Biomarker RIPK3 Is Silenced by Hypermethylation in Melanoma and Epigenetic Editing Reestablishes Its Tumor Suppressor Function
Abstract
:1. Introduction
2. Materials and Methods
3. Results
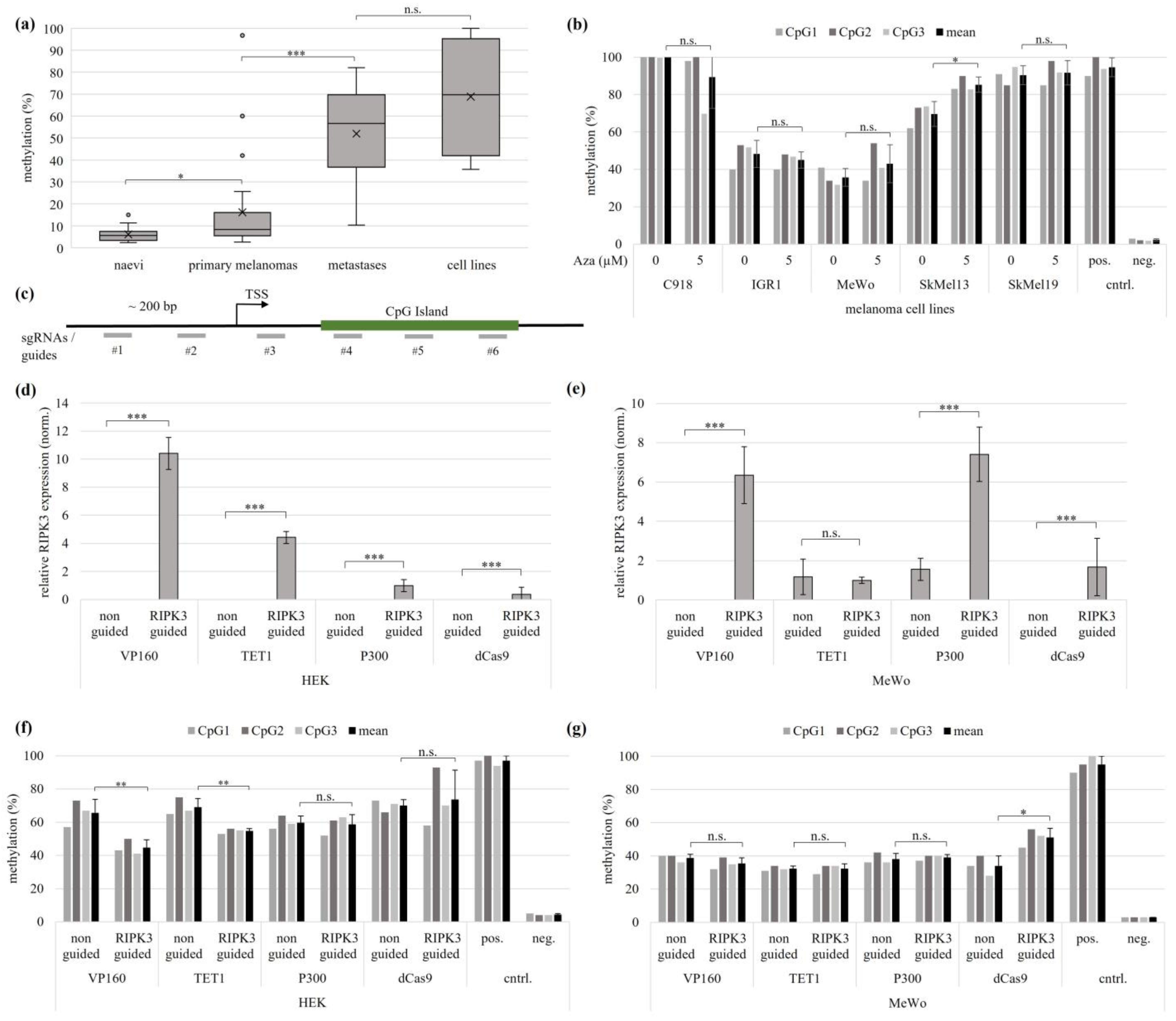
3.1. Epigenetic Silencing of RIPK3 in Melanomas
3.2. Epigenetic Editing by CRISPR-dCas9 (CRISPRa) System
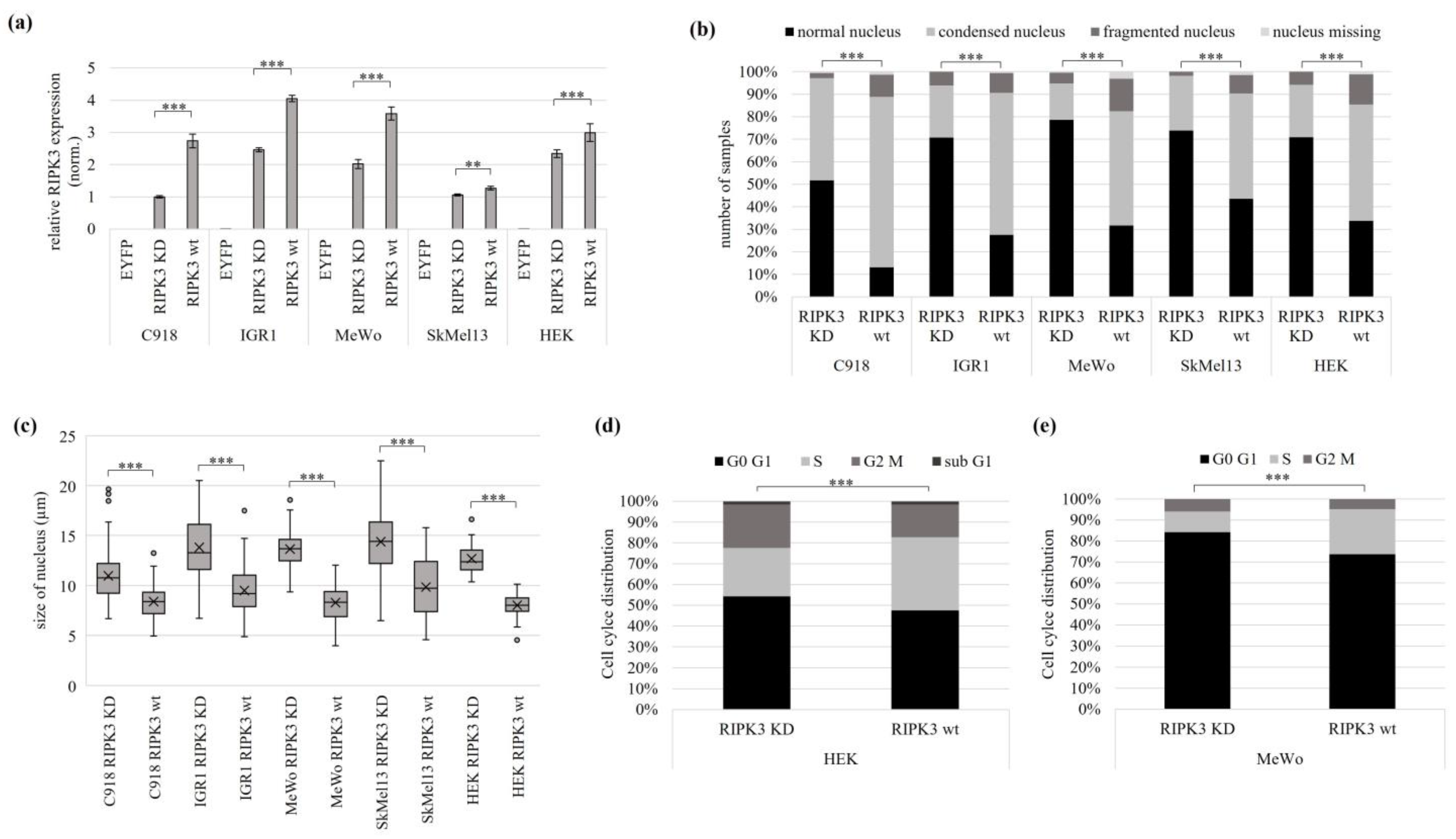
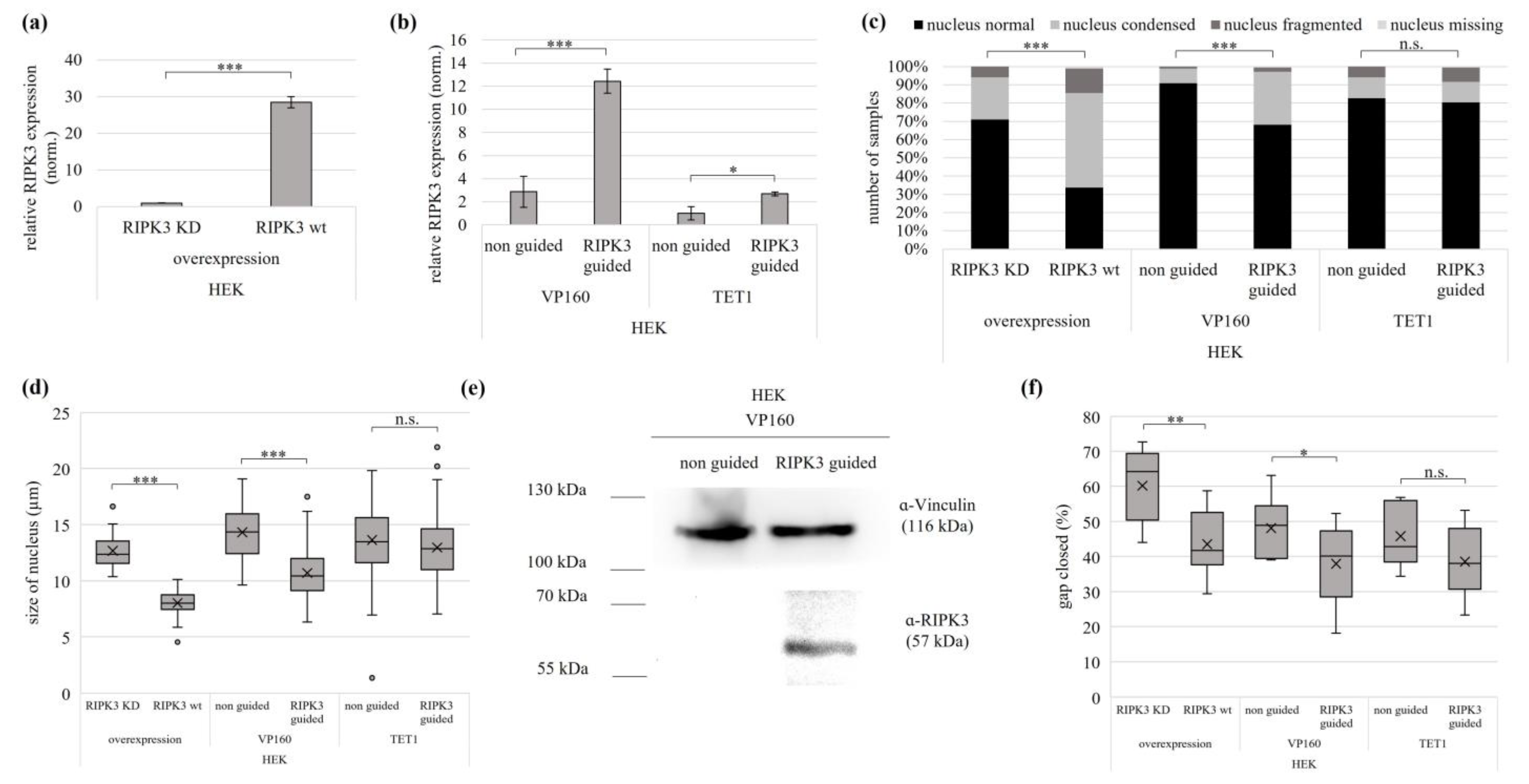
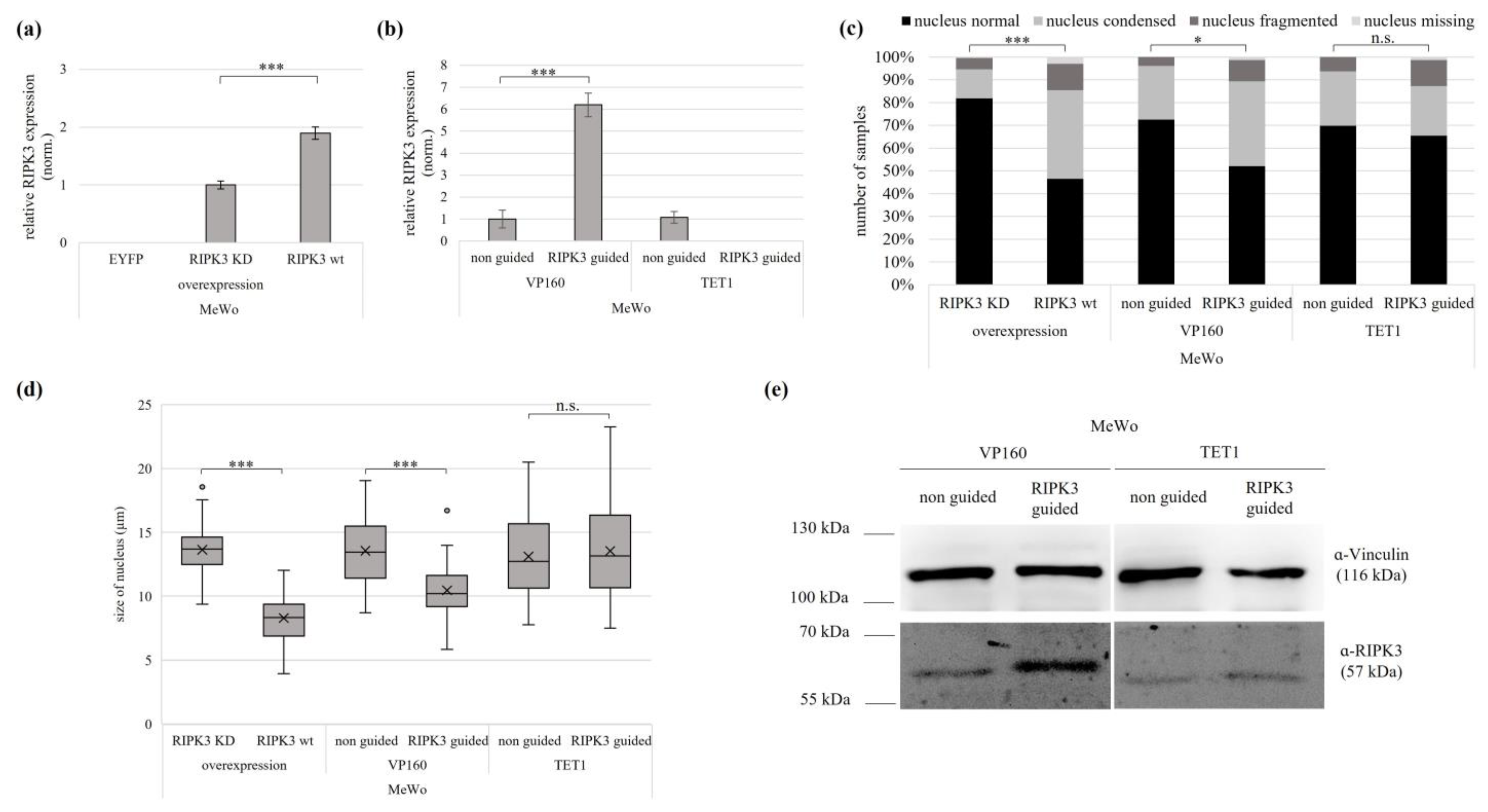
3.3. Reactivation of the Tumor Suppressive Function of RIPK3 using Overexpression and Epigenetic Editing
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Berwick, M.; Wiggins, C. The current epidemiology of cutaneous malignant melanoma. Front. Biosci. 2006, 11, 1244–1254. [Google Scholar] [CrossRef]
- Zhang, L.L.Q.; Chang, C. Epigenetics in Health and Disease. Adv. Exp. Med. Biol. 2020, 1253, 3–55. [Google Scholar]
- Pfalzer, A.C.; Choi, S.W.; Tammen, S.A.; Park, L.K.; Bottiglieri, T.; Parnell, L.D.; Lamon-Fava, S. S-adenosylmethionine mediates inhibition of inflammatory response and changes in DNA methylation in human macrophages. Physiol. Genom. 2014, 46, 617–623. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Weber, M.; Hellmann, I.; Stadler, M.B.; Ramos, L.; Paabo, S.; Rebhan, M.; Schübeler, D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 2007, 39, 457–466. [Google Scholar] [CrossRef] [PubMed]
- Saxonov, S.; Berg, P.; Brutlag, D.L. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc. Natl. Acad. Sci. USA 2006, 103, 1412–1417. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.L.; Grant, P.A. The role of DNA methylation and histone modifications in transcriptional regulation in humans. Subcell. Biochem. 2013, 61, 289–317. [Google Scholar] [PubMed]
