Polymorphism of Genes Potentially Affecting Growth and Body Size Suggests Genetic Divergence in Wild and Domestic Reindeer (Rangifer tarandus) Populations
Highlights
- Reindeer are the only species in which wild and domesticated animals regularly interbreed; studying their genetic variation will aid conservation efforts, improve meat productivity, reduce feed costs, and increase disease resistance.
- In this study, we identified particularly informative genetic markers that can be used for breeding purposes in wild and domestic reindeer (Nenets and Evenk breeds) by sequencing the genes GH, GHR, LCORL, and BMP2.
- Parts of some of the genes (loci) were not very variable in wild reindeer but were more so in domestic breeds, suggesting limited evolutionary divergence post domestication.
- Genetic markers associated with economically important traits in reindeer can be further developed using the data generated in this paper, reducing the time spent on selection efforts to enhance meat performance and encourage conservation efforts.
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Genomic DNA Extraction
2.2. PCR, Gene Sequencing and Computational Analyses
3. Results and Discussion
3.1. GHR and GH Gene Polymorphisms
3.2. LCORL and BMP2 Gene Polymorphisms
3.3. Divergence Estimation and Other General Considerations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gunn, A.; Russell, D. Update on the global status of wild reindeer and caribou. Deer Spec. Group Newsl. 2022, 33, 14–29. Available online: https://www.deerspecialistgroup.org/wp-content/uploads/2022/04/DSGNews33.pdf (accessed on 27 November 2024).
- Tryland, M.; Ravolainen, V.; Pedersen, Å.Ø. Climate change: Potential impacts on pasture resources, health and diseases of reindeer and caribou. In Reindeer and Caribou; Tryland, M., Kutz, S., Eds.; CRC Press: Boca Raton, FL, USA, 2018; pp. 493–514. [Google Scholar] [CrossRef]
- Dance, M. Genetic Legacies of Past Climate Change on Arctic Species: How Past Responses Shape Future Impacts. Ph.D. Dissertation, University of Oxford, Oxford, UK, 2023. Available online: https://ora.ox.ac.uk/objects/uuid:7e2e2273-73d8-4b2e-bc5a-72430830ebf6 (accessed on 27 November 2024).
- Mallory, C.D.; Boyce, M.S. Observed and predicted effects of climate change on Arctic caribou and reindeer. Environ. Rev. 2018, 26, 13–25. [Google Scholar] [CrossRef]
- Tahmin, K.K. Parasitic Infection Risk for the Svalbard Reindeer (Rangifer tarandus platyrhynchus) in Relation to Temperature, Host Density, and Grazing Behaviors. Master’s Thesis, Norwegian University of Life Sciences, Ås, Norway, 2023. Available online: https://nmbu.brage.unit.no/nmbu-xmlui/handle/11250/3148260 (accessed on 27 November 2024).
- Mustonen, T. Wild reindeer as a keystone cultural and ecological species in the Eurasian North. Glob. Change Biol. 2022, 28, 4225–4228. [Google Scholar] [CrossRef] [PubMed]
- Holand, Ø.; Mizin, I.; Weladji, R.B. Reindeer Rangifer tarandus (Linnaeus, 1758). In Handbook of the Mammals of Europe; Hackländer, K., Zachos, F.E., Eds.; Springer Nature: Cham, Switzerland, 2022; pp. 1–30. [Google Scholar] [CrossRef]
- Mitrofanova, O.V.; Dementieva, N.V.; Tyshchenko, V.I.; Krutikova, A.A.; Goncharov, V.V. Assessment of genetic polymorphisms of several populations reindeer (Rangifer tarandus). Genet. I Razved. Zivotn. [Anim. Genet. Breed.] 2017, 1, 49–52. Available online: https://www.vniigenjournal.ru/jour/article/view/61/46 (accessed on 27 November 2024). (In Russian with English summary).
- Kharzinova, V.R.; Dotsev, A.V.; Deniskova, T.E.; Solovieva, A.D.; Fedorov, V.I.; Layshev, K.A.; Romanenko, T.M.; Okhlopkov, I.M.; Wimmers, K.; Reyer, H.; et al. Genetic diversity and population structure of domestic and wild reindeer (Rangifer tarandus L. 1758): A novel approach using BovineHD BeadChip. PLoS ONE 2018, 13, e0207944. [Google Scholar] [CrossRef] [PubMed]
- Kholodova, M.V.; Kolpashchikov, L.A.; Kuznetsova, M.V.; Baranova, A.I. Genetic diversity of wild reindeer (Rangifer tarandus) of Taimyr: Analysis of polymorphism of the control region of mitochondrial DNA. Biol. Bull. 2011, 38, 42–49. [Google Scholar] [CrossRef]
- Klokov, K. Reindeer husbandry in Russia. Int. J. Entrep. Small Bus. 2007, 4, 726–784. [Google Scholar] [CrossRef]
- Yuzhakov, A.A.; Romanenko, T.M.; Layshev, K.A. Phenogeographic variability of the Nenets reindeer breed. Izv. St. Peterbg. Gos. Agrar. Univ. [Izv. St. Petersburg State Agrar. Univ.] 2017, 47, 115–122. Available online: http://archive.today/2024.12.14-141815/https://elibrary.ru/item.asp?id=29757902 (accessed on 11 December 2024). (In Russian).
