Comparative Transcriptome Analysis Reveals Mechanisms of Differential Salinity Tolerance Between Suaeda glauca and Suaeda salsa
Highlights
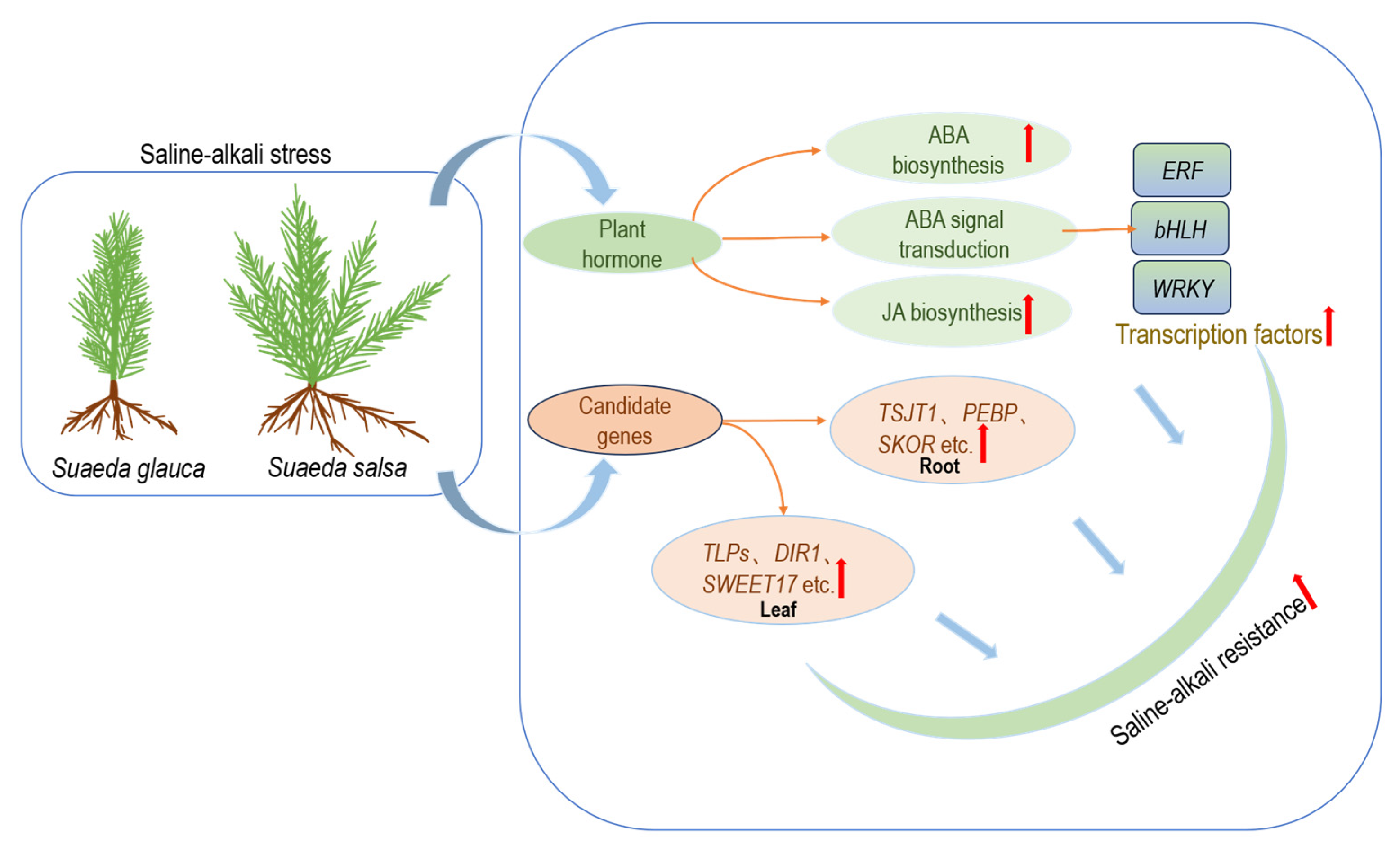
- A comparative transcriptome analysis revealed differences in the saline–alkali tolerance mechanisms between Suaeda glauca and Suaeda salsa.
