Validation of Endogenous Control Genes by Real-Time Quantitative Reverse Transcriptase Polymerase Chain Reaction for Acute Leukemia Gene Expression Studies
Abstract
:1. Introduction
2. Materials and Methods
2.1. Biological Samples
2.2. RNA Extraction and Reverse Transcription of RNA to cDNA
2.3. Identification of Gene Expression by Quantitative Real-Time Polymerase Chain Reaction (qPCR)
2.4. Data Analysis Based on the Gene Expression Omnibus (GEO) Database
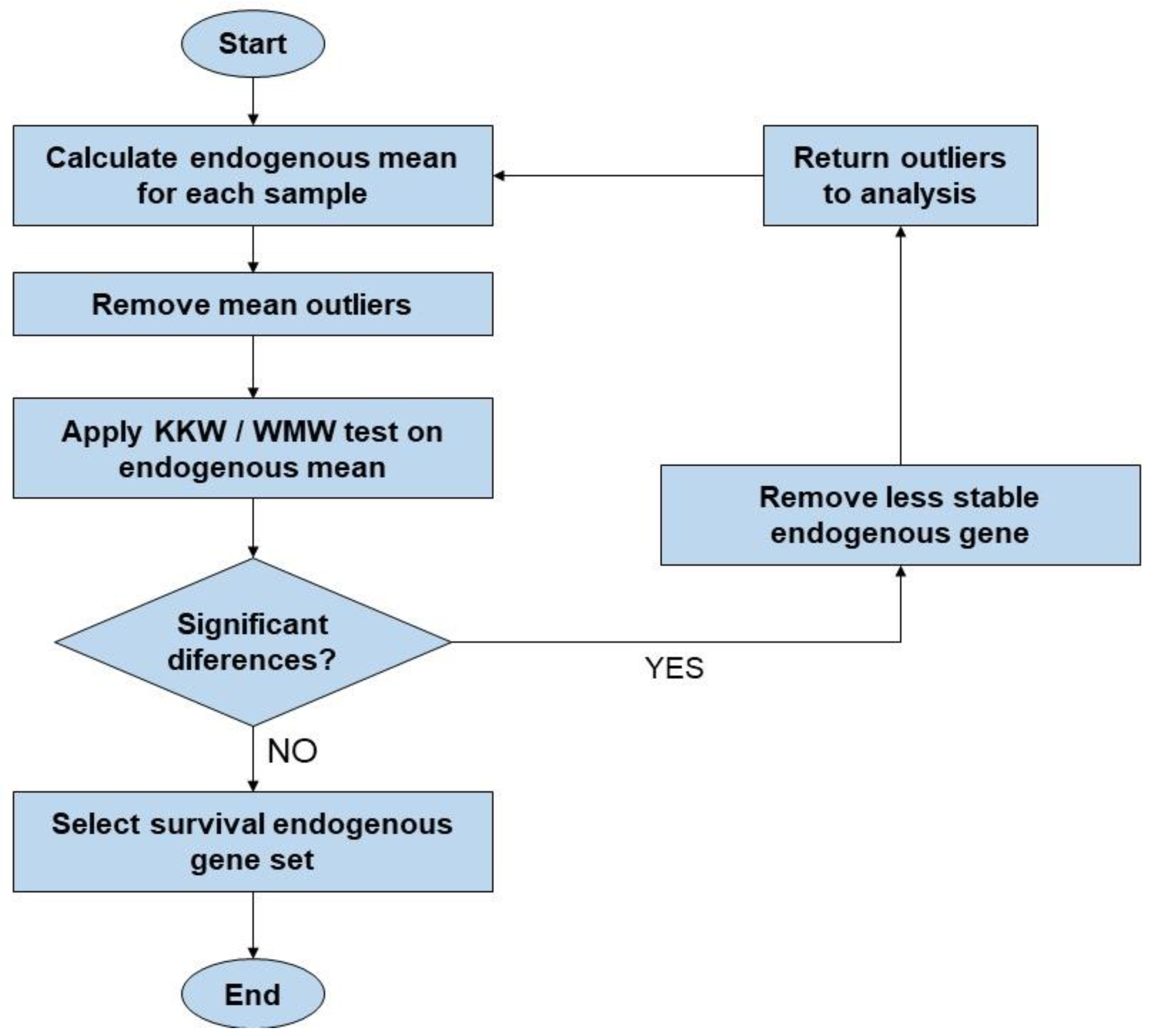
2.5. Statistical Analysis
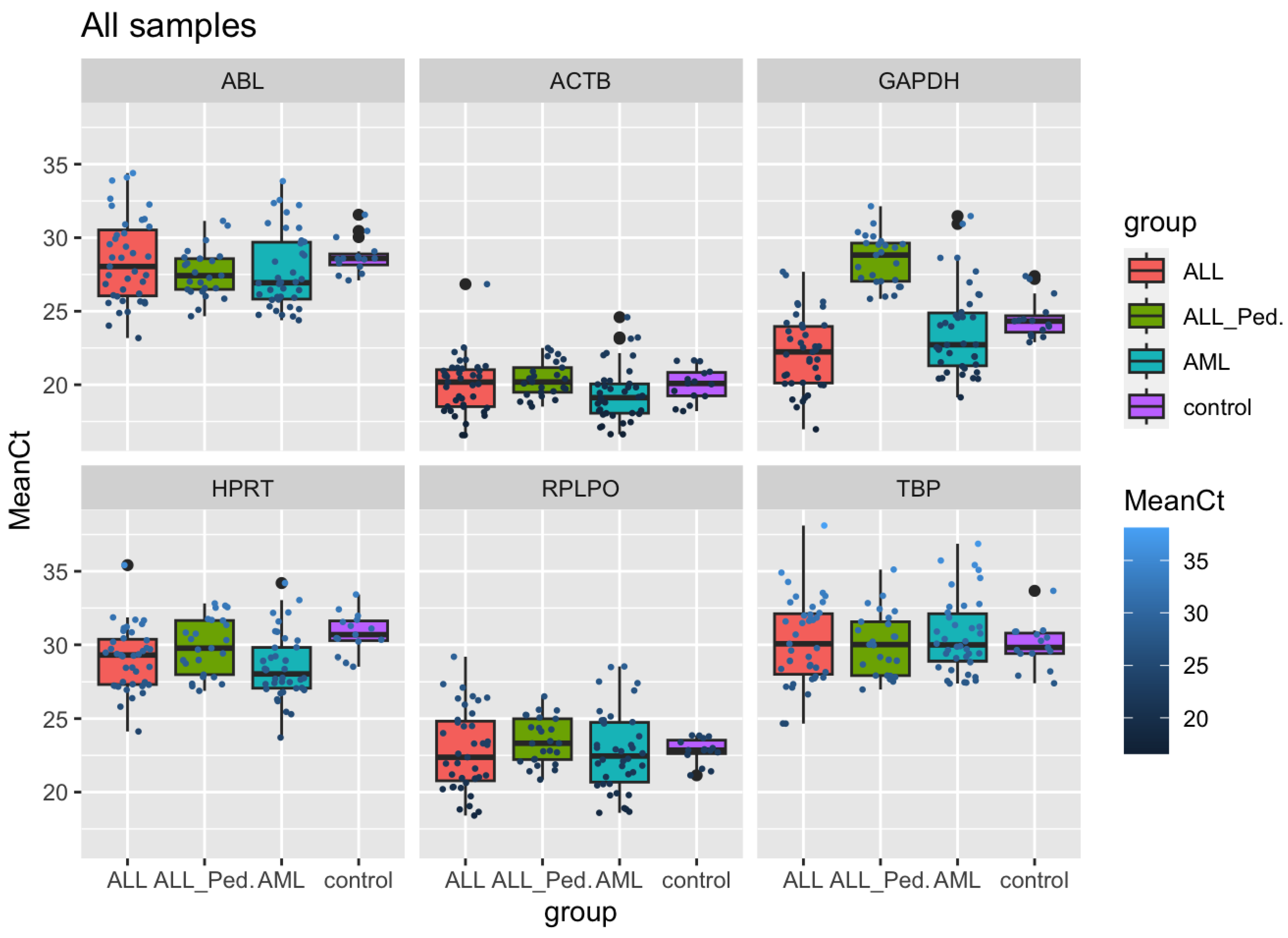
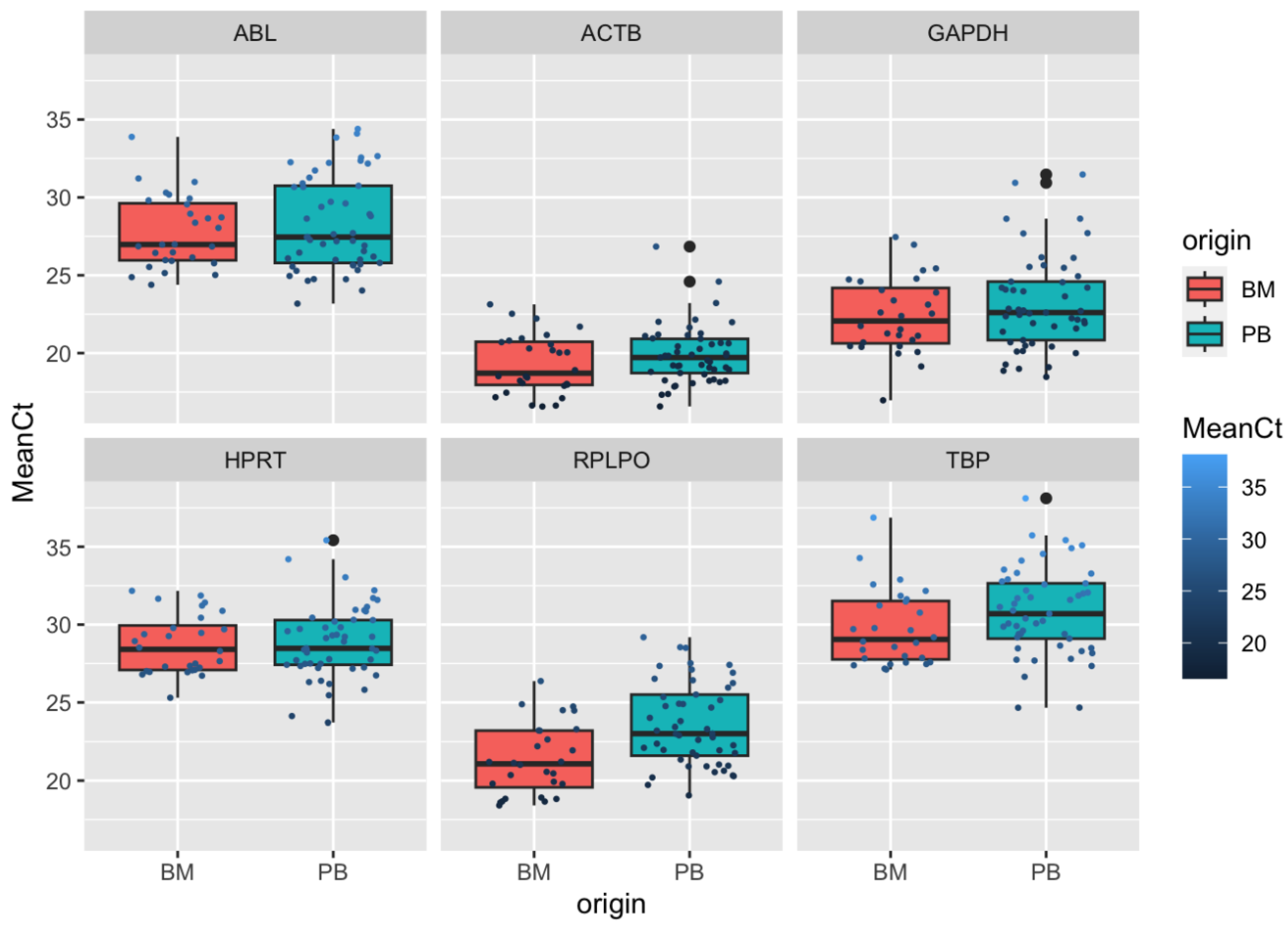
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Arya, M.; Shergill, I.S.; Williamson, M.; Gommersall, L.; Arya, N.; Patel, H.R.H. Basic principles of real-time quantitative PCR. Expert. Rev. Mol. Diagn. 2005, 5, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Jalali, M.; Zaborowska, J.; Jalali, M. The Polymerase Chain Reaction: PCR, qPCR, and RT-PCR; Elsevier Inc.: Amsterdam, The Netherlands, 2017; pp. 1–18. [Google Scholar] [CrossRef]
- Singh, J.; Birbian, N.; Sinha, S.; Goswami, A. A critical review on PCR, its types and applications. Int. J. Adv. Res. Biol. Sci. 2014, 1, 65–80. [Google Scholar]
- Seifi, M.; Ghasemi, A.; Heidarzadeh, S.; Khosravi, M.; Namipashaki, A.; Soofiany, V.M.; Khosroshahi, A.A.; Danaei, N. Overview of Real-Time PCR Principles. In Polymerase Chain Reaction; IntechOpen: London, UK, 2012. [Google Scholar]
- Loftis, A.D.; Reeves, W.K. Principles of Real-Time PCR. In Veterinary PCR Diagnosis; Bentham Science Publishers: Bussum, The Netherlands, 2012; pp. 3–17. [Google Scholar]
- Heid, C.A.; Stevens, J.; Livak, K.J.; Williams, P.M. Real Time Quantitative PCR. Genome Res. 1996, 6, 986–994. [Google Scholar] [CrossRef] [PubMed]
- Harshitha, R.; Arunraj, D.R. Real-time quantitative PCR: A tool for absolute and relative quantification. Biochem. Mol. Biol. Educ. 2021, 49, 800–812. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, R.; Dollinger, G.; Walsh, P.S.; Griffith, R. Simultaneous amplification and detection of specific DNA sequences. Biotechnology 1992, 10, 413–417. [Google Scholar] [CrossRef]
- Vilà, M.R.; Nicolás, A.; Morote, J.; De, I.; Meseguer, A. Increased glyceraldehyde-3-phosphate dehydrogenase expression in renal cell carcinoma identified by RNA-based, arbitrarily primed polymerase chain reaction. Cancer 2000, 89, 154–164. [Google Scholar] [CrossRef]
- Le, P.U.; Nguyen, T.N.; Drolet-Savoie, P.; Leclerc, N.; Nabi, I.R. Increased β-actin expression in an invasive Moloney sarcoma virus- transformed MDCK cell variant concentrates to the tips of multiple pseudopodia. Cancer Res. 1998, 58, 1631–1635. [Google Scholar]
- Wierschke, S.; Gigout, S.; Horn, P.; Lehmann, T.-N.; Dehnicke, C.; Bräuer, A.U.; Deisz, R.A. Evaluating reference genes to normalize gene expression in human epileptogenic brain tissues. Biochem. Biophys. Res. Commun. 2010, 403, 385–390. [Google Scholar] [CrossRef]
- Deindl, E.; Boengler, K.; van Royen, N.; Schaper, W. Differential expression of GAPDH and β-actin in growing collateral arteries. Mol. Cell Biochem. 2002, 236, 139–146. [Google Scholar] [CrossRef]
- Huggett, J.; Dheda, K.; Bustin, S.; Zumla, A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005, 6, 279–284. [Google Scholar] [CrossRef]
- Valasek, M.A.; Repa, J.J. The power of real-time PCR. Adv. Physiol. Educ. 2005, 29, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Fraga, D.; Meulia, T.; Fenster, S. Real-Time PCR. Curr. Protoc. Essent. Lab. Tech. 2014, 8, 10.3.1–10.3.40. [Google Scholar] [CrossRef]
- Jia, Y. Real-Time PCR; Elsevier: Amsterdam, The Netherlands, 2012; pp. 55–68. [Google Scholar] [CrossRef]
- Wilhelm, J.; Pingoud, A. Real-Time Polymerase Chain Reaction. ChemBioChem 2003, 4, 1120–1128. [Google Scholar] [CrossRef] [PubMed]
- Kozera, B.; Rapacz, M. Reference genes in real-time PCR. J. Appl. Genet. 2013, 54, 391–406. [Google Scholar] [CrossRef] [PubMed]
- Jacob, F.; Guertler, R.; Naim, S.; Nixdorf, S.; Fedier, A.; Hacker, N.F.; Heinzelmann-Schwarz, V. Careful Selection of Reference Genes Is Required for Reliable Performance of RT-qPCR in Human Normal and Cancer Cell Lines. PLoS ONE 2013, 8, e59180. [Google Scholar] [CrossRef]