- Røed, K.H.; Kvie, K.S.; Losey, R.J.; Kosintsev, P.A.; Hufthammer, A.K.; Dwyer, M.J.; Goncharov, V.; Klokov, K.B.; Arzyutov, D.V.; Plekhanov, A.; et al. Temporal and structural genetic variation in reindeer (Rangifer tarandus) associated with the pastoral transition in Northwestern Siberia. Ecol. Evol. 2020, 10, 9060–9072. [Google Scholar] [CrossRef]
- Davydov, A.V.; Morgunov, N.A.; Chugreev, M.K.; Tkacheva, I.S. Reindeer of the taiga zone of Western Siberia. Vestn. APK Verhn. [Her. Agroindustrial Complex Up. Volga Reg.] 2022, 3, 10–21, (In Russian with English summary). [Google Scholar] [CrossRef]
- Davydov, A.V.; Morgunov, N.A.; Chugreev, M.K.; Tkacheva, I.S. Reindeer of the taiga zone of Eastern Siberia. Vestn. APK Verhn. [Her. Agroindustrial Complex Up. Volga Reg.] 2022, 4, 74–87, (In Russian with English summary). [Google Scholar] [CrossRef]
- Pokharel, K.; Weldenegodguad, M.; Dudeck, S.; Honkatukia, M.; Lindeberg, H.; Mazzullo, N.; Paasivaara, A.; Peippo, J.; Soppela, P.; Stammler, F.; et al. Whole-genome sequencing provides novel insights into the evolutionary history and genetic adaptation of reindeer populations in northern Eurasia. Sci. Rep. 2023, 13, 23019. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Lin, Z.; Ba, H.; Chen, L.; Yang, Y.; Wang, K.; Qiu, Q.; Wang, W.; Li, G. Draft genome of the reindeer (Rangifer tarandus). Gigascience 2017, 6, gix102. [Google Scholar] [CrossRef] [PubMed]
- Ponomareva, E.S.; Yildirim, E.A.; Filippova, V.A.; Ilina, L.A.; Dubrowin, A.V.; Laptev, G.Y.; Kalitkina, K.A.; Dunyashev, T.P.; Tiurina, D.G. Comparison of the composition and metabolic potential of the reindeer’s rumen microbiome in the Yamal-Nenets and Nenets autonomous district of the Russian Arctic. Acta Biomed. Sci. 2022, 7, 30–37, (In Russian with English summary). [Google Scholar] [CrossRef]
- Hold, K.; Lord, E.; Brealey, J.C.; Le Moullec, M.; Bieker, V.C.; Ellegaard, M.R.; Rasmussen, J.A.; Kellner, F.L.; Guschanski, K.; Yannic, G.; et al. Ancient reindeer mitogenomes reveal island-hopping colonisation of the Arctic archipelagos. Sci. Rep. 2024, 14, 4143. [Google Scholar] [CrossRef]
- NCBI. Genome Assembly mRanTar1.h1.1. Sequence ID: GCA_949782905.1.; National Center for Biotechnology Information, National Library of Medicine: Bethesda, MD, USA, 2023. Available online: https://ncbi.nlm.nih.gov/datasets/genome/GCA_949782905.1/ (accessed on 27 November 2024).
- NCBI. Rangifer tarandus Growth Hormone Receptor (GHR) Gene, Exon 10 and Partial cds. Sequence ID: DQ062724.1; National Center for Biotechnology Information, National Library of Medicine: Bethesda, MD, USA, 2016. Available online: https://www.ncbi.nlm.nih.gov/nuccore/DQ062724.1 (accessed on 27 November 2024).
- Varvio, S.L.; Iso-Touru, T.; Kantanen, J.; Viitala, S.; Tapio, I.; Mäki-Tanila, A.; Zerabruk, M.; Vilkki, J. Molecular anatomy of the cytoplasmic domain of bovine growth hormone receptor, a quantitative trait locus. Proc. R. Soc. B Biol. Sci. 2008, 275, 1525–1534. [Google Scholar] [CrossRef]
- Krutikova, A.; Dementieva, N.; Mitrofanova, O.; Nikitkina, E. Polymorphic variants of the locus of the growth hormone gene and linkage disequilibrium in the populations of wild and domestic reindeer. Genet. I Razved. Zivotn. [Anim. Genet. Breed.] 2018, 1, 11–16, (In Russian with English summary). [Google Scholar] [CrossRef]
- Krutikova, A.; Barkova, O. Analysis of the ligand-dependent nuclear receptor gene of corepressor type polymorphism in reindeer. Meždunarodnyj Vestn. Vet. [Int. Bull. Vet. Med.] 2020, 4, 111–115, (In Russian with English summary). [Google Scholar] [CrossRef]
- Krutikova, A.A.; Peglivanyan, G.K. Analysis of BMP2 gene polymorphism of bone morphogenetic protein-2 in reindeer. Meždunarodnyj Vestn. Vet. [Int. Bull. Vet. Med.] 2023, 2, 161–170, (In Russian with English summary). [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- MEGA: Molecular Evolutionary Genetics Analysis. Available online: https://www.megasoftware.net/ (accessed on 27 November 2024).
- NCBI. Rangifer tarandus. Available online: https://www.ncbi.nlm.nih.gov/search/all/?term=Rangifer%20tarandus%20 (accessed on 27 November 2024).