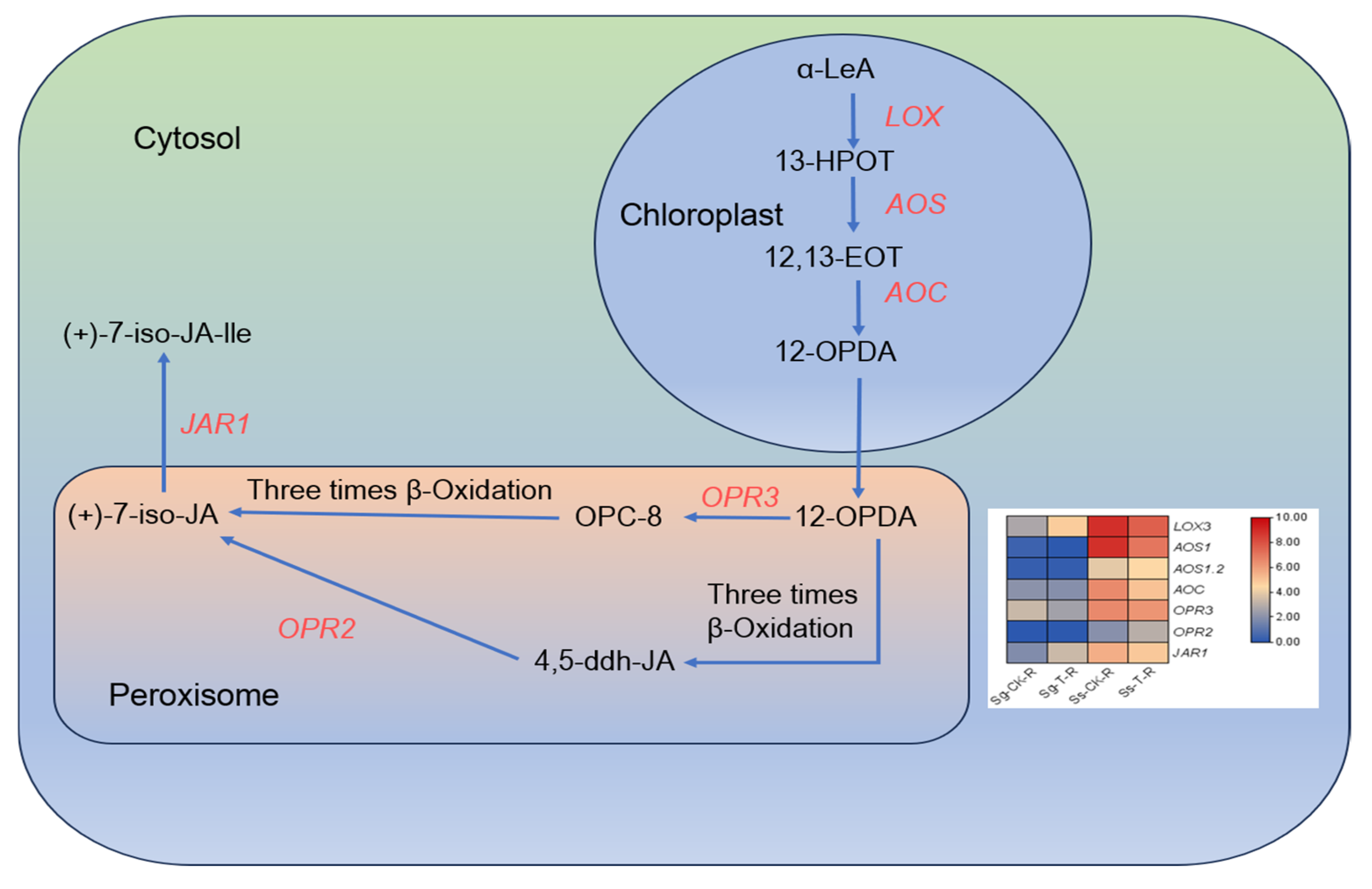
- Suaeda salsa showed a higher expression of genes in the ABA (abscisic acid) and JA (jasmonic acid) synthesis pathways compared to Suaeda glauca, indicating that these hormones may influence their differing saline–alkali tolerance.
- Twenty-four stress-resistance genes were identified in Suaeda glauca and Suaeda salsa, providing significant candidate genes for research on the saline–alkali tolerance abilities of the two plants.
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Extraction and Quality Testing of RNA
2.3. Construction of RNA-Seq Sequencing Library and Transcriptome Sequencing
2.4. Transcriptome Assembly and Functional Annotation
2.5. Differential Expression Gene Analysis
2.6. Statistical Analysis
3. Results
3.1. Morphological Analysis of S. glauca and S. salsa
3.2. Quantification of Sequencing Libraries and Transcriptome Assembly
3.3. Functional Annotation of Unigenes
3.4. Statistics of Unigene Gene Expression
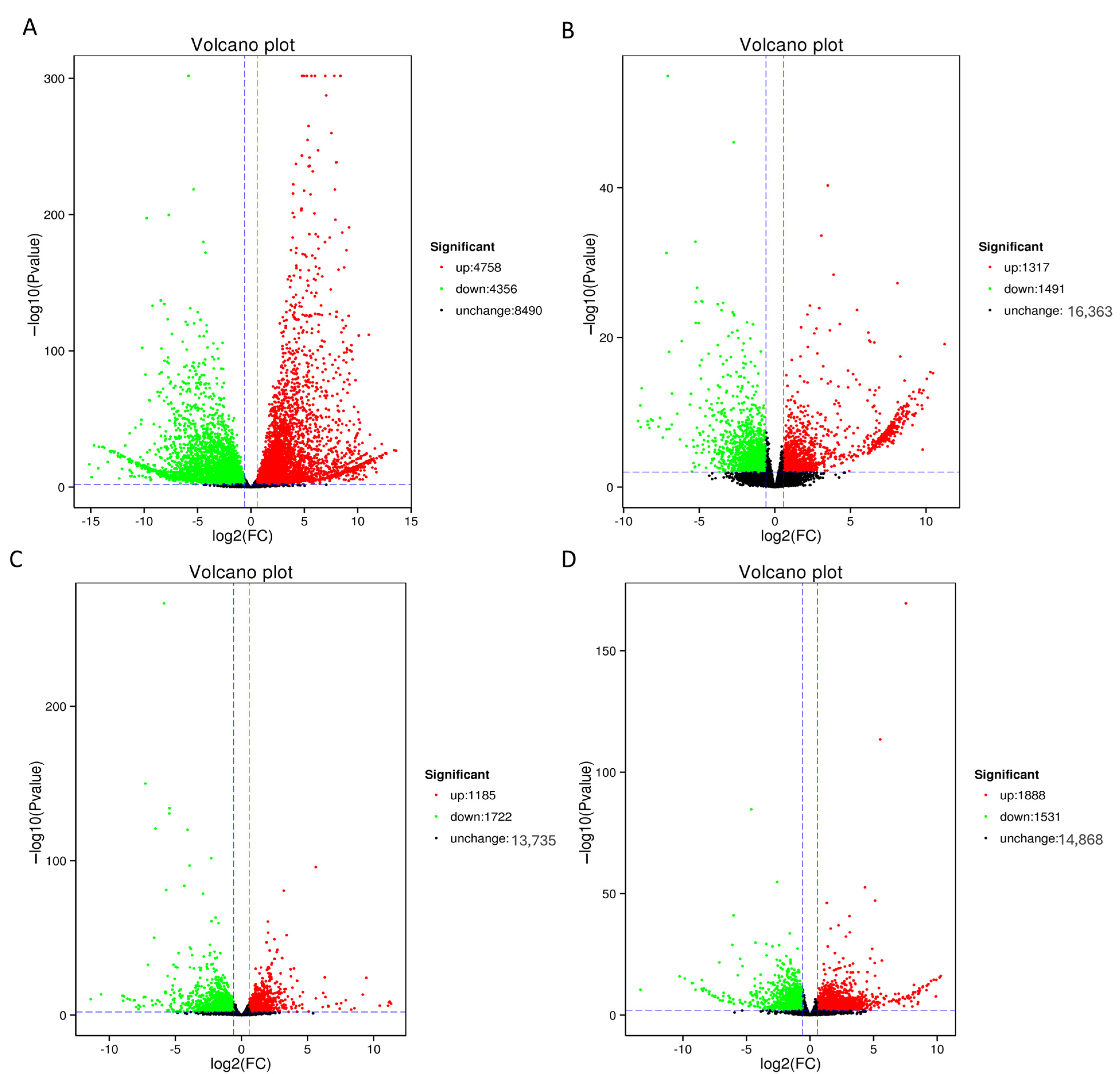
3.5. Screening of Differentially Expressed Genes (DEGs)
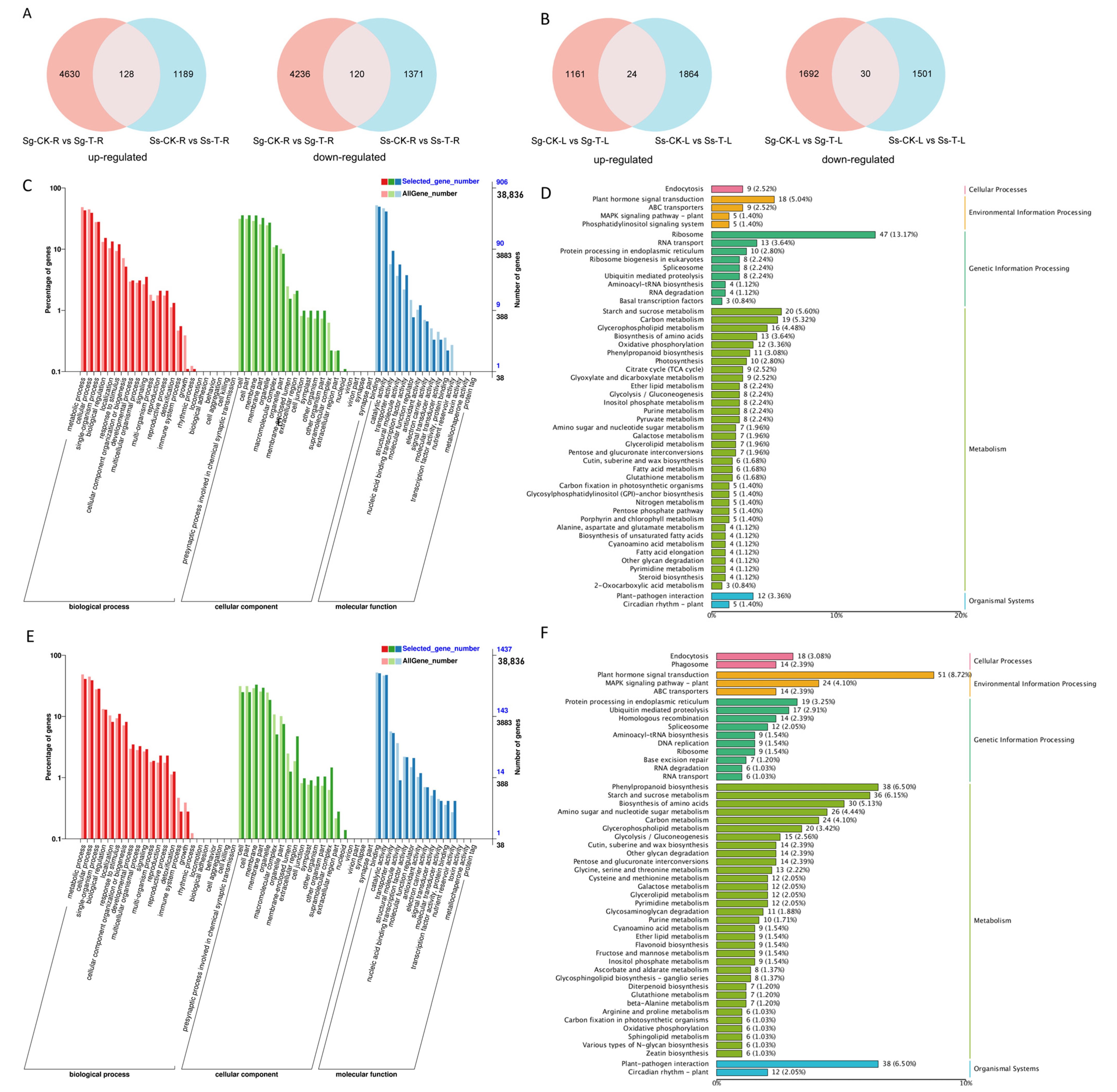
3.6. DEG Analysis
3.7. The Important Role of Plant Hormone-Related Genes in Saline–Alkali Stress
3.8. Identification of Candidate Genes for Saline–Alkali Tolerance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shrivastava, P.; Kumar, R. Soil salinity: A serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J. Biol. Sci. 2015, 22, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Machado, R.; Serralheiro, R. Soil Salinity: Effect on Vegetable Crop Growth. Management Practices to Prevent and Mitigate Soil Salinization. Horticulturae 2017, 3, 30. [Google Scholar] [CrossRef]
- Herbert, E.R.; Boon, P.; Burgin, A.J.; Neubauer, S.C.; Franklin, R.B.; Ardón, M.; Hopfensperger, K.N.; Lamers, L.P.M.; Gell, P. A global perspective on wetland salinization: Ecological consequences of a growing threat to freshwater wetlands. Ecosphere 2015, 6, 206–243. [Google Scholar] [CrossRef]
- Mazhar, S.; Pellegrini, E.; Contin, M.; Bravo, C.; De Nobili, M. Impacts of salinization caused by sea level rise on the biological processes of coastal soils—A review. Front. Environ. Sci. 2022, 10, 909415. [Google Scholar] [CrossRef]
- Hassani, A.; Azapagic, A.; Shokri, N. Global predictions of primary soil salinization under changing climate in the 21st century. Nat. Commun. 2021, 12, 6663. [Google Scholar] [CrossRef]
- Li, J.; Pu, L.; Han, M.; Zhu, M.; Zhang, R.; Xiang, Y. Soil salinization research in China: Advances and prospects. J. Geogr. Sci. 2014, 24, 943–960. [Google Scholar] [CrossRef]
- Zhang, J.T.N.N.; Mu, C.S. Effects of saline and alkaline stresses on the germination, growth, photosynthesis, ionic balance and anti-oxidant system in an alkali-tolerant leguminous forage Lathyrus quinquenervius. Soil Sci. Plant Nutr. 2009, 55, 685–697. [Google Scholar] [CrossRef]
- Munns, R.; James, R.A.; Lauchli, A. Approaches to increasing the salt tolerance of wheat and other cereals. J. Exp. Bot. 2006, 57, 1025–1043. [Google Scholar] [CrossRef]