- Bustin, S.A. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): Trends and problems. J. Mol. Endocrinol. 2002, 29, 23–39. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Nolan, T.; Pfaffl, M.W. Quantitative real-time RT-PCR—A perspective. J. Mol. Endocrinol. 2005, 34, 597–601. [Google Scholar] [CrossRef]
- Balogh, A.; Paragh, G., Jr.; Juhász, A.; Köbling, T.; Törőcsik, D.; Mikó, E.; Varga, V.; Emri, G.; Horkay, I.; Scholtz, B.; et al. Reference genes for quantitative real time PCR in UVB irradiated keratinocytes. J. Photochem. Photobiol. B Biol. 2008, 93, 133–139. [Google Scholar] [CrossRef]
- Hendriks-Balk, M.C.; Michel, M.C.; Alewijnse, A.E. Pitfalls in the normalization of real-time polymerase chain reaction data. Basic Res. Cardiol. 2007, 102, 195–197. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Zakrajsek, B.A. Effect of experimental treatment on housekeeping gene expression: Validation by real-time, quantitative RT-PCR. J. Biochem. Biophys. Methods 2000, 46, 69–81. [Google Scholar] [CrossRef]
- Galiveti, C.R.; Rozhdestvensky, T.S.; Brosius, J.; Lehrach, H.; Konthur, Z. Application of housekeeping npcRNAs for quantitative expression analysis of human transcriptome by real-time PCR. Rna 2010, 16, 450–461. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Zhang, R.; Gu, C.; Wu, J.; Wan, H.; Zhang, S.; Zhang, S. Evaluation of candidate reference genes for real time quantitative PCR normalization in pear fruit. Afr. J. Agric. Res. 2012, 7, 3701–3704. [Google Scholar]
- Chapman, J.R.; Waldenström, J. With reference to reference genes: A systematic review of endogenous controls in gene expression studies. PLoS ONE 2015, 10, e0141853. [Google Scholar] [CrossRef] [PubMed]
- Hellemans, J.; Mortier, G.; De Paepe, A.; Speleman, F.; Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2008, 8, R19. [Google Scholar] [CrossRef]
- Chervoneva, I.; Li, Y.; Schulz, S.; Croker, S.; Wilson, C.; Waldman, S.A.; Hyslop, T. Selection of optimal reference genes for normalization in quantitative RT-PCR. BMC Bioinform. 2010, 11, 253. [Google Scholar] [CrossRef] [PubMed]
- Harris, M.H.; Czuchlewski, D.R.; Arber, D.A.; Czader, M. Genetic Testing in the Diagnosis and Biology of Acute Leukemia. Am. J. Clin. Pathol. 2019, 152, 322–346. [Google Scholar] [CrossRef] [PubMed]
- Shakeel, M.; Rodriguez, A.; Tahir, U.B.; Jin, F. Gene expression studies of reference genes for quantitative real-time PCR: An overview in insects. Biotechnol. Lett. 2018, 40, 227–236. [Google Scholar] [CrossRef]
- Guénin, S.; Mauriat, M.; Pelloux, J.; Van Wuytswinkel, O.; Bellini, C.; Gutierrez, L. Normalization of qRT-PCR data: The necessity of adopting a systematic, experimental conditions-specific, validation of references. J. Exp. Bot. 2009, 60, 487–493. [Google Scholar] [CrossRef]
- Feuer, R.; Vlaic, S.; Arlt, J.; Sawodny, O.; Dahmen, U.; Zanger, U.M.; Thomas, M.; Kaderali, L. LEMming: A linear error model to normalize parallel quantitative real-time PCR (qPCR) data as an alternative to reference gene based methods. PLoS ONE 2015, 10, e0135852. [Google Scholar] [CrossRef]
- Richly, E.; Chinnery, P.F.; Leister, D. Evolutionary diversification of mitochondrial proteomes: Implications for human disease. Trends Genet. 2003, 19, 356–362. [Google Scholar] [CrossRef]
- Micheva, K.D.; Vallée, A.; Beaulieu, C.; Herman, I.M.; Leclerc, N. Β-Actin Is Confined To Structures Having High Capacity of Remodelling in Developing and Adult Rat Cerebellum. Eur. J. Neurosci. 1998, 10, 3785–3798. [Google Scholar] [CrossRef] [PubMed]
- McLysaght, A. Evolutionary steps of sex chromosomes are reflected in retrogenes. Trends Genet. 2008, 24, 478–481. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, E.; Levanon, E.Y. Human housekeeping genes, revisited. Trends Genet. 2013, 29, 569–574. [Google Scholar] [CrossRef] [PubMed]
- Chan, B.K.C. Data Analysis Using R Programming. In Biostatistics for Human Genetic Epidemiology; Advances in Experimental Medicine and Biology: New York, NY, USA, 2018; pp. 47–122. [Google Scholar] [CrossRef]
- Dheda, K.; Huggett, J.F.; Bustin, S.A.; Johnso, M.A.; Rook, G.; Zumla, A. Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques 2004, 37, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, E.; Levanon, E.Y. Human housekeeping genes are compact. Trends Genet. 2003, 19, 362–365. [Google Scholar] [CrossRef]
- Zhu, J.; He, F.; Hu, S.; Yu, J. On the nature of human housekeeping genes. Genome Anal. 2008, 24, 481–484. [Google Scholar] [CrossRef] [PubMed]