- Zenkova, D.; Kamenev, V.; Sablina, R.; Artyomov, M.; Sergushichev, A. Phantasus: Visual and Interactive Gene Expression Analysis, Bioconductor version 3.20; Fred Hutchinson Cancer Center: Seattle, WA, USA, 2018. [Google Scholar] [CrossRef]
- Nei, M. Estimation of average heterozygosity and genetic distance from small number of individuals. Genetics 1978, 89, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Kalinowski, S.T. Counting alleles with rarefaction: Private alleles and hierarchical sampling designs. Conserv. Genet. 2004, 5, 539–543. [Google Scholar] [CrossRef]
- Keenan, K.; McGinnity, P.; Cross, T.F.; Crozier, W.W.; Prodohl, P.A. diveRsity: An R package for the estimation of population genetics parameters and their associated errors. Methods Ecol. Evol. 2013, 4, 782–788. [Google Scholar] [CrossRef]
- Tanihara, F.; Hirata, M.; Namula, Z.; Wittayarat, M.; Do, L.T.K.; Lin, Q.; Takebayashi, K.; Hara, H.; Nagahara, M.; Otoi, T. GHR-mutant pig derived from domestic pig and microminipig hybrid zygotes using CRISPR/Cas9 system. Mol. Biol. Rep. 2023, 50, 5049–5057. [Google Scholar] [CrossRef]
- Wallis, M. Mammalian genome projects reveal new growth hormone (GH) sequences: Characterization of the GH-encoding genes of armadillo (Dasypus novemcinctus), hedgehog (Erinaceus europaeus), bat (Myotis lucifugus), hyrax (Procavia capensis), shrew (Sorex araneus), ground squirrel (Spermophilus tridecemlineatus), elephant (Loxodonta africana), cat (Felis catus) and opossum (Monodelphis domestica). Gen. Comp. Endocrinol. 2008, 155, 271–279. [Google Scholar] [CrossRef]
- Menzies, B.R.; Pask, A.J.; Renfree, M.B. Placental expression of pituitary hormones is an ancestral feature of therian mammals. EvoDevo 2011, 2, 16. [Google Scholar] [CrossRef]
- Lioupis, A.; Wallis, O.C.; Wallis, M. Cloning and characterisation of the gene encoding red deer (Cervus elaphus) growth hormone: Implications for the molecular evolution of growth hormone in artiodactyls. J. Mol. Endocrinol. 1997, 19, 259–266. [Google Scholar] [CrossRef]
- Majeres, L.E.; Dilger, A.C.; Shike, D.W.; McCann, J.C.; Beever, J.E. Defining a haplotype encompassing the LCORL-NCAPG Locus associated with increased lean growth in beef cattle. Genes 2024, 15, 576. [Google Scholar] [CrossRef]
- Larkina, T.A.; Barkova, O.Y.; Peglivanyan, G.K.; Mitrofanova, O.V.; Dementieva, N.V.; Stanishevskaya, O.I.; Vakhrameev, A.B.; Makarova, A.V.; Shcherbakov, Y.S.; Pozovnikova, M.V.; et al. Evolutionary subdivision of domestic chickens: Implications for local breeds as assessed by phenotype and genotype in comparison to commercial and fancy breeds. Agriculture 2021, 11, 914. [Google Scholar] [CrossRef]
- Larkina, T.A.; Romanov, M.N.; Barkova, O.Y.; Peglivanyan, G.K.; Mitrofanova, O.V.; Dementieva, N.V.; Stanishevskaya, O.I.; Vakhrameev, A.B.; Makarova, A.V.; Shcherbakov, Y.S.; et al. Genetic Variation of the NCAPG-LCORL Locus in Chickens of Local Breeds Based on SNP Genotyping Data. In Molecular Genetic Technologies for Analysis of Gene Expression Related to Animal Productivity and Disease Resistance, Materials of the 3rd International Scientific and Practical Conference, Moscow, Russia, September 30, 2021; Pozyabin, S.V., Kochish, I.I., Romanov, M.N., Eds.; Ministry of Agriculture of the Russian Federation; Federal State Budgetary Educational Institution of Higher Education “Moscow State Academy of Veterinary Medicine and Biotechnology—MVA named after K.I. Scriabin”; Sel’skokhozyaistvennye Tekhnologii: Moscow, Russia, 2021; pp. 133–146, (In Russian with English summary). [Google Scholar] [CrossRef]
- Feng, J.Q.; Chen, D.; Ghosh-Choudhury, N.; Esparza, J.; Mundy, G.R.; Harris, S.E. Bone morphogenetic protein 2 transcripts in rapidly developing deer antler tissue contain an extended 5’ non-coding region arising from a distal promoter. Biochim. Biophys. Acta 1997, 1350, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Davydov, A.V.; Mizin, I.A.; Sipko, T.P.; Gruzdev, A.R. Reindeer of the Arctic islands of Russia. Vestn. Ohotovedeniâ [Bull. Game Sci.] 2017, 14, 253–271. Available online: http://archive.today/2024.12.14-142914/https://elibrary.ru/item.asp?id=32501476 (accessed on 11 December 2024). (In Russian).