- Rahman, M.M.; Mostofa, M.G.; Keya, S.S.; Siddiqui, M.N.; Ansary, M.; Das, A.K.; Rahman, M.A.; Tran, L.S. Adaptive Mechanisms of Halophytes and Their Potential in Improving Salinity Tolerance in Plants. Int. J. Mol. Sci. 2021, 22, 10733. [Google Scholar] [CrossRef]
- Hasegawa, P.M.; Bressan, R.A.; Zhu, J.K.; Bohnert, H.J. Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000, 51, 463–499. [Google Scholar] [CrossRef]
- Toscano, S.; Romano, D.; Ferrante, A. Molecular Responses of Vegetable, Ornamental Crops, and Model Plants to Salinity Stress. Int. J. Mol. Sci. 2023, 24, 3190. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Ishitani, M.; Kim, C.; Zhu, J.K. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. USA 2000, 97, 6896–6901. [Google Scholar] [CrossRef] [PubMed]
- Jaime-Perez, N.; Pineda, B.; Garcia-Sogo, B.; Atares, A.; Athman, A.; Byrt, C.S.; Olias, R.; Asins, M.J.; Gilliham, M.; Moreno, V.; et al. The sodium transporter encoded by the HKT1;2 gene modulates sodium/potassium homeostasis in tomato shoots under salinity. Plant Cell Environ. 2017, 40, 658–671. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zeng, H.; Xu, F.; Yan, F.; Xu, W. H (+)-ATPases in Plant Growth and Stress Responses. Annu. Rev. Plant Biol. 2022, 73, 495–521. [Google Scholar] [CrossRef]
- Krishnamurthy, P.; Ranathunge, K.; Nayak, S.; Schreiber, L.; Mathew, M.K. Root apoplastic barriers block Na+ transport to shoots in rice (Oryza sativa L.). J. Exp. Bot. 2011, 62, 4215–4228. [Google Scholar] [CrossRef]
- Azeem, M.; Pirjan, K.; Qasim, M.; Mahmood, A.; Javed, T.; Muhammad, H.; Yang, S.; Dong, R.; Ali, B.; Rahimi, M. Salinity stress improves antioxidant potential by modulating physio-biochemical responses in Moringa oleifera Lam. Sci. Rep. 2023, 13, 2895. [Google Scholar] [CrossRef]
- Zhang, Y.; Qin, C.; Liu, S.; Xu, Y.; Li, Y.; Zhang, Y.; Song, Y.; Sun, M.; Fu, C.; Qin, Z.; et al. Establishment of an efficient Agrobacterium-mediated genetic transformation system in halophyte Puccinellia tenuiflora. Mol. Breed. 2021, 41, 55. [Google Scholar] [CrossRef]
- Jin, T.; Sun, Y.; Shan, Z.; He, J.; Wang, N.; Gai, J.; Li, Y. Natural variation in the promoter of GsERD15B affects salt tolerance in soybean. Plant Biotechnol. J. 2021, 19, 1155–1169. [Google Scholar] [CrossRef]
- Li, H.; Tang, X.; Yang, X.; Zhang, H. Comprehensive transcriptome and metabolome profiling reveal metabolic mechanisms of Nitraria sibirica Pall. to salt stress. Sci. Rep. 2021, 11, 12878. [Google Scholar] [CrossRef]
- Yu, W.; Wu, W.; Zhang, N.; Wang, L.; Wang, Y.; Wang, B.; Lan, Q.; Wang, Y. Research Advances on Molecular Mechanism of Salt Tolerance in Suaeda. Biology 2022, 11, 1273. [Google Scholar] [CrossRef]
- Li, Q.; Liu, R.; Li, Z.; Fan, H.; Song, J. Positive effects of NaCl on the photoreaction and carbon assimilation efficiency in Suaeda salsa. Plant Physiol. Biochem. 2022, 177, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Hu, Y.; Liu, M.; Li, C.; Chang, Y. Evolutions of 30-Year Spatio-Temporal Distribution and Influencing Factors of Suaeda salsa in Bohai Bay, China. Remote Sens. 2022, 14, 138. [Google Scholar] [CrossRef]