- Kuster, M.M.; Schneider, M.A.; Richter, A.M.; Richtmann, S.; Winter, H.; Kriegsmann, M.; Pullamsetti, S.S.; Stiewe, T.; Savai, R.; Muley, T. Epigenetic Inactivation of the Tumor Suppressor IRX1 Occurs Frequently in Lung Adenocarcinoma and Its Silencing Is Associated with Impaired Prognosis. Cancers 2020, 12, 3528. [Google Scholar] [CrossRef] [PubMed]
- Dammann, R.; Li, C.; Yoon, J.H.; Chin, P.L.; Bates, S.; Pfeifer, G.P. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat. Genet. 2000, 25, 315–319. [Google Scholar] [CrossRef]
- Deutschmeyer, V.; Breuer, J.; Walesch, S.K.; Sokol, A.M.; Graumann, J.; Bartkuhn, M.; Boettger, T.; Rossbach, O.; Richter, A.M. Epigenetic therapy of novel tumour suppressor ZAR1 and its cancer biomarker function. Clin. Epigenetics 2019, 11, 182. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Samir, P.; Malireddi, R.K.S.; Kanneganti, T.D. The PANoptosome: A Deadly Protein Complex Driving Pyroptosis, Apoptosis, and Necroptosis (PANoptosis). Front. Cell Infect. Microbiol. 2020, 10, 238. [Google Scholar] [CrossRef] [PubMed]
- Christgen, S.; Zheng, M.; Kesavardhana, S.; Karki, R.; Malireddi, R.K.S.; Banoth, B.; Place, D.E.; Briard, B.; Sharma, B.R.; Tuladhar, S. Identification of the PANoptosome: A Molecular Platform Triggering Pyroptosis, Apoptosis, and Necroptosis (PANoptosis). Front. Cell. Infect. Microbiol. 2020, 10, 237. [Google Scholar] [CrossRef] [PubMed]