- Glazov, P.M.; Loshchagina, Y.A.; Shmatova, A.G.; Gnedenko, A.E.; Tishkov, A.A. Kolguev Island as an object for monitoring the biota of the western sector of the Russian Arctic. Arktika Èkologiâ I Èkonomika [Arctic Ecol. Econ.] 2024, 14, 261–273, (In Russian with English summary). [Google Scholar] [CrossRef]
- Wiener, P.; Wilkinson, S. Deciphering the genetic basis of animal domestication. Proc. R. Soc. B Biol. Sci. 2011, 278, 3161–3170. [Google Scholar] [CrossRef]
- Wang, G.D.; Xie, H.B.; Peng, M.S.; Irwin, D.; Zhang, Y.P. Domestication genomics: Evidence from animals. Annu. Rev. Anim. Biosci. 2014, 2, 65–84. [Google Scholar] [CrossRef]
- Frantz, L.A.; Bradley, D.G.; Larson, G.; Orlando, L. Animal domestication in the era of ancient genomics. Nat. Rev. Genet. 2020, 21, 449–460. [Google Scholar] [CrossRef]
- Romanov, M.N.; Sölkner, J.; Zinovieva, N.A.; Wimmers, K.; Weigend, S. Editorial: Traditional and up-to-date genomic insights into domestic animal diversity. Front. Genet. 2023, 13, 1117708. [Google Scholar] [CrossRef]
- Seebacher, F. The evolution of metabolic regulation in animals. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2018, 224, 195–203. [Google Scholar] [CrossRef]
- Thompson, S.; Romanov, M.N.; Griffin, D.K. Study of animal myosins in a comparative genomic aspect. In Molecular Genetic Technologies for Analysis of Gene Expression Related to Animal Productivity and Disease Resistance, Materials of the 3rd International Scientific and Practical Conference, Moscow, Russia, September 30, 2021; Pozyabin, S.V., Kochish, I.I., Romanov, M.N., Eds.; Ministry of Agriculture of the Russian Federation; Federal State Budgetary Educational Institution of Higher Education “Moscow State Academy of Veterinary Medicine and Biotechnology—MVA named after K.I. Scriabin”; Sel’skokhozyaistvennye tekhnologii: Moscow, Russia, 2021; pp. 444–449, (In Russian with English summary). [Google Scholar] [CrossRef]
- Rejduch, B. Genes associated with production and health in farm animals. J. Cent. Eur. Agric. 2008, 9, 829–835. Available online: https://jcea.agr.hr/en/issues/article/682 (accessed on 27 November 2024).
- Pritchard, T.; Coffey, M.; Mrode, R.; Wall, E. Genetic parameters for production, health, fertility and longevity traits in dairy cows. Animal 2013, 7, 34–46. [Google Scholar] [CrossRef]
- Laville, E.; Bouix, J.; Sayd, T.; Bibé, B.; Elsen, J.M.; Larzul, C.; Eychenne, F.; Marcq, F.; Georges, M. Effects of a quantitative trait locus for muscle hypertrophy from Belgian Texel sheep on carcass conformation and muscularity. J. Anim. Sci. 2004, 82, 3128–3137. [Google Scholar] [CrossRef] [PubMed]
- Cockett, N.E.; Smit, M.A.; Bidwell, C.A.; Segers, K.; Hadfield, T.L.; Snowder, G.D.; Georges, M.; Charlier, C. The callipyge mutation and other genes that affect muscle hypertrophy in sheep. Genet. Sel. Evol. 2005, 37 (Suppl. 1), S65. [Google Scholar] [CrossRef] [PubMed]
- Stinckens, A.; Van den Maagdenberg, K.; Luyten, T.; Georges, M.; De Smet, S.; Buys, N. The RYR1 g.1843C>T mutation is associated with the effect of the IGF2 intron3-g.3072G>A mutation on muscle hypertrophy. Anim. Genet. 2007, 38, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Freeman, L.M.; Rush, J.E.; Stern, J.A.; Huggins, G.S.; Maron, M.S. Feline hypertrophic cardiomyopathy: A spontaneous large animal model of human HCM. Cardiovasc. Res. 2017, 8, 139–142. [Google Scholar] [CrossRef]
- Aiello, D.; Patel, K.; Lasagna, E. The myostatin gene: An overview of mechanisms of action and its relevance to livestock animals. Anim. Genet. 2018, 49, 505–519. [Google Scholar] [CrossRef]
- Moiseyeva, I.G.; Nikiforov, A.A.; Romanov, M.N.; Balanovsky, O.P. Dynamics of domestic animal diversity as influenced by anthropogenic factor. In Integration of Archaeological and Ethnographic Studies: Collection of Scientific Papers; Korusenko, M.A., Tikhonov, S.S., Tomilov, N.A., Eds.; Nauka-Omsk: Omsk, Russia, 2003; pp. 284–287. Available online: https://kar.kent.ac.uk/46429/ (accessed on 27 November 2024).