- Xu, K.; Cui, X.; Ren, X.; Meng, J.; Fu, X.; Xia, Q. Discovery of natural polyphenols from the wild vegetable Suaeda salsa L. with potential cardioprotective functions. Food Chem. 2023, 405, 134968. [Google Scholar] [CrossRef] [PubMed]
- Joshi, A.; Rajput, V.D.; Verma, K.K.; Minkina, T.; Ghazaryan, K.; Arora, J. Potential of Suaeda nudiflora and Suaeda fruticosa to Adapt to High Salinity Conditions. Horticulturae 2023, 9, 74. [Google Scholar] [CrossRef]
- Zhao, H.; Ma, Y.; Li, J.; Piao, L. Study on Edible Value of Suaeda salsa (L.) Pall. J. Anhui Agric. Sci. 2010, 38, 14350–14351. [Google Scholar]
- Lu, C.; Qiu, N.; Wang, B.; Zhang, J. Salinity treatment shows no effects on photosystem II photochemistry, but increases the resistance of photosystem II to heat stress in halophyte Suaeda salsa. J. Exp. Bot. 2003, 54, 851–860. [Google Scholar] [CrossRef]
- Wang, B.; Luttge, U.; Ratajczak, R. Specific regulation of SOD isoforms by NaCl and osmotic stress in leaves of the C3 halophyte Suaeda salsa L. J. Plant Physiol. 2004, 161, 285–293. [Google Scholar] [CrossRef]
- Zhang, L.; Ma, X.L.; Zhang, Q.; Ma, C.L.; Wang, P.P.; Sun, Y.F.; Zhao, Y.X.; Zhang, H. Expressed sequence tags from a NaCl-treated Suaeda salsa cDNA library. Gene 2001, 267, 193–200. [Google Scholar] [CrossRef]
- Li, H.; Wang, H.; Wen, W.; Yang, G. The antioxidant system in Suaeda salsa under salt stress. Plant Signal. Behav. 2020, 15, 1771939. [Google Scholar] [CrossRef]
- Li, P.-H.; Wang, Z.-L.; Zhang, H.; Wang, B.-S. Cloning and Expression Analysis of the B Subunit of V-H+-ATPase in Leaves of Halophyte Suaeda salsa Under Salt Stress. Acta Bot. Sin. 2004, 46, 93–99. [Google Scholar]
- Yang, C.; Shi, D.; Wang, D. Comparative effects of salt and alkali stresses on growth, osmotic adjustment and ionic balance of an alkali-resistant halophyte Suaeda glauca (Bge.). Plant Growth Regul. 2008, 56, 179–190. [Google Scholar] [CrossRef]
- Wu, Y.; Feng, X.; Zang, R.; Li, J.; Liu, X. Comparative study on the growth and Cd uptake of Suaeda salsa and Suaeda glauca under the stress of salt and Cd and their interaction. Chin. J. Eco-Agric. 2022, 30, 1186–1193. [Google Scholar]
- Qi, C.; Chen, M.; Song, J.; Wang, B. Increase in aquaporin activity is involved in leaf succulence of the euhalophyte Suaeda salsa, under salinity. Plant Sci. 2009, 176, 200–205. [Google Scholar] [CrossRef]
- Song, X.; Yang, N.; Su, Y.; Lu, X.; Liu, J.; Liu, Y.; Zhang, Z.; Tang, Z. Suaeda glauca and Suaeda salsa Employ Different Adaptive Strategies to Cope with Saline–Alkali Environments. Agronomy 2022, 12, 2496. [Google Scholar] [CrossRef]
- Wang, W.; Liu, Y.; Duan, H.; Yin, X.; Cui, Y.; Chai, W.; Song, X.; Flowers, T.J.; Wang, S. SsHKT1;1 is coordinated with SsSOS1 and SsNHX1 to regulate Na+ homeostasis in Suaeda salsa under saline conditions. Plant Soil. 2020, 449, 117–131. [Google Scholar] [CrossRef]
- Qiu, N.; Chen, M.; Guo, J.; Bao, H.; Ma, X.; Wang, B. Coordinate up-regulation of V-H+-ATPase and vacuolar Na+/H+ antiporter as a response to NaCl treatment in a C3 halophyte Suaeda salsa. Plant Sci. 2007, 172, 1218–1225. [Google Scholar] [CrossRef]
- Mori, S.; Suzuki, K.; Oda, R.; Higuchi, K.; Maeda, Y.; Yoshiba, M.; Tadano, T. Characteristics of Na+ and K+ absorption in Suaeda salsa (L.) Pall. Soil Sci. Plant Nutr. 2011, 57, 377–386. [Google Scholar] [CrossRef]
