Guan, Y.J.; Yang, X.; Wei, L.; Chen, Q. MiR-365: A mechanosensitive microRNA stimulates chondrocyte differentiation through targeting histone deacetylase 4. FASEB J. 2011, 25, 4457–4466. [Google Scholar] [CrossRef]
- Xue, Y.; Chen, C.; Tan, R.; Zhang, J.; Fang, Q.; Jin, R.; Mi, X.; Sun, D.; Xue, Y.; Wang, Y.; et al. Artificial Intelligence-Assisted Bioinformatics, Microneedle, and Diabetic Wound Healing: A “New Deal” of an Old Drug. ACS Appl. Mater. Interfaces 2022, 14, 37396–37409. [Google Scholar] [CrossRef]
- Zhu, K.; Wang, H.; Gul, Y.; Zhao, Y.; Wang, W.; Liu, S.; Wang, M. Expression characterization and the promoter activity analysis of zebrafish hdac4. Fish Physiol. Biochem. 2012, 38, 585–593. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, L.; Hou, M.; Wang, C.; Wang, L.; Sun, T.; He, H.; Zhong, J. Cloning and Activity Analysis of Bovine Natural Resistance Associated Macrophage Protein 1 (Nramp1) Gene Promoter. Sci. Agric. Sin. 2011, 44, 1022–1028. [Google Scholar]
- Khan, R.; Ali, Z.; Niazi, A.K.; Carolan, J.C.; Wilkinson, T.L. In silico Characterization of a Candidate Protein from Aphid Gelling Saliva with Potential for Aphid Control in Plants. Protein Pept. Lett. 2020, 27, 158–167. [Google Scholar] [CrossRef]
- Chen, Y.; He, X.; Cheng, F.; Li, M.; Wu, X.; Zhang, C.; Li, J.; Huang, B.; Qi, M. Angiotensin II promotes EMT of hepatocellular carcinoma cells through high mobility group protein B1 mediated by E4F1. Biochem. Biophys. Res. Commun. 2021, 547, 198–203. [Google Scholar] [CrossRef]
- Solovyev, V.; Kosarev, P.; Seledsov, I.; Vorobyev, D. Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol. 2006, 7 (Suppl. S1), S10–S11. [Google Scholar] [CrossRef]
- Liu, X.; Chen, J.G.; Munshi, M.; Hunter, Z.R.; Xu, L.; Kofides, A.; Tsakmaklis, N.; Demos, M.G.; Guerrera, M.L.; Chan, G.G.; et al. Expression of the prosurvival kinase HCK requires PAX5 and mutated MYD88 signaling in MYD88-driven B-cell lymphomas. Blood Adv. 2020, 4, 141–153. [Google Scholar] [CrossRef]
- Li, S.; Wei, X.; He, J.; Cao, Q.; Du, D.; Zhan, X.; Zeng, Y.; Yuan, S.; Sun, L. The comprehensive landscape of miR-34a in cancer research. Cancer Metastasis Rev. 2021, 40, 925–948. [Google Scholar] [CrossRef]
- Rana, N.K.; Singh, P.; Koch, B. CoCl2 simulated hypoxia induce cell proliferation and alter the expression pattern of hypoxia associated genes involved in angiogenesis and apoptosis. Biol. Res. 2019, 52, 12. [Google Scholar] [CrossRef]
- Oudelaar, A.M.; Higgs, D.R. The relationship between genome structure and function. Nat. Rev. Genet. 2021, 22, 154–168. [Google Scholar] [CrossRef]
- Li, M.; Xing, W.; Wang, B.; Qi, N.; Liu, J.; Yu, S. Analysis of core promoter and transcription factors screening of porcine ATG7 gene. Gene 2024, 899, 148138. [Google Scholar] [CrossRef]
- Gao, Q.; Wei, A.; Chen, F.; Chen, X.; Ding, W.; Ding, Z.; Wu, Z.; Du, R.; Cao, W. Enhancing PPARgamma by HDAC inhibition reduces foam cell formation and atherosclerosis in ApoE deficient mice. Pharmacol. Res. 2020, 160, 105059. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Lv, W.; He, M.; Deng, H.; Li, H.; Wu, W.; Rao, Y. Plasticity in designing PROTACs for selective and potent degradation of HDAC6. Chem. Commun. 2019, 55, 14848–14851. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Yang, K.; Zhang, Z.; Leisten, E.D.; Li, Z.; Xie, H.; Liu, J.; Smith, K.A.; Novakova, Z.; Barinka, C.; et al. Development of Multifunctional Histone Deacetylase 6 Degraders with Potent Antimyeloma Activity. J. Med. Chem. 2019, 62, 7042–7057. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Zhang, L.; Zhuang, S.; Qin, G.J.; Zhao, T.C. HDAC4 degradation mediates HDAC inhibition-induced protective effects against hypoxia/reoxygenation injury. J. Cell. Physiol. 2015, 230, 1321–1331. [Google Scholar] [CrossRef]