- Druet, T.; Ahariz, N.; Cambisano, N.; Tamma, N.; Michaux, C.; Coppieters, W.; Charlier, C.; Georges, M. Selection in action: Dissecting the molecular underpinnings of the increasing muscle mass of Belgian Blue Cattle. BMC Genom. 2014, 15, 796. [Google Scholar] [CrossRef]
- Kristensen, T.N.; Hoffmann, A.A.; Pertoldi, C.; Stronen, A.V. What can livestock breeders learn from conservation genetics and vice versa? Front. Genet. 2015, 6, 38. [Google Scholar] [CrossRef]
- Georges, M.; Charlier, C.; Hayes, B. Harnessing genomic information for livestock improvement. Nat. Rev. Genet. 2019, 20, 135–156. [Google Scholar] [CrossRef]
- Romanov, M.N.; Kochish, I.I.; Sharafetdinov, G.R.; Myasnikova, O.V.; Nikonov, I.N.; Selina, M.V.; Surai, P.F. Towards Advanced Biotechnological Developments to Realize the Genetic Potential of Egg-type Poultry. In Molecular Genetic Technologies for Analysis of Gene Expression Related to Animal Productivity and Disease Resistance, Materials of the 3rd International Scientific and Practical Conference, Moscow, 29 September 2021; Sel’skokhozyaistvennye tekhnologii: Moscow, Russia, 2021; pp. 40–51, (In Russian with English summary). [Google Scholar] [CrossRef]
- Røed, K.H.; Flagstad, Ø.; Nieminen, M.; Holand, Ø.; Dwyer, M.J.; Røv, N.; Vilà, C. Genetic analyses reveal independent domestication origins of Eurasian reindeer. Proc. R. Soc. B Biol. Sci. 2008, 275, 1849–1855. [Google Scholar] [CrossRef]
- Røed, K.H.; Flagstad, Ø.; Bjørnstad, G.; Hufthammer, A.K. Elucidating the ancestry of domestic reindeer from ancient DNA approaches. Quat. Int. 2011, 238, 83–88. [Google Scholar] [CrossRef]
- Røed, K.H.; Bjørklund, I.; Olsen, B.J. From wild to domestic reindeer—Genetic evidence of a non-native origin of reindeer pastoralism in northern Fennoscandia. J. Archaeol. Sci. Rep. 2018, 19, 279–286. [Google Scholar] [CrossRef]