- Wang, B.; Tseng, E.; Regulski, M.; Clark, T.A.; Hon, T.; Jiao, Y.; Lu, Z.; Olson, A.; Stein, J.C.; Ware, D. Unveiling the complexity of the maize transcriptome by single-molecule long-read sequencing. Nat. Commun. 2016, 7, 11708. [Google Scholar] [CrossRef]
- Ozsolak, F.; Milos, P.M. RNA sequencing: Advances, challenges and opportunities. Nat. Rev. Genet. 2011, 12, 87–98. [Google Scholar] [CrossRef]
- Stark, R.; Grzelak, M.; Hadfield, J. RNA sequencing: The teenage years. Nat. Rev. Genet. 2019, 20, 631–656. [Google Scholar] [CrossRef]
- Qian, G.; Wang, M.; Wang, X.; Liu, K.; Li, Y.; Bu, Y.; Li, L. Integrated Transcriptome and Metabolome Analysis of Rice Leaves Response to High Saline-Alkali Stress. Int. J. Mol. Sci. 2023, 24, 4062. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Xiao, B.; Kaderbek, T.; Lin, Z.; Tan, K.; Wu, Q.; Yuan, L.; Lai, J.; Zhao, H.; Song, W. Dynamic transcriptome landscape of developing maize ear. Plant. J. 2023, 116, 1856–1870. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Xu, Y.; Lu, P.; Wang, X.; Li, Z.; Cai, X.; Zhou, Z.; Wang, Y.; Zhang, Z.; Lin, Z.; et al. Salt stress responsiveness of a wild cotton species (Gossypium klotzschianum) based on transcriptomic analysis. PLoS ONE 2017, 12, e178313. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, A.; Zhao, J.; Shang, J.; Zhu, Z.; Wu, X.; Zha, D. Transcriptome profiling reveals potential genes involved in browning of fresh-cut eggplant (Solanum melongena L.). Sci. Rep. 2021, 11, 16081. [Google Scholar] [CrossRef]
- Chen, P.; Ran, H.; Li, J.; Zong, J.; Luo, Q.; Zhao, T.; Liao, Z.; Tang, Y.; Fu, Y. Antioxidant Activity of Phenolic Extraction from Different Sweetpotato (Ipomoea batatas (L.) Lam.) Blades and Comparative Transcriptome Analysis Reveals Differentially Expressed Genes of Phenolic Metabolism in Two Genotypes. Genes 2022, 13, 1078. [Google Scholar] [CrossRef]
- Liu, X.; Liu, F.; Zhao, S.; Guo, B.; Ling, P.; Han, G.; Cui, Z. Purification of an acidic polysaccharide from Suaeda salsa plant and its anti-tumor activity by activating mitochondrial pathway in MCF-7 cells. Carbohydr. Polym. 2019, 215, 99–107. [Google Scholar] [CrossRef]
- Gao, Y.Q.; Yan, B.W.; Zhao, Y.; Wang, F.; Dong, J.J.; He, L.; Zhao, C.J.; Li, Z.T.; Xu, J.Y. Transcriptome analysis of Suaeda salsa and expression profiles of genes related to oil synthesis. Chin. J. Oil Crop Sci. 2018, 40, 801–811. [Google Scholar]
- Xu, Y.; Zhao, Y.; Duan, H.; Sui, N.; Yuan, F.; Song, J. Transcriptomic profiling of genes in matured dimorphic seeds of euhalophyte Suaeda salsa. BMC Genomics 2017, 18, 727. [Google Scholar] [CrossRef]
- Ma, P.; Li, J.; Sun, G.; Zhu, J. Comparative transcriptome analysis reveals the adaptive mechanisms of halophyte Suaeda dendroides encountering high saline environment. Front. Plant Sci. 2024, 15, 1283912. [Google Scholar] [CrossRef]
- Guo, S.M.; Tan, Y.; Chu, H.J.; Sun, M.X.; Xing, J.C. Transcriptome sequencing revealed molecular mechanisms underlying tolerance of Suaeda salsa to saline stress. PLoS ONE 2019, 14, e219979. [Google Scholar] [CrossRef]