- Iyer, G.; Wang, A.R.; Brennan, S.R.; Bourgeois, S.; Armstrong, E.; Shah, P.; Harari, P.M. Identification of stable housekeeping genes in response to ionizing radiation in cancer research. Sci. Rep. 2017, 7, 43763. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef]
- Shen, Y.; Li, Y.; Ye, F.; Wang, F.; Lu, W.; Xie, X. Identification of suitable reference genes for measurement of gene expression in human cervical tissues. Anal. Biochem. 2010, 405, 224–229. [Google Scholar] [CrossRef]
- Taube, M.; Andersson-Assarsson, J.C.; Lindberg, K.; Pereira, M.J.; Gäbel, M.; Svensson, M.K.; Eriksson, J.W.; Svensson, P.A. Evaluation of reference genes for gene expression studies in human brown adipose tissue. Adipocyte 2015, 4, 280–285. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- McNeill, R.E.; Miller, N.; Kerin, M.J. Evaluation and validation of candidate endogenous control genes for real-time quantitative PCR studies of breast cancer. BMC Mol. Biol. 2007, 8, 107. [Google Scholar] [CrossRef] [PubMed]
- Weaver, S.; Dube, S.; Mir, A.; Qin, J.; Sun, G.; Ramakrishnan, R.; Jones, R.C.; Livak, K.J. Taking qPCR to a higher level: Analysis of CNV reveals the power of high throughput qPCR to enhance quantitative resolution. Methods 2010, 50, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Janssens, N.; Janicot, M.; Perera, T.; Bakker, A. Housekeeping genes as internal standards in cancer research. Mol. Diagnosis 2004, 8, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Khimani, A.H.; Mhashilkar, A.M.; Mikulskis, A.; O’Malley, M.; Liao, J.; Golenko, E.E.; Mayer, P.; Chada, S.; Killian, J.B.; Lott, S.T. Housekeeping genes in cancer: Normalization of array data. Biotechniques 2005, 38, 739–745. [Google Scholar] [CrossRef] [PubMed]
- Ayakannu, T.; Taylor, A.H.; Konje, J.C. Selection of Endogenous Control Reference Genes for Studies on Type 1 or Type 2 Endometrial Cancer. Sci. Rep. 2020, 10, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 14 September 2023).
- Ostertagová, E.; Ostertag, O.; Kováč, J. Methodology and application of the Kruskal-Wallis test. Appl. Mech. Mater. 2014, 611, 115–120. [Google Scholar] [CrossRef]
- Frey, B.B. Kruskal-Wallis Test. In The SAGE Encyclopedia of Educational Research, Measurement, and Evaluation; SAGE Publications, Inc.: Thousand Oaks, CA, USA, 2018; Voluem 10, p. n377. [Google Scholar]
- Almonroeder, T.G. Introduction to analysis of variance. In Advanced Statistics for Physical and Occupational Therapy; Routledge: London, UK, 2022; pp. 77–97. [Google Scholar] [CrossRef]
- Sthle, L.; Wold, S. Analysis of variance (ANOVA). Chemom. Intell. Lab. Syst. 1989, 6, 259–272. [Google Scholar] [CrossRef]
- Fujikoshi, Y. Two-way ANOVA models with unbalanced data. Discrete Math. 1993, 116, 315–334. [Google Scholar] [CrossRef]
- Song, Q.; Dou, L.; Zhang, W.; Peng, Y.; Huang, M.; Wang, M. Public transcriptome database-based selection and validation of reliable reference genes for breast cancer research. Biomed. Eng. Online 2021, 20, 124. [Google Scholar] [CrossRef]
- Martin, J.L. Validation of reference genes for oral cancer detection panels in a prospective blinded cohort. PLoS ONE 2016, 11, e0158462. [Google Scholar] [CrossRef] [PubMed]
- de Campos, R.P.; Schultz, I.C.; de Andrade Mello, P.; Davies, S.; Gasparin, M.S.; Bertoni, A.P.S.; Buffon, A.; Wink, M.R. Cervical cancer stem-like cells: Systematic review and identification of reference genes for gene expression. Cell Biol. Int. 2018, 42, 139–152. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002, 30, e36. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Jelinek, D.F.; Tschumper, R.C.; Stolovitzky, G.A.; Iturria, S.J.; Tu, Y.; Lepre, J.; Shah, N.; Kay, N. Identification of a global geneexpression signature of B-chronic lymphocytic leukemia. Mol. Cancer Res. 2003, 1, 346–361. [Google Scholar]
- Joshi, A.D.; Hegde, G.V.; Dickinson, J.D.; Mittal, A.K.; Lynch, J.C.; Eudy, J.D.; Armitage, J.O.; Bierman, P.J.; Bociek, R.G.; Devetten, M.P.; et al. ATM, CTLA4, MNDA, andHEM1 in high versus low CD38 expressing B-cell chronic lym-phocytic leukemia. Clin. Cancer Res. 2007, 13, 5295–5304. [Google Scholar] [CrossRef]
- Ghani, M.; Sato, C.; Rogaeva, E. Segmental duplications in genome-wide significant loci and housekeeping genes; warning for GAPDH and ACTB. Neurobiol. Aging 2013, 34, e1–e1710. [Google Scholar] [CrossRef]