- Green, D.R.; Ferguson, T.; Zitvogel, L.; Kroemer, G. Immunogenic and tolerogenic cell death. Nat. Rev. Immunol. 2009, 9, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Basavaraju, S.; Mishra, S.; Jindal, R.; Kesavardhana, S. Emerging Role of ZBP1 in Z-RNA Sensing, Influenza Virus-Induced Cell Death, and Pulmonary Inflammation. mBio 2022, 13, e0040122. [Google Scholar] [CrossRef] [PubMed]
- Ermine, K.; Yu, J.; Zhang, L. Role of Receptor Interacting Protein (RIP) kinases in cancer. Genes. Dis. 2022, 9, 1579–1593. [Google Scholar] [CrossRef]
- Park, S.H.; Lee, H.C.; Jeong, H.M.; Lee, J.S.; Cha, H.J.; Kim, C.H.; Kim, J.; Song, K.S. Inhibition of Urban Particulate Matter-Induced Airway Inflammation by RIPK3 through the Regulation of Tight Junction Protein Production. Int. J. Mol. Sci. 2023, 24, 13320. [Google Scholar] [CrossRef]
- Conev, N.V.; Dimitrova, E.G.; Bogdanova, M.K.; Kashlov, Y.K.; Chaushev, B.G.; Radanova, M.A.; Petrov, D.P.; Georgiev, K.D. RIPK3 expression as a potential predictive and prognostic marker in metastatic colon cancer. Clin. Investig. Med. 2019, 42, E31–E38. [Google Scholar] [CrossRef]
- Hockendorf, U.; Yabal, M.; Herold, T.; Munkhbaatar, E.; Rott, S.; Jilg, S.; Kauschinger, J.; Magnani, G.; Reisinger, F.; Heuser, M. RIPK3 Restricts Myeloid Leukemogenesis by Promoting Cell Death and Differentiation of Leukemia Initiating Cells. Cancer Cell 2016, 30, 75–91. [Google Scholar] [CrossRef]
- Geserick, P.; Wang, J.; Schilling, R.; Horn, S.; Harris, P.A.; Bertin, J.; Gough, P.J.; Feoktistova, M.; Leverkus, M. Absence of RIPK3 predicts necroptosis resistance in malignant melanoma. Cell Death Dis. 2015, 6, e1884. [Google Scholar] [CrossRef] [PubMed]