- Chen, C.; Shang, X.; Sun, M.; Tang, S.; Khan, A.; Zhang, D.; Yan, H.; Jiang, Y.; Yu, F.; Wu, Y.; et al. Comparative Transcriptome Analysis of Two Sweet Sorghum Genotypes with Different Salt Tolerance Abilities to Reveal the Mechanism of Salt Tolerance. Int. J. Mol. Sci. 2022, 23, 2272. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Hou, L.; Meng, J.; You, H.; Li, Z.; Gong, Z.; Yang, S.; Shi, Y. The Antagonistic Action of Abscisic Acid and Cytokinin Signaling Mediates Drought Stress Response in Arabidopsis. Mol. Plant 2018, 11, 970–982. [Google Scholar] [CrossRef] [PubMed]
- Shao, X.; Gai, D.; Gao, D.; Geng, Y.; Guo, L. Effects of Salt-Alkaline Stress on Carbohydrate Metabolism in Rice Seedlings. Phyton-Ann. REI Bot 2022, 91, 745–759. [Google Scholar]
- Cuin, T.A.; Shabala, S. Amino acids regulate salinity-induced potassium efflux in barley root epidermis. Planta 2007, 225, 753–761. [Google Scholar] [CrossRef]
- Jalmi, S.K.; Sinha, A.K. ROS mediated MAPK signaling in abiotic and biotic stress- striking similarities and differences. Front. Plant Sci. 2015, 6, 769. [Google Scholar] [CrossRef]
- Dong, N.Q.; Lin, H.X. Contribution of phenylpropanoid metabolism to plant development and plant-environment interactions. J. Integr. Plant Biol. 2021, 63, 180–209. [Google Scholar] [CrossRef]
- McDowell, N.G. Mechanisms linking drought, hydraulics, carbon metabolism, and vegetation mortality. Plant Physiol. 2011, 155, 1051–1059. [Google Scholar] [CrossRef]
- Flexas, J.; Bota, J.; Loreto, F.; Cornic, G.; Sharkey, T.D. Diffusive and metabolic limitations to photosynthesis under drought and salinity in C(3) plants. Plant Biol. 2004, 6, 269–279. [Google Scholar] [CrossRef]
- Saddhe, A.A.; Manuka, R.; Penna, S. Plant sugars: Homeostasis and transport under abiotic stress in plants. Physiol. Plant. 2021, 171, 739–755. [Google Scholar] [CrossRef]
- Liang, Y.; Li, X.; Lei, F.; Yang, R.; Bai, W.; Yang, Q.; Zhang, D. Transcriptome Profiles Reveals ScDREB10 from Syntrichia caninervis Regulated Phenylpropanoid Biosynthesis and Starch/Sucrose Metabolism to Enhance Plant Stress Tolerance. Plants 2024, 13, 205. [Google Scholar] [CrossRef]
- Wang, X.; Chang, L.; Wang, B.; Wang, D.; Li, P.; Wang, L.; Yi, X.; Huang, Q.; Peng, M.; Guo, A. Comparative proteomics of The llungiella halophila leaves from plants subjected to salinity reveals the importance of chloroplastic starch and soluble sugars in halophyte salt tolerance. Mol. Cell. Proteom. 2013, 12, 2174–2195. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Meng, Y.; Bai, Y.; Yu, H.; Qian, Y.; Zhang, D.; Zhou, Y. Starch and Sucrose Metabolism and Plant Hormone Signaling Pathways Play Crucial Roles in Aquilegia Salt Stress Adaption. Int. J. Mol. Sci. 2023, 24, 3948. [Google Scholar] [CrossRef] [PubMed]
- Waadt, R.; Seller, C.A.; Hsu, P.K.; Takahashi, Y.; Munemasa, S.; Schroeder, J.I. Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 680–694. [Google Scholar] [CrossRef] [PubMed]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 2009, 324, 1064–1068. [Google Scholar] [CrossRef]