- Kienle, D.; Benner, A.; Laufle, C.; Winkler, D.; Schneider, C.; Buhler, A.; Zenz, T.; Habermann, A.; Jager, U.; Lichter, P.; et al. Gene expression factors as predictors of genetic risk and survival in chronic lymphocytic leukemia. Haematologica 2010, 95, 102–109. [Google Scholar] [CrossRef]
- Nuckel, H.; Collins, C.H.; Frey, U.H.; Sellmann, L.; Durig, J.; Siffert, W.; Duhrsen, U. FCRL2 mRNA expression is inverselyassociated with clinical progression in chronic lymphocytic leukemia. Eur. J. Haematol. 2009, 83, 541–549. [Google Scholar] [CrossRef] [PubMed]
- Glare, E.M.; Divjak, M.; Bailey, M.J.; Walters, E.H. β-actin and GAPDH housekeeping gene expression in asthmatic airways is variable and not suitable for normalising mRNA levels. Thorax 2002, 57, 765–770. [Google Scholar] [CrossRef] [PubMed]
- Valceckiene, V.; Kontenyte, R.; Jakubauskas, A.; Griskevicius, L. Selection of reference genes for quantitative polymerase chain reaction studies in purified B cells from B cell chronic lymphocytic leukaemia patients. Br. J. Haematol. 2010, 151, 232–238. [Google Scholar] [CrossRef] [PubMed]
- Tong, Z.; Gao, Z.; Wang, F.; Zhou, J.; Zhang, Z. Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol. Biol. 2009, 10, 71. [Google Scholar] [CrossRef]
- Bednarz-Misa, I.; Neubauer, K.; Zacharska, E.; Kapturkiewicz, B.; Krzystek-Korpacka, M. Whole blood ACTB, B2M and GAPDH expression reflects activity of inflammatory bowel disease, advancement of colorectal cancer, and correlates with circulating inflammatory and angiogenic factors: Relevance for real-time quantitative PCR. Adv. Clin. Exp. Med. 2020, 29, 547–556. [Google Scholar] [CrossRef]
- Ullmannová, V.; Haškovec, C. The use of housekeeping genes (HKG) as an internal control for the detection of gene expression by quantitative real-time RT-PCR. Folia Biol. 2003, 49, 211–216. [Google Scholar]
- Tan, S.C.; Carr, C.A.; Yeoh, K.K.; Schofield, C.J.; Davies, K.E.; Clarke, K. Identification of valid housekeeping genes for quantitative RT-PCR analysis of cardiosphere-derived cells preconditioned under hypoxia or with prolyl-4-hydroxylase inhibitors. Mol. Biol. Rep. 2012, 39, 4857–4867. [Google Scholar] [CrossRef]
- Barber, R.D.; Harmer, D.W.; Coleman, R.A.; Clark, B.J. GAPDH as a housekeeping gene: Analysis of GAPDH mRNA expression in a panel of 72 human tissues. Physiol. Genomics 2005, 21, 389–395. [Google Scholar] [CrossRef]
- Albertini, R.J. HPRT mutations in humans: Biomarkers for mechanistic studies. Mutat. Res./Rev. Mutat. Res. 2001, 489, 1–16. [Google Scholar] [CrossRef]
- Stout, J.T.; Caskey, C.T. Hprt: Gene structure, expression, and mutation. Annu. Rev. Genet. 1985, 19, 127–148. [Google Scholar] [CrossRef]
- Everaert, B.R.; Boulet, G.A.; Timmermans, J.P.; Vrints, C.J. Importance of suitable reference gene selection for quantitative real-time PCR: Special reference to mouse myocardial infarction studies. PLoS ONE 2011, 6, e23793. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Wang, X.Y.; Fan, J.; Qiu, S.J.; Zhou, J.; Shi, Y.H.; Xiao, Y.S.; Xu, Y.; Huang, X.W.; Sun, J. Selection of reference genes for real-time PCR in human hepatocellular carcinoma tissues. J. Cancer Res. Clin. Oncol. 2008, 134, 979–986. [Google Scholar] [CrossRef] [PubMed]
- Maurer-Morelli, C.V.; de Vasconcellos, J.F.; Reis-Pinto, F.C.; Rocha, C.d.S.; Domingues, R.R.; Yasuda, C.L.; Tedeschi, H.; Oliveira, E.D.; Cendes, F.; Lopes-Cendes, I. A comparison between different reference genes for expression studies in human hippocampal tissue. J. Neurosci. Methods 2012, 208, 44–47. [Google Scholar] [CrossRef] [PubMed]
- Ropenga, A.; Chapel, A.; Vandamme, M.; Griffiths, N.M. Use of reference gene expression in rat distal colon after radiation exposure: A caveat. Radiat. Res. 2004, 161, 597–602. [Google Scholar] [CrossRef] [PubMed]
- de Kok, J.B.; Roelofs, R.W.; Giesendorf, B.A.; Pennings, J.L.; Waas, E.T.; Feuth, T.; Swinkels, D.W.; Span, P.N. Normalization of gene expression measurements in tumor tissues: Comparison of 13 endogenous control genes. Lab. Investig. 2005, 85, 154–159. [Google Scholar] [CrossRef]
- Townsend, M.H.; Felsted, A.M.; Ence, Z.E.; Piccolo, S.R.; Robison, R.A.; O’Neill, K.L. Falling from grace: HPRT is not suitable as an endogenous control for cancer-related studies. Mol. Cell Oncol. 2019, 6, 1–10. [Google Scholar] [CrossRef]