- Røed, K.H.; Kvie, K.S.; Bårdsen, B.J. Genetic structure and origin of semi-domesticated reindeer. In Reindeer Husbandry and Global Environmental Change: Pastoralism in Fennoscandia; Horstkotte, T., Holand, Ø., Kumpula, J., Moen, J., Eds.; Routledge: London, UK, 2022; pp. 48–60. [Google Scholar] [CrossRef]
- Sampels, S.; Pickova, J.; Wiklund, E. Influence of production system, age an sex on carcass parameters and some biochemical meat quality characteristics of reindeer (Rangifer tarandus tarandus L.). Rangifer 2005, 25, 85–96. [Google Scholar] [CrossRef]
- Wiklund, E.; Farouk, M.; Finstad, G. Venison: Meat from red deer (Cervus elaphus) and reindeer (Rangifer tarandus tarandus). Anim. Front. 2014, 4, 55–61. [Google Scholar] [CrossRef]
- Plemyashov, K.V.; Smaragdov, M.G.; Romanov, M.N. Molecular Genetic Polymorphism in Animal Populations and Its Application in Intensive Breeding of Dairy Cattle—A Review. In Molecular Genetic Technologies for Analysis of Gene Expression Related to Animal Productivity and Disease Resistance, Materials of the 3rd International Scientific and Practical Conference, Moscow, Russia, September 30, 2021; Pozyabin, S.V., Kochish, I.I., Romanov, M.N., Eds.; Ministry of Agriculture of the Russian Federation; Federal State Budgetary Educational Institution of Higher Education “Moscow State Academy of Veterinary Medicine and Biotechnology—MVA named after K.I. Scriabin”; Sel’skokhozyaistvennye tekhnologii: Moscow, Russia, 2021; pp. 368–378, (In Russian with English summary). [Google Scholar] [CrossRef]
- Konorov, E.A.; Kurbakov, K.A.; Semina, M.T.; Stolpovsky, Y.A.; Layshev, K.A. Analysis of the calpastatin (CAST) and androgen receptor (AR) gene polymorphisms as biomarkers for meat quality traits in reindeer Rangifer tarandus. Russ. J. Genet. 2024, 60, 1040–1045. [Google Scholar] [CrossRef]