- Fujii, H.; Chinnusamy, V.; Rodrigues, A.; Rubio, S.; Antoni, R.; Park, S.Y.; Cutler, S.R.; Sheen, J.; Rodriguez, P.L.; Zhu, J.K. In vitro reconstitution of an abscisic acid signalling pathway. Nature 2009, 462, 660–664. [Google Scholar] [CrossRef]
- Wang, P.; Xue, L.; Batelli, G.; Lee, S.; Hou, Y.J.; Van Oosten, M.J.; Zhang, H.; Tao, W.A.; Zhu, J.K. Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc. Natl. Acad. Sci. USA 2013, 110, 11205–11210. [Google Scholar] [CrossRef]
- Liao, Y.; Zou, H.F.; Wang, H.W.; Zhang, W.K.; Ma, B.; Zhang, J.S.; Chen, S.Y. Soybean GmMYB76, GmMYB92, and GmMYB177 genes confer stress tolerance in transgenic Arabidopsis plants. Cell Res. 2008, 18, 1047–1060. [Google Scholar] [CrossRef]
- Zhang, G.; Chen, M.; Li, L.; Xu, Z.; Chen, X.; Guo, J.; Ma, Y. Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J. Exp. Bot. 2009, 60, 3781–3796. [Google Scholar] [CrossRef]
- Song, Y.; Li, J.; Sui, Y.; Han, G.; Zhang, Y.; Guo, S.; Sui, N. The sweet sorghum SbWRKY50 is negatively involved in salt response by regulating ion homeostasis. Plant Mol. Biol. 2020, 102, 603–614. [Google Scholar] [CrossRef]
- Chen, H.C.; Cheng, W.H.; Hong, C.Y.; Chang, Y.S.; Chang, M.C. The transcription factor OsbHLH035 mediates seed germination and enables seedling recovery from salt stress through ABA-dependent and ABA-independent pathways, respectively. Rice 2018, 11, 50. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, Z.; Sun, S.; Dai, Z.; Zhang, J.; Wang, W.; Peng, K.; Geng, W.; Xia, S.; Liu, Q.; et al. IbNIEL-mediated degradation of IbNAC087 regulates jasmonic acid-dependent salt and drought tolerance in sweet potato. J. Integr. Plant Biol. 2024, 66, 176–195. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Snyder, C.L.; Truksa, M.; Shah, S.; Weselake, R.J. sn-Glycerol-3-phosphate acyltransferases in plants. Plant Signal. Behav. 2011, 6, 1695–1699. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, H.; Gao, S.; Zhai, H.; He, S.; Zhao, N.; Liu, Q. An ABA-inducible gene IbTSJT1 positively regulates drought tolerance in transgenic sweetpotato. J. Integr. Agric. 2023, in press. [Google Scholar] [CrossRef]
- Perez-Salamo, I.; Papdi, C.; Rigo, G.; Zsigmond, L.; Vilela, B.; Lumbreras, V.; Nagy, I.; Horvath, B.; Domoki, M.; Darula, Z.; et al. The heat shock factor A4A confers salt tolerance and is regulated by oxidative stress and the mitogen-activated protein kinases MPK3 and MPK6. Plant Physiol. 2014, 165, 319–334. [Google Scholar] [CrossRef]
- Zhao, C.; Zhu, M.; Guo, Y.; Sun, J.; Ma, W.; Wang, X. Genomic Survey of PEBP Gene Family in Rice: Identification, Phylogenetic Analysis, and Expression Profiles in Organs and under Abiotic Stresses. Plants 2022, 11, 1576. [Google Scholar] [CrossRef]
- Chen, Y.; Peng, X.; Cui, J.; Zhang, H.; Jiang, J.; Song, Z. Isolation and Functional Determination of SKOR Potassium Channel in Purple Osier Willow, Salix purpurea. Int. J. Genomics 2021, 2021, 6669509. [Google Scholar] [CrossRef]
- Guo, W.J.; Nagy, R.; Chen, H.Y.; Pfrunder, S.; Yu, Y.C.; Santelia, D.; Frommer, W.B.; Martinoia, E. SWEET17, a facilitative transporter, mediates fructose transport across the tonoplast of Arabidopsis roots and leaves. Plant Physiol. 2014, 164, 777–789. [Google Scholar] [CrossRef]