- Radonić, A.; Thulke, S.; Mackay, I.M.; Landt, O.; Siegert, W.; Nitsche, A. Guideline to reference gene selection for quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2004, 313, 856–862. [Google Scholar] [CrossRef]
- Popov, A.; Nowak, D.; Malicka-Błaszkiewicz, M. Actin-cytoskeleton and b-actin expression in correlation with higher invasiveness of selected hepatoma Morris 5123 cells. J. Physiol. Pharmacol. 2006, 57, 111–123. [Google Scholar]
- Guo, C.; Liu, S.; Wang, J.; Sun, M.Z.; Greenaway, F.T. ACTB in cancer. Clin. Chim. Acta 2013, 417, 39–44. [Google Scholar] [CrossRef]
- Morse, D.L.; Carroll, D.; Weberg, L.; Borgstrom, M.C.; Ranger-Moore, J.; Gillies, R.J. Determining suitable internal standards for mRNA quantification of increasing cancer progression in human breast cells by real-time reverse transcriptase polymerase chain reaction. Anal. Biochem. 2005, 342, 69–77. [Google Scholar] [CrossRef]
- Majidzadeh, A.K.; Esmaeili, R.; Abdoli, N. TFRC and ACTB as the best reference genes to quantify Urokinase Plasminogen Activator in breast cancer. BMC Res. Notes 2011, 4, 215. [Google Scholar] [CrossRef]
- Gur-Dedeoglu, B.; Konu, O.; Bozkurt, B.; Ergul, G.; Seckin, S.; Yulug, I.G. Identification of endogenous reference genes for qRT-PCR analysis in normal matched breast tumor tissues. Oncol. Res. 2009, 17, 353–365. [Google Scholar] [CrossRef] [PubMed]
- Goidin, D.; Mamessier, A.; Staquet, M.; Schmitt, D.; Berthier-Vergnes, O. Ribosomal 18S RNA prevails over glyceraldehyde-3-phosphate dehydrogenase and beta-actin genes as internal standard for quantitative comparison of mRNA levels in invasive and noninvasive human melanoma cell subpopulations. Anal. Biochem. 2001, 295, 17–21. [Google Scholar] [CrossRef] [PubMed]
- Jung, M.; Ramankulov, A.; Roigas, J.; Johannsen, M.; Ringsdorf, M.; Kristiansen, G.; Jung, K. In search of suitable reference genes for gene expression studies of human renal cell carcinoma by real-time PCR. BMC Mol. Biol. 2007, 8, 47. [Google Scholar] [CrossRef] [PubMed]
- Lemma, S.; Avnet, S.; Salerno, M.; Chano, T.; Baldini, N. Identification and Validation of Housekeeping Genes for Gene Expression Analysis of Cancer Stem Cells. PLoS ONE 2016, 11, e0149481. [Google Scholar] [CrossRef]
- Gu, Y.; Tang, S.; Wang, Z.; Cai, L.; Lian, H.; Shen, Y.; Zhou, Y. A pan-cancer analysis of the prognostic and immunological role of β-actin (ACTB) in human cancers. Bioengineered 2021, 12, 6166–6185. [Google Scholar] [CrossRef]
- Gupta, D.G.; Varma, N.; Abdulkadir, S.A.; Singh, P.; Sachdeva, M.U.S.; Naseem, S.; Siddiqui, M.R.; Bose, P.; Binota, J.; Malhotra, P.; et al. Identifcation and validation of the optimal reference genes for standardizing the gene expression profling diagnostic panel of Ph-like B-lineage acute lymphoblastic leukemia. Clin. Exp. Med. 2023, 23, 4539–4551. [Google Scholar] [CrossRef]
- Wang, J.Y.J. The Capable ABL: What Is Its Biological Function? Mol. Cell Biol. 2014, 34, 1188–1197. [Google Scholar] [CrossRef]
- Van Etten, R.A. Cycling, stressed-out and nervous: Cellular functions of c-Abl. Trends Cell Biol. 1999, 9, 179–186. [Google Scholar] [CrossRef]
- Ramakrishnan, L.; Rosenberg, N. Abl Genes. Biochim. Biophys. Acta (BBA)-Rev. Cancer 1989, 989, 209–224. [Google Scholar] [CrossRef]
- Davidson, I. The genetics of TBP and TBP-related factors. Trends Biochem. Sci. 2003, 28, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Emig, M.; Saußele, S.; Wittor, H.; Weißer, A.; Reiter, A.; Willer, A.; Berger, U.; Hehlmann, R.; Cross, N.C.P.; Hochhaus, A. Accurate and rapid analysis of residual disease in patients with CML using specific fluorescent hybridization probes for real time quantitative RT-PCR. Leukemia 1999, 13, 1825–1832. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.C.; Erben, P.; Saglio, G.; Gottardi, E.; Nyvold, C.G.; Schenk, T.; Ernst, T.; Lauber, S.; Kruth, J.; Hehlmann, R.; et al. Harmonization of BCR-ABL mRNA quantification using a uniform multifunctional control plasmid in 37 international laboratories. Leukemia 2008, 22, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Schoch, C.; Schnittger, S.; Bursch, S.; Gerstner, D.; Hochhaus, A.; Berger, U.; Hehlmann, R.; Hiddemann, W.; Haferlach, T. Comparison of chromosome banding analysis, interphase- and hypermetaphase-FISH, qualitative and quantitative PCR for diagnosis and for follow-up in chronic myeloid leukemia: A study on 350 cases. Leukemia 2002, 16, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Weisser, M.; Haferlach, T.; Schoch, C.; Hiddeman, W.; Schnittger, S. The use of housekeeping genes for real-time PCR-based quantification of fusion gene transcripts in acute myeloid leukemia. Leukemia 2004, 18, 1551–1553. [Google Scholar] [CrossRef] [PubMed]