- Davydov, A.V.; Morgunov, N.A.; Chugreev, M.K.; Tkacheva, I.S. Reindeer of Taimyr. Vestn. APK Verhn. [Her. Agroindustrial Complex Up. Volga Reg.] 2023, 3, 5–20, (In Russian with English summary). [Google Scholar] [CrossRef]
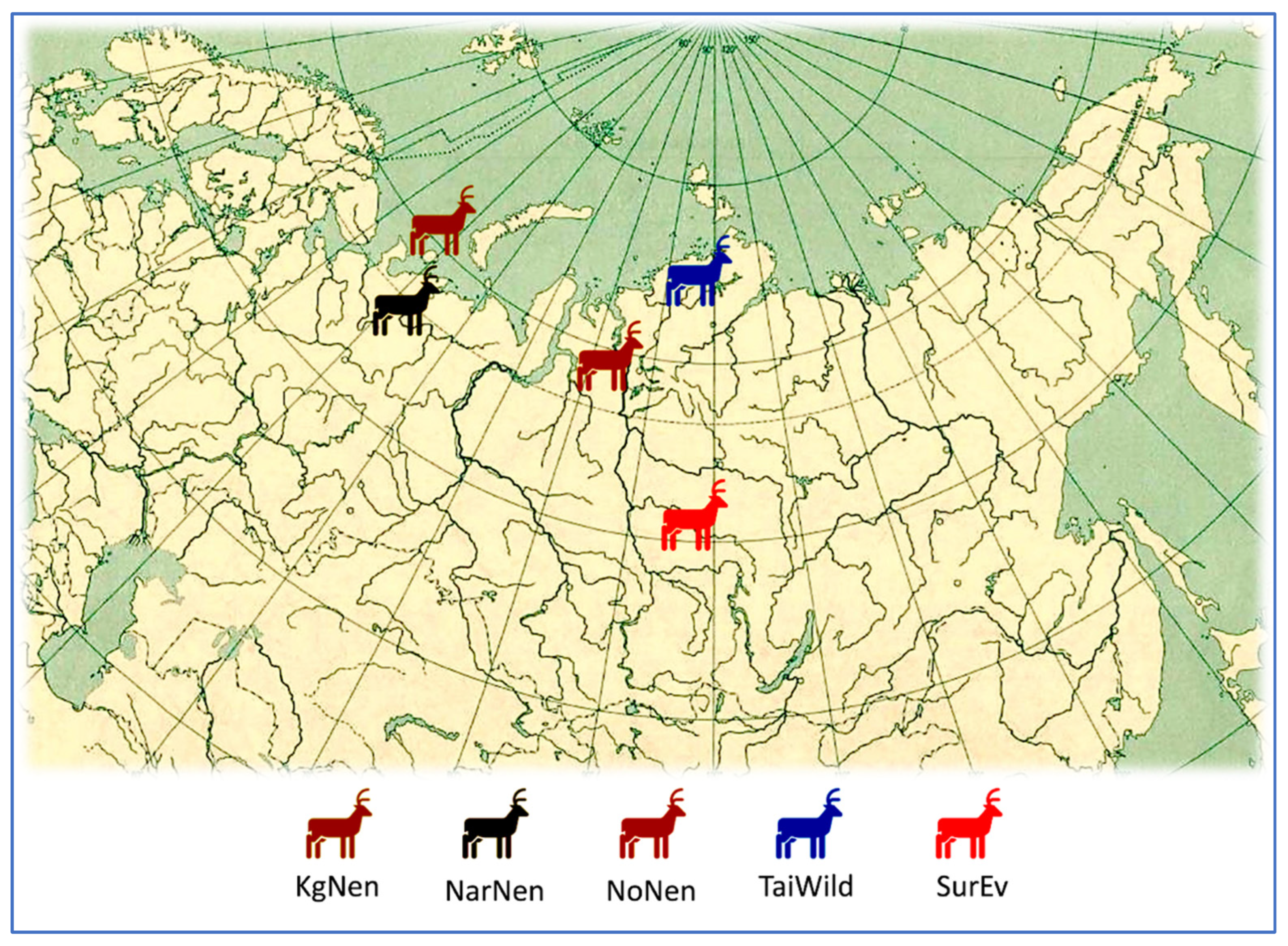


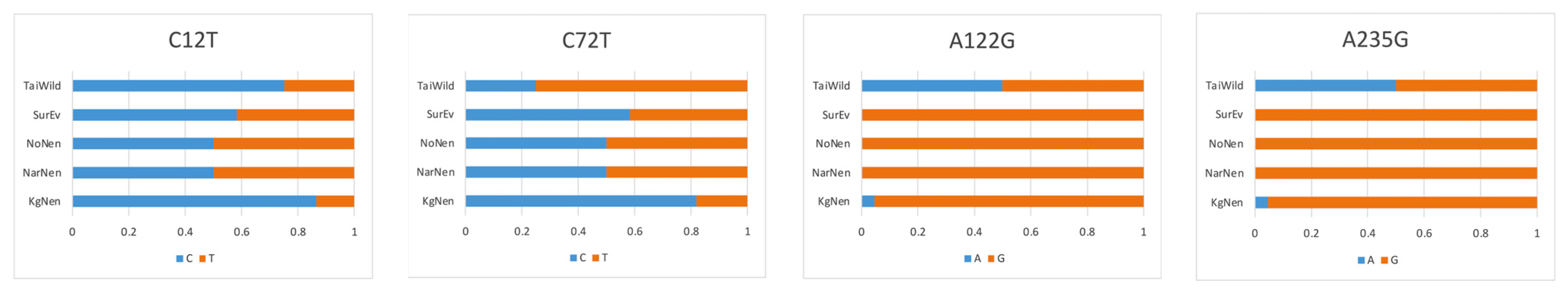

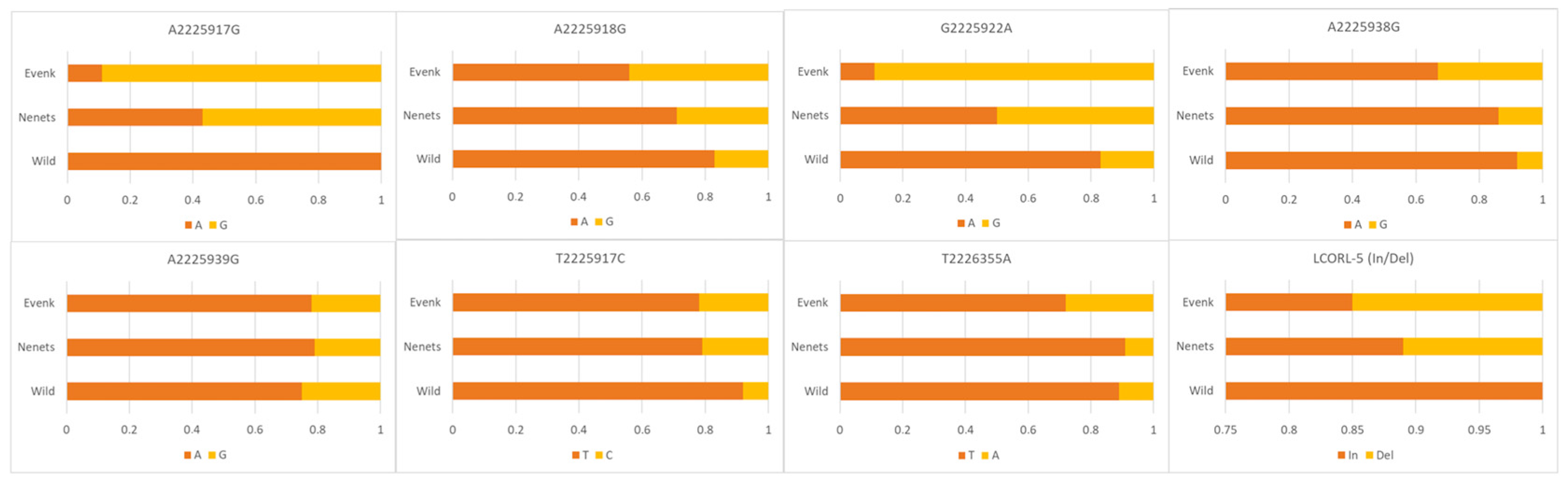
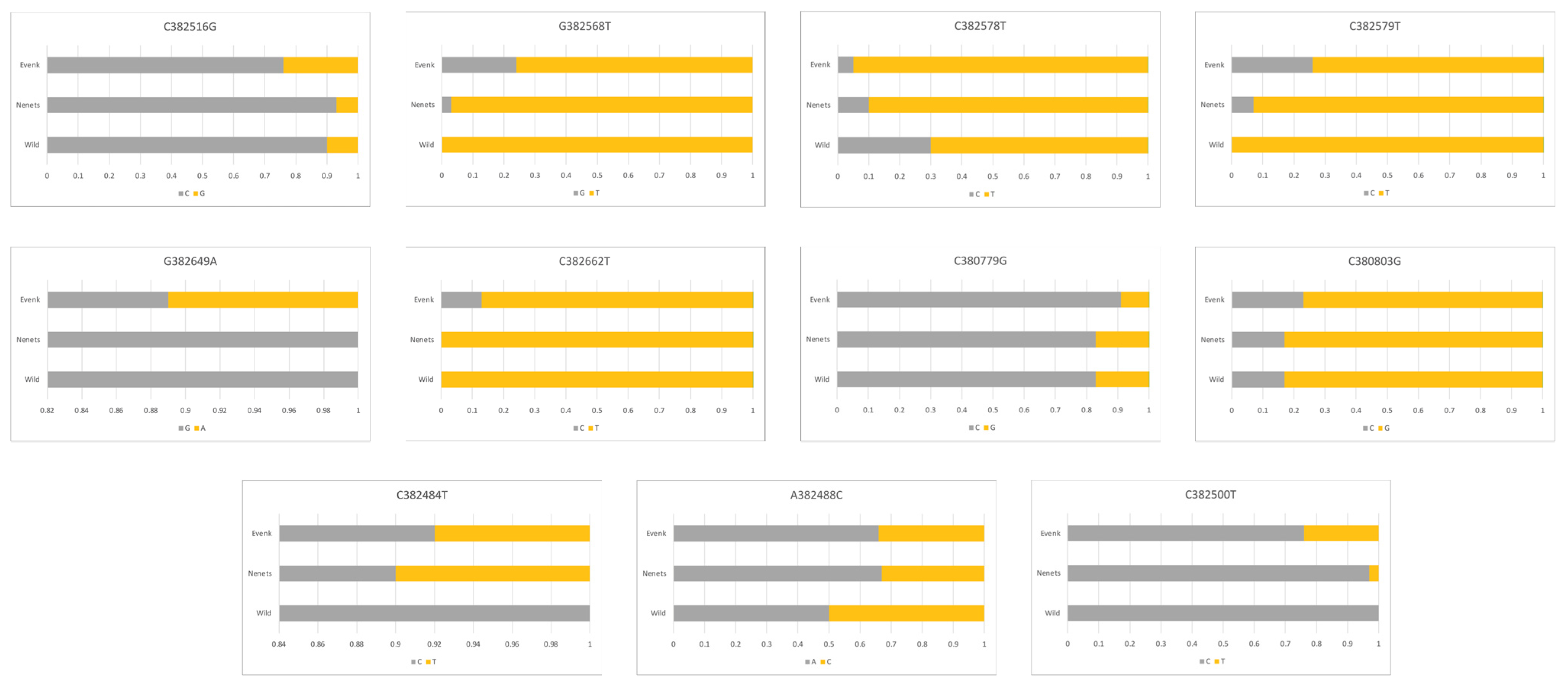

| Gene/Locus | Primers (F, Forward; R, Reverse) | Fragment Size, bp |
|---|---|---|
| GHR | Exon 10 F: TTTGTTAAATCAATTGTTGTGAG R: GTCGCATTGAGTACAAGGC | |
| 844 | ||
| GH | Exons 2 and 3 F: GGAGAAGCAGAAGGCAACC R: CTCTGCCTGCCCTGGACT | |
| 382 | ||
| LCORL | Exon 7 regions | |
| LCORL-5 | F: CATCCAAGAAATTGATAGAA R: TTTCACAACCTGGGGACCTA | 682, 646 |
| LCORL-9 | F: TTTTGAGTAAGACTGAGGGA R: GTGGTCTTCCATGGTGGTCT | 657 |
| LCORL-10 | F: TCTTAGCAAACTGAACAAAA R: GCCAAGAAATTAGATTGTCCA | 640 |
| BMP2 | Regions of exons 1 and 2 | |
| BMP2-1 (exon 1) | F: TCGCGGATTACTAGGGACTCA R: GCGCAAGTTATTCTCCCTGC | 705 |
| BMP2-1 (exon 2) | F: GCGCTGTGTGTTTGGGTTAG R: AAAGCCAGGTTCGGAAAGGT | 872 |
| SNPs and Alleles | Populations 1 | ||||
|---|---|---|---|---|---|
| KgNen | NarNen | NoNen | SurEv | TaiWild | |
| GHR1 | |||||
| A | 0.55 | 0.59 | 0.65 | 0.58 | 0.80 |
| G | 0.45 | 0.41 | 0.35 | 0.42 | 0.20 |
| GHR2 | |||||
| C | 0.64 | 0.55 | 0.85 | 0.83 | 0.70 |
| T | 0.36 | 0.45 | 0.15 | 0.17 | 0.30 |
| GHR3 | |||||
| C | 0.41 | 0.41 | 0.20 | 0.08 | 0.30 |
| T | 0.59 | 0.59 | 0.80 | 0.92 | 0.70 |
| Populations | HO | HE | UHE | AR | |
|---|---|---|---|---|---|
| KgNen | 0.26 ± 0.05 | 0.31 ± 0.07 | 0.32 ± 0.07 | 1.77 ± 0.09 | |
| NarNen | 0.44 ± 0.16 | 0.35 ± 0.09 | 0.37 ± 0.09 | 1.71 ± 0.18 | |
| NoNen | 0.37 ± 0.17 | 0.29 ± 0.08 | 0.30 ± 0.09 | 1.65 ± 0.17 | |
| SurEv | 0.28 ± 0.14 | 0.27 ± 0.08 | 0.29 ± 0.09 | 1.60 ± 0.17 | |
| TaiWild | 0.23 ± 0.02 | 0.41 ± 0.02 | 0.44 ± 0.03 | 1.95 ± 0.01 | |
| Ranks | M | ||||
| SurEv | 3 | 1 | 1 | 1 | 1.5 |
| NoNen | 4 | 2 | 2 | 2 | 2.5 |