- Molenaar, J.J.; Koster, J.; Zwijnenburg, D.A.; van Sluis, P.; Valentijn, L.J.; van der Ploeg, I.; Hamdi, M.; Van Nes, J.; Westerman, B.A.; Van Arkel, J. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature 2012, 483, 589–593. [Google Scholar] [CrossRef]
- Diez-Villanueva, A.; Mallona, I.; Peinado, M.A. Wanderer, an interactive viewer to explore DNA methylation and gene expression data in human cancer. Epigenetics Chromatin 2015, 8, 22. [Google Scholar] [CrossRef]
- Xiong, Z.; Laird, P.W. COBRA: A sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997, 25, 2532–2534. [Google Scholar] [CrossRef]
- Helmbold, P.; Richter, A.M.; Walesch, S.; Skorokhod, A.; Marsch, W.; Enk, A.; Dammann, R.H. RASSF10 promoter hypermethylation is frequent in malignant melanoma of the skin but uncommon in nevus cell nevi. J. Investig. Dermatol. 2012, 132, 687–694. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef] [PubMed]
- Adli, M. The CRISPR tool kit for genome editing and beyond. Nat. Commun. 2018, 9, 1911. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, L.A.; Larson, M.H.; Morsut, L.; Liu, Z.; Brar, G.A.; Torres, S.E.; Stern-Ginossar, N.; Brandman, O.; Whitehead, E.H.; Doudna, J.A. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 2013, 154, 442–451. [Google Scholar] [CrossRef] [PubMed]
- Hirai, H.; Tani, T.; Kikyo, N. Structure and functions of powerful transactivators: VP16, MyoD and FoxA. Int. J. Dev. 2010, 54, 1589–1596. [Google Scholar] [CrossRef] [PubMed]
- Cheng, A.W.; Wang, H.; Yang, H.; Shi, L.; Katz, Y.; Theunissen, T.W.; Rangarajan, S.; Shivalila, C.S.; Dadon, D.B.; Jaenisch, R. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 2013, 23, 1163–1171. [Google Scholar] [CrossRef]