- Lim, H.J.; Kim, J.; Park, C.H.; Lee, S.A.; Lee, M.R.; Kim, K.S.; Kim, J.; Bae, Y.S. Regulation of c-Myc Expression by Ahnak Promotes Induced Pluripotent Stem Cell Generation. J. Biol. Chem. 2016, 291, 752–761. [Google Scholar] [CrossRef]
- Lu, Y.; Yang, L.; Shen, M.; Zhang, Z.; Wang, S.; Chen, F.; Chen, N.; Xu, Y.; Zeng, H.; Chen, M.; et al. Tespa1 facilitates hematopoietic and leukemic stem cell maintenance by restricting c-Myc degradation. Leukemia 2023, 37, 1039–1047. [Google Scholar] [CrossRef]
- Braydich-Stolle, L.; Kostereva, N.; Dym, M.; Hofmann, M.C. Role of Src family kinases and N-Myc in spermatogonial stem cell proliferation. Dev. Biol. 2007, 304, 34–45. [Google Scholar] [CrossRef]
- Kanatsu-Shinohara, M.; Tanaka, T.; Ogonuki, N.; Ogura, A.; Morimoto, H.; Cheng, P.F.; Eisenman, R.N.; Trumpp, A.; Shinohara, T. Myc/Mycn-mediated glycolysis enhances mouse spermatogonial stem cell self-renewal. Genes Dev. 2016, 30, 2637–2648. [Google Scholar] [CrossRef]
- Kanatsu-Shinohara, M.; Onoyama, I.; Nakayama, K.I.; Shinohara, T. Skp1-Cullin-F-box (SCF)-type ubiquitin ligase FBXW7 negatively regulates spermatogonial stem cell self-renewal. Proc. Natl. Acad. Sci. USA 2014, 111, 8826–8831. [Google Scholar] [CrossRef]
- Tilekar, K.; Hess, J.D.; Upadhyay, N.; Bianco, A.L.; Schweipert, M.; Laghezza, A.; Loiodice, F.; Meyer-Almes, F.J.; Aguilera, R.J.; Lavecchia, A.; et al. Thiazolidinedione “Magic Bullets” Simultaneously Targeting PPARgamma and HDACs: Design, Synthesis, and Investigations of their In Vitro and In Vivo Antitumor Effects. J. Med. Chem. 2021, 64, 6949–6971. [Google Scholar] [CrossRef]
- Lu, D.; Bao, D.; Dong, W.; Liu, N.; Zhang, X.; Gao, S.; Ge, W.; Gao, X.; Zhang, L. Dkk3 prevents familial dilated cardiomyopathy development through Wnt pathway. Lab. Investig. 2016, 96, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Davy, P.M.; Gardner, L.H.; Mathews, J.; Yamazaki, Y.; Allsopp, R.C. Hypoxia Inducible Factor 1 Alpha Is Expressed in Germ Cells throughout the Murine Life Cycle. PLoS ONE 2016, 11, e0154309. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, H.; Yamamoto, T.; Miyazaki, T.; Ogonuki, N.; Ogura, A.; Tanaka, T.; Kanatsu-Shinohara, M.; Yabe-Nishimura, C.; Zhang, H.; Pommier, Y.; et al. An interplay of NOX1-derived ROS and oxygen determines the spermatogonial stem cell self-renewal efficiency under hypoxia. Genes Dev. 2021, 35, 250–260. [Google Scholar] [CrossRef]
- Qian, D.Z.; Kachhap, S.K.; Collis, S.J.; Verheul, H.M.; Carducci, M.A.; Atadja, P.; Pili, R. Class II histone deacetylases are associated with VHL-independent regulation of hypoxia-inducible factor 1 alpha. Cancer Res. 2006, 66, 8814–8821. [Google Scholar] [CrossRef]

| Gene | Gene ID | Primer (5′–3′) |
|---|---|---|
| ACTB | NM_205518.2 | qF: ACCAACTGGGATGATATGGAGAA |
| qR:TTGGCTTTGGGGTTCAGG | ||
| HDAC4 | NM_204313.2 | qF: GTTGGAGCAGCAGCGCATTC |
| qR: TGGCTTAGTGGGTGGCTCCT | ||
| MYC | NM_001030952.2 | qF: GGTCTTCCCCTACCCGCTCA |
| qR:CGGACTGTGGTGGGGCTTAC | ||
| HIF1A | NM_001396327.1 | qF: CAAGAGCAACCAACCAGCCCT |
| qR: TGATCAAAGGAGCGTAGCTGGA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Kong, R.; Xie, K.; Wang, X.; Wu, H.; Zhang, Y. The Chicken HDAC4 Promoter and Its Regulation by MYC and HIF1A. Genes 2024, 15, 1518. https://doi.org/10.3390/genes15121518
Wang Y, Kong R, Xie K, Wang X, Wu H, Zhang Y. The Chicken HDAC4 Promoter and Its Regulation by MYC and HIF1A. Genes. 2024; 15(12):1518. https://doi.org/10.3390/genes15121518
Chicago/Turabian StyleWang, Yingjie, Ruihong Kong, Ke Xie, Xu Wang, Han Wu, and Yani Zhang. 2024. "The Chicken HDAC4 Promoter and Its Regulation by MYC and HIF1A" Genes 15, no. 12: 1518. https://doi.org/10.3390/genes15121518
APA StyleWang, Y., Kong, R., Xie, K., Wang, X., Wu, H., & Zhang, Y. (2024). The Chicken HDAC4 Promoter and Its Regulation by MYC and HIF1A. Genes, 15(12), 1518. https://doi.org/10.3390/genes15121518