- Lossos, I.S.; Czerwinski, D.K.; Wechser, M.A.; Levy, R. Optimization of quantitative real-time RT-PCR parameters for the study of lymphoid malignancies. Leukemia 2003, 17, 789–795. [Google Scholar] [CrossRef] [PubMed]
- Meller, M.; Vadachkoira, S.; Luthy, D.A.; Williams, M.A. Evaluation of housekeeping genes in placental comparative expression studies. Placenta 2005, 26, 601–607. [Google Scholar] [CrossRef]
- Ohl, F.; Jung, M.; Radonić, A.; Sachs, M.; Loening, S.A.; Jung, K. Identification and Validation of Suitable Endogenous Reference Genes for Gene Expression Studies of Human Bladder Cancer. J. Urol. 2006, 175, 1915–1920. [Google Scholar] [CrossRef]
- Turabelidze, A.; Guo, S.; Dipietro, L.A. Importance of housekeeping gene selection for accurate reverse transcription-quantitative polymerase chain reaction in a wound healing model. Wound Repair. Regen. 2010, 18, 460–466. [Google Scholar] [CrossRef]
- Valente, V.; Teixeira, S.A.; Neder, L.; Okamoto, O.K.; Oba-Shinjo, S.M.; Marie, S.K.N.; Scrideli, C.A.; Paçó-Larson, M.L.; Carlotti, C.G. Selection of suitable housekeeping genes for expression analysis in glioblastoma using quantitative RT-PCR. BMC Mol. Biol. 2009, 10, 17. [Google Scholar] [CrossRef]
- Wang, C.H.; Wang, L.K.; Wu, C.C.; Chen, M.L.; Lee, M.C.; Lin, Y.Y.; Tsai, F.M. The Ribosomal Protein RPLP0 Mediates PLAAT4-induced Cell Cycle Arrest and Cell Apoptosis. Cell Biochem. Biophys. 2019, 77, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.L.; Zhao, W.W.; Bai, S.M.; Ma, Y.; Yin, X.K.; Feng, L.L.; Zeng, G.D.; Wang, F.; Feng, W.X.; Zheng, J.; et al. DNA damage-induced paraspeckle formation enhances DNA repair and tumor radioresistance by recruiting ribosomal protein P0. Cell Death Dis. 2022, 13, 709. [Google Scholar] [CrossRef] [PubMed]
- Artero-Castro, A.; Castellvi, J.; García, A.; Hernández, J.; Ramón y Cajal, S.; LLeonart, M.E. Expression of the ribosomal proteins Rplp0, Rplp1, and Rplp2 in gynecologic tumors. Hum. Pathol. 2011, 42, 194–203. [Google Scholar] [CrossRef] [PubMed]
- Stern-Straeter, J.; Bonaterra, G.A.; Hörmann, K.; Kinscherf, R.; Goessler, U.R. Identification of valid reference genes during the differentiation of human myoblasts. BMC Mol. Biol. 2009, 10, 66. [Google Scholar] [CrossRef]
- Ali, H.; Du, Z.; Li, X.; Yang, Q.; Zhang, Y.C.; Wu, M.; Li, Y.; Zhang, G. Identification of suitable reference genes for gene expression studies using quantitative polymerase chain reaction in lung cancer in vitro. Mol. Med. Rep. 2015, 11, 3767–3773. [Google Scholar] [CrossRef]
- Bakhashab, S.; Lary, S.; Ahmed, F.; Schulten, H.J.; Bashir, A.; Ahmed, F.W.; Al-Malki, A.L.; Jamal, H.S.; Gari, M.A.; Weaver, J.U. Reference genes for expression studies in hypoxia and hyperglycemia models in human umbilical vein endothelial cells. G3 Genes Genomes Genet. 2014, 4, 2159–2165. [Google Scholar] [CrossRef]
- Wang, H.; Yang, B.; Geng, T.; Li, B.; Dai, P.; Chen, C. Tissue-specific selection of optimal reference genes for expression analysis of anti-cancer drug-related genes in tumor samples using quantitative real-time RT-PCR. Exp. Mol. Pathol. 2015, 98, 375–381. [Google Scholar] [CrossRef]
| Number of Patients | Bone Marrow | Peripheral Blood | Mean Age (Range) | Median Age | Gender | |
|---|---|---|---|---|---|---|
| AML | 24 | 14 | 24 | 51.1 (19–96) | 47.5 | F: 9 M: 15 |
| ALL | 25 | 14 | 25 | 42.5 (19–96) | 36 | F: 8 M: 17 |
| ALL_Ped | 25 | - | 25 | 8 (0–18) * | 7 * | F: 8 M: 14 NA: 3 |
| Control | 15 | - | 15 | 33.7 (20–47) | 33 | F: 10 M: 5 |
| Total | 89 | 28 | 89 |
| Gene Symbol | Gene Name | Chromosome Location | Function | Amplicon Size | Assay Number |
|---|---|---|---|---|---|
| ABL1 | Abelson murine leukemia viral oncogene human homolog 1 | Chr.9: 130713881–130887675 | Protein tyrosine kinase involved in a variety of cellular processes | 60 | Hs01104728_m1 |
| ACTB | β-actin | Chr.7: 5527148–5530601 | Cytoskeletal structural protein | 63 | Hs01060665_g1 |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | Chr.12: 6534405–6538375 | Oxidoreductase in glycolysis and gluconeogenesis | 157 | Hs02786624_g1 |