- Klann, T.S.; Black, J.B.; Chellappan, M.; Safi, A.; Song, L.; Hilton, I.B.; Crawford, G.E.; Reddy, T.E.; Gersbach, C.A. CRISPR-Cas9 epigenome editing enables high-throughput screening for functional regulatory elements in the human genome. Nat. Biotechnol. 2017, 35, 561–568. [Google Scholar] [CrossRef]
- Amabile, A.; Migliara, A.; Capasso, P.; Biffi, M.; Cittaro, D.; Naldini, L.; Lombardo, A. Inheritable Silencing of Endogenous Genes by Hit-and-Run Targeted Epigenetic Editing. Cell 2016, 167, 219–232.e214. [Google Scholar] [CrossRef]
- Sapozhnikov, D.M.; Szyf, M. Enzyme-free targeted DNA demethylation using CRISPR-dCas9-based steric hindrance to identify DNA methylation marks causal to altered gene expression. Nat. Protoc. 2022, 17, 2840–2881. [Google Scholar] [CrossRef]
- Richter, A.M.; Walesch, S.K.; Wurl, P.; Taubert, H.; Dammann, R.H. The tumor suppressor RASSF10 is upregulated upon contact inhibition and frequently epigenetically silenced in cancer. Oncogenesis 2012, 1, e18. [Google Scholar] [CrossRef]
- Blackledge, N.P.; Klose, R.J. The molecular principles of gene regulation by Polycomb repressive complexes. Nat. Rev. Mol. Cell Biol. 2021, 22, 815–833. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Guo, J.; Cheng, X.; Liao, Y.; Bi, Y.; Gong, Y.; Zhang, X.; Guo, Y.; Wang, X.; Yu, W. RIPK3 Suppresses the Progression of Spontaneous Intestinal Tumorigenesis. Front. Oncol. 2021, 11, 664927. [Google Scholar] [CrossRef] [PubMed]
- Bozec, D.; Iuga, A.C.; Roda, G.; Dahan, S.; Yeretssian, G. Critical function of the necroptosis adaptor RIPK3 in protecting from intestinal tumorigenesis. Oncotarget 2016, 7, 46384–46400. [Google Scholar] [CrossRef] [PubMed]