| KgNen | 2 | 3 | 3 | 4 | 3 |
| NarNen | 5 | 4 | 4 | 3 | 4 |
| TaiWild | 1 | 5 | 5 | 5 | 4 |
| Populations | HO | HE | UHE | AR | |
|---|---|---|---|---|---|
| Wild | 0.32 ± 0.09 | 0.19 ± 0.05 | 0.19 ± 0.05 | 1.47 ± 0.12 | |
| Nenets | 0.16 ± 0.08 | 0.11 ± 0.05 | 0.11 ± 0.05 | 1.29 ± 0.11 | |
| Evenk | 0.28 ± 0.08 | 0.24 ± 0.05 | 0.24 ± 0.05 | 1.64 ± 0.12 | |
| Ranks | M | ||||
| Wild | 3 | 2 | 2 | 2 | 2.25 |
| Nenets | 1 | 1 | 1 | 1 | 1 |
| Evenk | 2 | 3 | 3 | 3 | 2.75 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krutikova, A.A.; Dementieva, N.V.; Shcherbakov, Y.S.; Goncharov, V.V.; Griffin, D.K.; Romanov, M.N. Polymorphism of Genes Potentially Affecting Growth and Body Size Suggests Genetic Divergence in Wild and Domestic Reindeer (Rangifer tarandus) Populations. Genes 2024, 15, 1629. https://doi.org/10.3390/genes15121629
Krutikova AA, Dementieva NV, Shcherbakov YS, Goncharov VV, Griffin DK, Romanov MN. Polymorphism of Genes Potentially Affecting Growth and Body Size Suggests Genetic Divergence in Wild and Domestic Reindeer (Rangifer tarandus) Populations. Genes. 2024; 15(12):1629. https://doi.org/10.3390/genes15121629
Chicago/Turabian StyleKrutikova, Anna A., Natalia V. Dementieva, Yuri S. Shcherbakov, Vasiliy V. Goncharov, Darren K. Griffin, and Michael N. Romanov. 2024. "Polymorphism of Genes Potentially Affecting Growth and Body Size Suggests Genetic Divergence in Wild and Domestic Reindeer (Rangifer tarandus) Populations" Genes 15, no. 12: 1629. https://doi.org/10.3390/genes15121629
APA StyleKrutikova, A. A., Dementieva, N. V., Shcherbakov, Y. S., Goncharov, V. V., Griffin, D. K., & Romanov, M. N. (2024). Polymorphism of Genes Potentially Affecting Growth and Body Size Suggests Genetic Divergence in Wild and Domestic Reindeer (Rangifer tarandus) Populations. Genes, 15(12), 1629. https://doi.org/10.3390/genes15121629