| HPRT1 | Hypoxanthine phosphoribosyl-transferase 1 | Chr.X: 134460165–134500668 | Purine synthesis in salvage pathway | 82 | Hs2800695_m1 |
| RPLP0 | Ribosomal protein lateral stalk subunit P0 | Chr.12: 120196700–120201211 | Ribosomal protein translation | 76 | Hs00420895_gH |
| TBP | TATA box binding protein | Chr.6: 170554333–170572870 | Regulation of transcription DNA and component of the DNA-binding protein complex TFIID | 91 | Hs00427620_m1 |
| ABL | ACTB | GAPDH | HPRT | TBP | RPLP0 | |
|---|---|---|---|---|---|---|
| ALL | 0.113 | 0.104 | 0.128 | 0.100 | 0.098 | 0.121 |
| AML | 0.087 | 0.083 | 0.090 | 0.098 | 0.073 | 0.091 |
| ALL_Ped | 0.059 | 0.066 | 0.082 | 0.059 | 0.060 | 0.071 |
| Control | 0.045 | 0.052 | 0.067 | 0.040 | 0.044 | 0.044 |
| Gene | Stability Value | |
|---|---|---|
| Overall * | ||
| 1 | ACTB | 0.47 |
| 2 | RPLP0 | 0.52 |
| 3 | HPRT | 0.72 |
| 4 | ABL | 0.86 |
| 5 | TBP | 0.91 |
| 6 | GAPDH | 1.78 |
| ABL | GAPDH | ACTB | HPRT | TBP | RPLP0 | |
|---|---|---|---|---|---|---|
| Kruskal–Wallis p-value | 0.176 | 0.000 | 0.046 | 0.000 | 0.914 | 0.368 |
| Gene Standard Deviation | 2.421 | 3.352 | 1.698 | 2.150 | 2.472 | 2.362 |
| Sum of Mean Square Difference | 0.893 | 22.500 | 0.487 | 3.256 | 0.286 | 0.385 |
| Sum of Standard Deviation Square Difference | 2.448 | 7.396 | 0.626 | 0.715 | 1.223 | 3.170 |
| Model (Tissue Type) | Number of Patients | Analyzed Sample | Endogenous Genes Tested | Endogenous Genes with Best Behavior | Year | Reference |
|---|---|---|---|---|---|---|
| Acute leukemias | 89 | Peripheral blood and bone marrow | ACTB, ABL, GAPDH, HPRT1, TBP and RPLP0 | ACTB, ABL, TBP and RPLP0 | 2024 | Present study |
| Acute myeloid Leukemia | 29 | Peripheral blood | ABL1, G6PDH, B2M and PBGD | G6PDH and ABL | 2004 | [103] |
| ALL, Ph-like B-lineage | 23 | Peripheral blood | ABL1, GUSB, EEF2, 18S, ACTB, GAPDH, TBP, PGK1, B2M, JCHAIN, SPATS2L, CA6, NRXN3, MUC4, CRLF2, ADGRF1 and BMPR1B | EEF2, GAPDH and PGK1 | 2023 | [95] |
| B-cell chronic lymphocytic leukemia | 30 | Peripheral blood | ACTB, B2M, GAPDH, GUSB, HMBS, HPRT1, MRPL19, TBP and UBC | B2M, HPRT1 and GUSB | 2010 | [71] |
| Bladder and colon cancer | 58 | Tumor biopsies | FLOT2, ATP5B, HSPCB, S100A6, TEGT, CFL1, FLJ20030, TPT1, UBB, TBC, RPS23, GAPD, ACTB, CLTC, NACA, SU11 and TUBA6 | UBC, GAPD and TPT1 for colon and HSPCB, TEGT and ATP5B for bladder | 2004 | [53] |
| Bladder cancer | 14 | Tissue biopsies | ACTB, ALAS1, G6PD, GAPD, HMBS, HPRT1, K-α-1, SDHA and TBP | SDH and TBP | 2006 | [106] |
| Breast cancer | 87 | Tumor biopsies and cell lines | SF1, TARDBP, THRAP3, QRICH1, TRA2B, SRSF3, YY1, DNAJC8, RNF10 and RHOA | SF1, TRA2B, THRAP3, RHOA and QRICH1 | 2021 | [60] |
| Breast cancer | - | Cell line (MCF-10A) | 18S, 28S, ACTB, PPIA, GAP and RPL32 | 18S and ACTB | 2005 | [88] |
| Breast cancer | 40 | Tumor biopsies | GAPDH, TFRC, RPLP0, GUSB, HPRT1, UPA and ACTB | ACTB and TFRC | 2011 | [89] |
| Breast cancer | 23 | Tumor biopsies | ACTB, GAPD, TBP, SDHA, HPRT, HMBS, B2M, PPIA, GUSB, YWHAZ, PGK1, RPL41, PUM1, RPLP0, MRPL19, TTC22, IL22RA1 and ZNF224 | ACTB and SDHA | 2009 | [90] |
| Breast, gastric, esophageal, colon, rectum, and lung carcinomas | 327 | Tissue biopsies | ACTB, GAPDH, GUSB, RPLPO and TFRC | The optimal reference genes were tissue-specific | 2014 | [115] |
| Brown adipose tissue | - | Tissue biopsies | 18S, B2M, GAPDH, LRP10, PPIA, RPLP0, UBC and YWHAZ | PSMB2, GNB2 and GNB1 | 2015 | [45] |
| Cancer stem cells | - | Cell lines (RD, MG63, HOS, Saos-2, A673, MDA-MB-231 and ACHN) | 18S, ACTB, B2M, G6PD, GAPDH, GUSB, HMBS, HPRT1, PGK1, PPIA, RPL13a, SDHA, TBP, TUBB and YWHAZ | GAPDH, TBP and PPIA | 2016 | [93] |
| Cervical cancer | - | Cell lines (SiHa, HeLa and ME180) | ACTB, B2M, GAPDH, HPRT1 and TBP | B2M, GAPDH, HPRT1 and TBP | 2018 | [62] |
| Colon | - | Rat tissue biopsies | GAPD, ACTB, Cyclophilin A, HPRT, AcRP0, L32, 18S and 28S | AcRP0 | 2004 | [82] |
| Colon, breast, prostate, skin, and bladder | 16 | Tissue biopsies | LRP, BACT, CYC, GAPDH, PGK, B2M, BGUS, HPRT, TBO, TfR, PBGD and ATP6 | HPRT | 2005 | [83] |
| Colon, liver, pancreas, rectum, lung, cervix, ovary, prostate, umbilical, breast, spleen, etc. | 72 | Tissue biopsies | GAPDH | GAPDH varies a lot between tissues | 2005 | [76] |