- Simonicova, K.; Janotka, L.; Kavcova, H.; Sulova, Z.; Messingerova, L.; Breier, A. Resistance of Leukemia Cells to 5-Azacytidine: Different Responses to the Same Induction Protocol. Cancers 2023, 15, 3063. [Google Scholar] [CrossRef]
- Xing, X.Q.; Li, B.; Xu, S.L.; Zhang, C.F.; Liu, J.; Deng, Y.S.; Yang, J. 5-Aza-2′-deoxycytidine, a DNA methylation inhibitor, attenuates hypoxic pulmonary hypertension via demethylation of the PTEN promoter. Eur. J. Pharmacol. 2019, 855, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Patra, S.K.; Bettuzzi, S. Epigenetic DNA-(cytosine-5-carbon) modifications: 5-aza-2′-deoxycytidine and DNA-demethylation. Biochemistry 2009, 74, 613–619. [Google Scholar] [CrossRef] [PubMed]
- Pappalardi, M.B.; Keenan, K.; Cockerill, M.; Kellner, W.A.; Stowell, A.; Sherk, C.; Wong, K.; Pathuri, S.; Briand, J.; Steidel, M. Discovery of a first-in-class reversible DNMT1-selective inhibitor with improved tolerability and efficacy in acute myeloid leukemia. Nat. Cancer 2021, 2, 1002–1017. [Google Scholar] [CrossRef] [PubMed]
- Sapozhnikov, D.M.; Szyf, M. Unraveling the functional role of DNA demethylation at specific promoters by targeted steric blockage of DNA methyltransferase with CRISPR/dCas9. Nat. Commun. 2021, 12, 5711. [Google Scholar] [CrossRef] [PubMed]
- Crump, N.T.; Hazzalin, C.A.; Bowers, E.M.; Alani, R.M.; Cole, P.A.; Mahadevan, L.C. Dynamic acetylation of all lysine-4 trimethylated histone H3 is evolutionarily conserved and mediated by p300/CBP. Proc. Natl. Acad. Sci. USA 2011, 108, 7814–7819. [Google Scholar] [CrossRef]