- Maldonado, A.M.; Doerner, P.; Dixon, R.A.; Lamb, C.J.; Cameron, R.K. A putative lipid transfer protein involved in systemic resistance signalling in Arabidopsis. Nature 2002, 419, 399–403. [Google Scholar] [CrossRef]
- de Jesus-Pires, C.; Ferreira-Neto, J.; Pacifico, B.J.; Kido, E.A.; de Oliveira, S.R.; Pandolfi, V.; Wanderley-Nogueira, A.C.; Binneck, E.; Da, C.A.; Pio-Ribeiro, G.; et al. Plant Thaumatin-like Proteins: Function, Evolution and Biotechnological Applications. Curr. Protein Pept. Sci. 2020, 21, 36–51. [Google Scholar] [CrossRef]
- Han, G.; Qiao, Z.; Li, Y.; Wang, C.; Wang, B. The Roles of CCCH Zinc-Finger Proteins in Plant Abiotic Stress Tolerance. Int. J. Mol. Sci. 2021, 22, 8327. [Google Scholar] [CrossRef] [PubMed]




| Length Range (bp) | All Unigenes | Sg | Ss |
|---|---|---|---|
| 200–300 | 32,346 (32.39%) | 23,823 (37.53%) | 31,004 (39.05%) |
| 300–500 | 25,655 (25.69%) | 14,152 (22.30%) | 17,542 (22.09%) |
| 500–1000 | 13,275 (13.29%) | 10,349 (16.30%) | 12,685 (15.98%) |
| 1000–2000 | 12,328 (12.34%) | 7032 (11.08%) | 9121 (11.49%) |
| 2000+ | 16,264 (16.29%) | 8117 (12.79%) | 9045 (11.39%) |
| Total Number | 99,868 | 63,473 | 79,397 |
| Total Length | 99,204,098 | 56,364,907 | 64,912,814 |
| N50 Length | 2205 | 1964 | 1716 |
| Mean Length | 993.35 | 888.01 | 817.57 |
| Annotated Database | Annotated Number | 300 bp ≤ Length | Length ≥ 1000 bp |
|---|---|---|---|
| COG_Annotation | 14,193 | 2688 | 9220 |
| GO_Annotation | 38,836 | 9731 | 21,949 |
| KEGG_Annotation | 31,422 | 7187 | 18,911 |
| KOG_Annotation | 26,358 | 5957 | 15,531 |
| Pfam_Annotation | 35,881 | 8355 | 21,966 |
| Swissprot_Annotation | 29,004 | 6160 | 19,207 |
| TrEMBL_Annotation | 45,556 | 11,737 | 26,045 |
| eggNOG_Annotation | 37,071 | 9066 | 22,573 |
| nr_Annotation | 50,760 | 13,778 | 26,301 |
| All_Annotated | 52,527 | 14,345 | 26,426 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, Q.; Gao, S.; Zhang, X.; Liu, G.; Chen, P.; Gao, X.; Yuan, L.; Tian, Y.; Li, D.; Zhang, X.; et al. Comparative Transcriptome Analysis Reveals Mechanisms of Differential Salinity Tolerance Between Suaeda glauca and Suaeda salsa. Genes 2024, 15, 1628. https://doi.org/10.3390/genes15121628
Yan Q, Gao S, Zhang X, Liu G, Chen P, Gao X, Yuan L, Tian Y, Li D, Zhang X, et al. Comparative Transcriptome Analysis Reveals Mechanisms of Differential Salinity Tolerance Between Suaeda glauca and Suaeda salsa. Genes. 2024; 15(12):1628. https://doi.org/10.3390/genes15121628
Chicago/Turabian StyleYan, Qidong, Shang Gao, Xianglun Zhang, Guoping Liu, Peitao Chen, Xuanyi Gao, Li Yuan, Yucheng Tian, Dapeng Li, Xuepeng Zhang, and et al. 2024. "Comparative Transcriptome Analysis Reveals Mechanisms of Differential Salinity Tolerance Between Suaeda glauca and Suaeda salsa" Genes 15, no. 12: 1628. https://doi.org/10.3390/genes15121628
APA StyleYan, Q., Gao, S., Zhang, X., Liu, G., Chen, P., Gao, X., Yuan, L., Tian, Y., Li, D., Zhang, X., & Zhang, H. (2024). Comparative Transcriptome Analysis Reveals Mechanisms of Differential Salinity Tolerance Between Suaeda glauca and Suaeda salsa. Genes, 15(12), 1628. https://doi.org/10.3390/genes15121628