| Colorectal cancer | 64 | Peripheral blood | ACTB, B2M, GAPDH, HPRT1, SDHA TBP, IL-1B and CCL4 | HPRT1, SDHA and TBP | 2020 | [73] |
| Endometrial cancer (type 1 or type 2) | 15 | Endometrial biopsies | RPL30, MT-ATP6, 18S, ACTB, TBP, RPLP0, PES1, POLR2A, TFRC, HPRT1, ABL1, GADD45A, HMBS, CDKN1A, RPL37A, UBC, GAPDH, CDKN1B, CASC3, POP4, PGK1, GUSB, YWHAZ, PPIA, RPS17, MRPL19, B2M, EIF2B1, ELF1, PSMC4, PUM1 and IPO8 | PSMC4, PUM1 and IPO8 for type 1 and UBC, MRPL19, PGK1 and PPIA for type 2 | 2020 | [51] |
| Glioblastoma | 30 | Tumor biopsies | ACTB, GAPDH, GUSB, HMBS, HPRT1, TBP, 18S, TG1, TG2, TG3, TG4, TG5, TG6, TG7, TG8, GT9, TG10, TG11 and TG12 | TBP and HPRT1 | 2009 | [108] |
| Hepatocellular carcinoma | 65 | Tumor biopsies | ACTB, GAPDH, B2M, HPRT1 and TBP | TBP and HPRT | 2008 | [80] |
| Hippocampal tissue | 25 | Tissue biopsies | ACTB, GAPDH, HPRT, NSE, SDHA and SYP | HPRT, NSE, SDHA and SYP | 2012 | [81] |
| Hypoxia and hyperglycemia model | - | Umbilical cords | RPLP0, GAPDH, GUSB, TFRC and ACTB | TFRC and RPLP0 | 2014 | [114] |
| Lung cancer | - | Cell lines (A549, NCI-H446 and NCI-H460) | 18S, GAPDH, RPLP0, ACTB, PPIA, PGK1, B2M, RPL13A, HPRT1 and TBP | ACTB, PPIA and PGK1 | 2015 | [113] |
| Lung, breast, colon, prostate, and pancreas | 326 | Tissue biopsies | HPRT | HPRT should no longer be used as an endogenous standard | 2019 | [84] |
| Lymphoid malignancies | 92 | Cell lines, tumor biopsies and peripheral blood | 18S, RPLP0, GAPD, PPIA, PRKG1, TBP, ACTB, B2M and GUSB | PRKG1 and TBP | 2003 | [104] |
| Melanoma | - | Cell lines (IC8 and T1C3) | GAPDH, 18S and ACTB | 18S | 2001 | [91] |
| Myoblasts | 15 | Tissue biopsies | ACTA1, MYOG, MYH3, ACTB, B2M, GAPDH, PPIA, RPLP0 and TBP | RPLP0 and TBP | 2009 | [112] |
| Myocardial infarction | - | Mouse myocardial infarction tissue sample | ACTB, B2M, EEF1A1, GAPDH, HPRT, POLR2A, PPIA, RPL13a, TBP and TPT1 | HPRT, RPL13A and TPT1 | 2011 | [79] |
| Oral cancer | 68 | Saliva | B2M, MT-ATP6, RPL30, RPL37A, RPL0, RPS17 and UBC | MT-ATP6, RPL30, RPL37A, RPLP0 and RPS17 | 2016 | [61] |
| Placenta | 20 | Placenta tissue | B2M, GAPDH, HMBS, HPRT, SDHA, TBP, YWHAZ and LEP | SDHA, TBP and YWHAZ | 2004 | [105] |
| Renal cell carcinoma | 25 | Tissue biopsies | ACTB, ALASI, GAPDH, HMBS, HPRT1, PPIA, RPLP0, SDHA, TBP, TUBB and ADAM9 | PPIA and TBP | 2007 | [92] |
| Wound healing model | - | Skin and wound samples from mice | B2M, TBP, GAPDH, GUSB, RPLP2, ACTB and 18S | GAPDH, TBP and B2M | 2010 | [107] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pessoa, F.M.C.d.P.; Viana, V.B.d.J.; de Oliveira, M.B.; Nogueira, B.M.D.; Ribeiro, R.M.; Oliveira, D.d.S.; Lopes, G.S.; Vieira, R.P.G.; de Moraes Filho, M.O.; de Moraes, M.E.A.; et al. Validation of Endogenous Control Genes by Real-Time Quantitative Reverse Transcriptase Polymerase Chain Reaction for Acute Leukemia Gene Expression Studies. Genes 2024, 15, 151. https://doi.org/10.3390/genes15020151
Pessoa FMCdP, Viana VBdJ, de Oliveira MB, Nogueira BMD, Ribeiro RM, Oliveira DdS, Lopes GS, Vieira RPG, de Moraes Filho MO, de Moraes MEA, et al. Validation of Endogenous Control Genes by Real-Time Quantitative Reverse Transcriptase Polymerase Chain Reaction for Acute Leukemia Gene Expression Studies. Genes. 2024; 15(2):151. https://doi.org/10.3390/genes15020151
Chicago/Turabian StylePessoa, Flávia Melo Cunha de Pinho, Vitória Beatriz de Jesus Viana, Marcelo Braga de Oliveira, Beatriz Maria Dias Nogueira, Rodrigo Monteiro Ribeiro, Deivide de Sousa Oliveira, Germison Silva Lopes, Ricardo Parente Garcia Vieira, Manoel Odorico de Moraes Filho, Maria Elisabete Amaral de Moraes, and et al. 2024. "Validation of Endogenous Control Genes by Real-Time Quantitative Reverse Transcriptase Polymerase Chain Reaction for Acute Leukemia Gene Expression Studies" Genes 15, no. 2: 151. https://doi.org/10.3390/genes15020151
APA StylePessoa, F. M. C. d. P., Viana, V. B. d. J., de Oliveira, M. B., Nogueira, B. M. D., Ribeiro, R. M., Oliveira, D. d. S., Lopes, G. S., Vieira, R. P. G., de Moraes Filho, M. O., de Moraes, M. E. A., Khayat, A. S., Moreira, F. C., & Moreira-Nunes, C. A. (2024). Validation of Endogenous Control Genes by Real-Time Quantitative Reverse Transcriptase Polymerase Chain Reaction for Acute Leukemia Gene Expression Studies. Genes, 15(2), 151. https://doi.org/10.3390/genes15020151