- Ragvin, A.; Valvatne, H.; Erdal, S.; Arskog, V.; Tufteland, K.R.; Breen, K.; Øyan, A.M.; Eberharter, A.; Gibson, T.J.; Becker, P.B. Nucleosome binding by the bromodomain and PHD finger of the transcriptional cofactor p300. J. Mol. Biol. 2004, 337, 773–788. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arroyo Villora, S.; Castellanos Silva, P.; Zenz, T.; Kwon, J.S.; Schlaudraff, N.; Nitaj, D.; Meckbach, C.; Dammann, R.; Richter, A.M. Biomarker RIPK3 Is Silenced by Hypermethylation in Melanoma and Epigenetic Editing Reestablishes Its Tumor Suppressor Function. Genes 2024, 15, 175. https://doi.org/10.3390/genes15020175
Arroyo Villora S, Castellanos Silva P, Zenz T, Kwon JS, Schlaudraff N, Nitaj D, Meckbach C, Dammann R, Richter AM. Biomarker RIPK3 Is Silenced by Hypermethylation in Melanoma and Epigenetic Editing Reestablishes Its Tumor Suppressor Function. Genes. 2024; 15(2):175. https://doi.org/10.3390/genes15020175
Chicago/Turabian StyleArroyo Villora, Sarah, Paula Castellanos Silva, Tamara Zenz, Ji Sun Kwon, Nico Schlaudraff, Dafina Nitaj, Cornelia Meckbach, Reinhard Dammann, and Antje M. Richter. 2024. "Biomarker RIPK3 Is Silenced by Hypermethylation in Melanoma and Epigenetic Editing Reestablishes Its Tumor Suppressor Function" Genes 15, no. 2: 175. https://doi.org/10.3390/genes15020175
APA StyleArroyo Villora, S., Castellanos Silva, P., Zenz, T., Kwon, J. S., Schlaudraff, N., Nitaj, D., Meckbach, C., Dammann, R., & Richter, A. M. (2024). Biomarker RIPK3 Is Silenced by Hypermethylation in Melanoma and Epigenetic Editing Reestablishes Its Tumor Suppressor Function. Genes, 15(2), 175. https://doi.org/10.3390/genes15020175







