Transcriptome Analysis Reveals the Molecular Mechanisms Underlying Growth Superiority in a Novel Gymnocypris Hybrid, Gymnocypris przewalskii ♀ × Gymnocypris eckloni ♂
Abstract
:1. Introduction
2. Material and Method
2.1. Fish and Sample Preparation
2.2. RNA Extraction, Library Construction, and High-Throughput Sequencing
2.3. RNA Sequencing Analysis
2.4. Quantitative Real-Time PCR (qRT-PCR) Analysis
2.5. Statistical Analyses
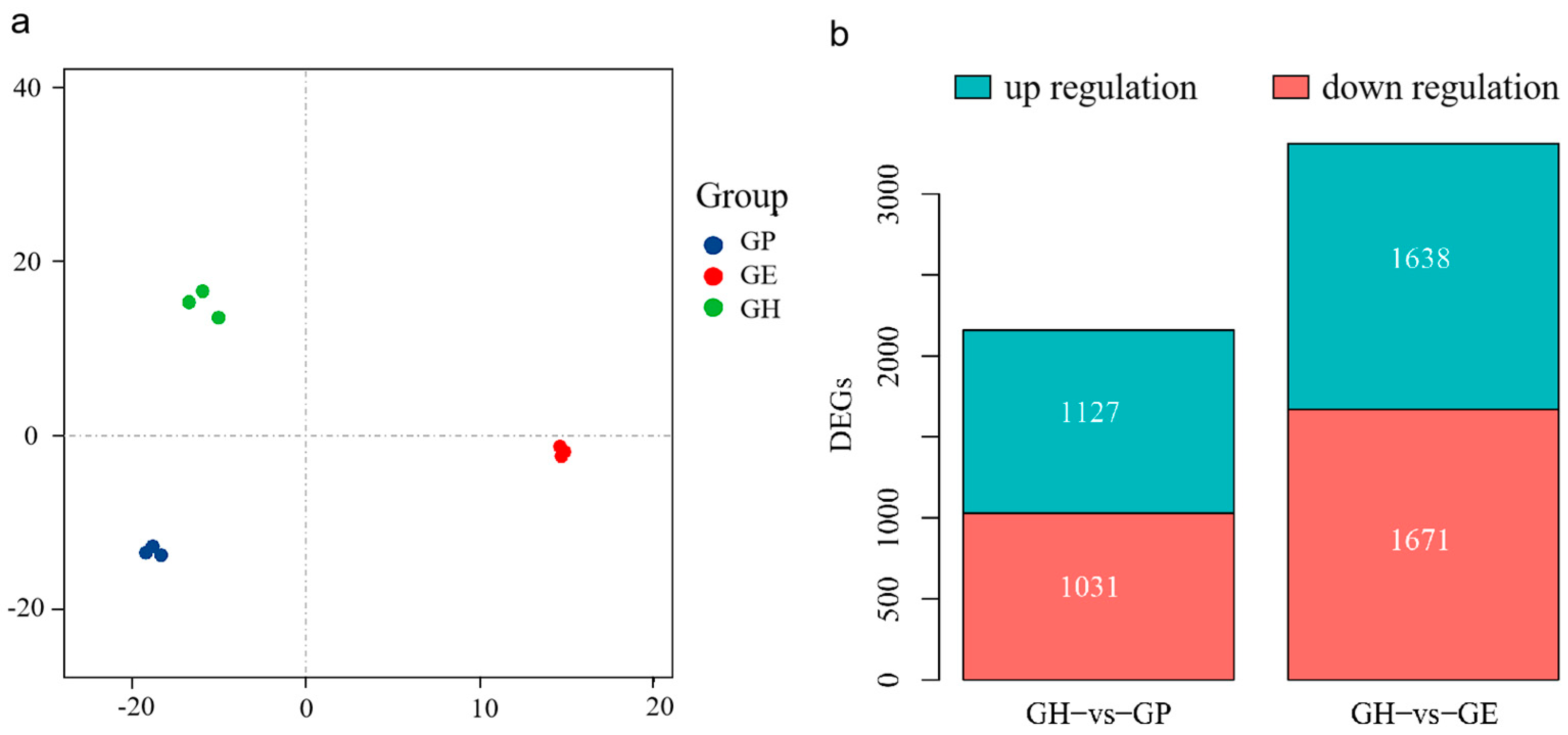
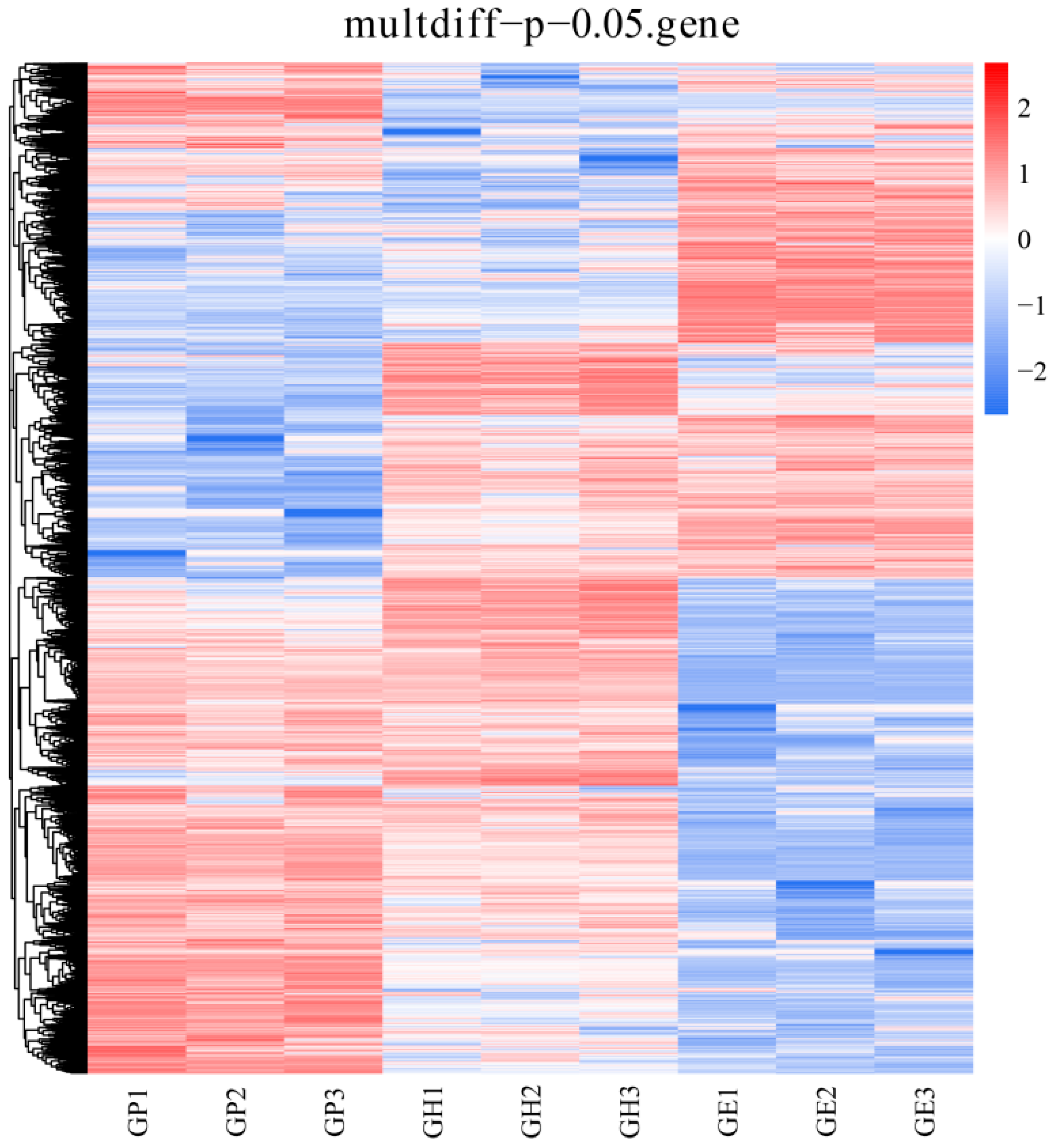
3. Results
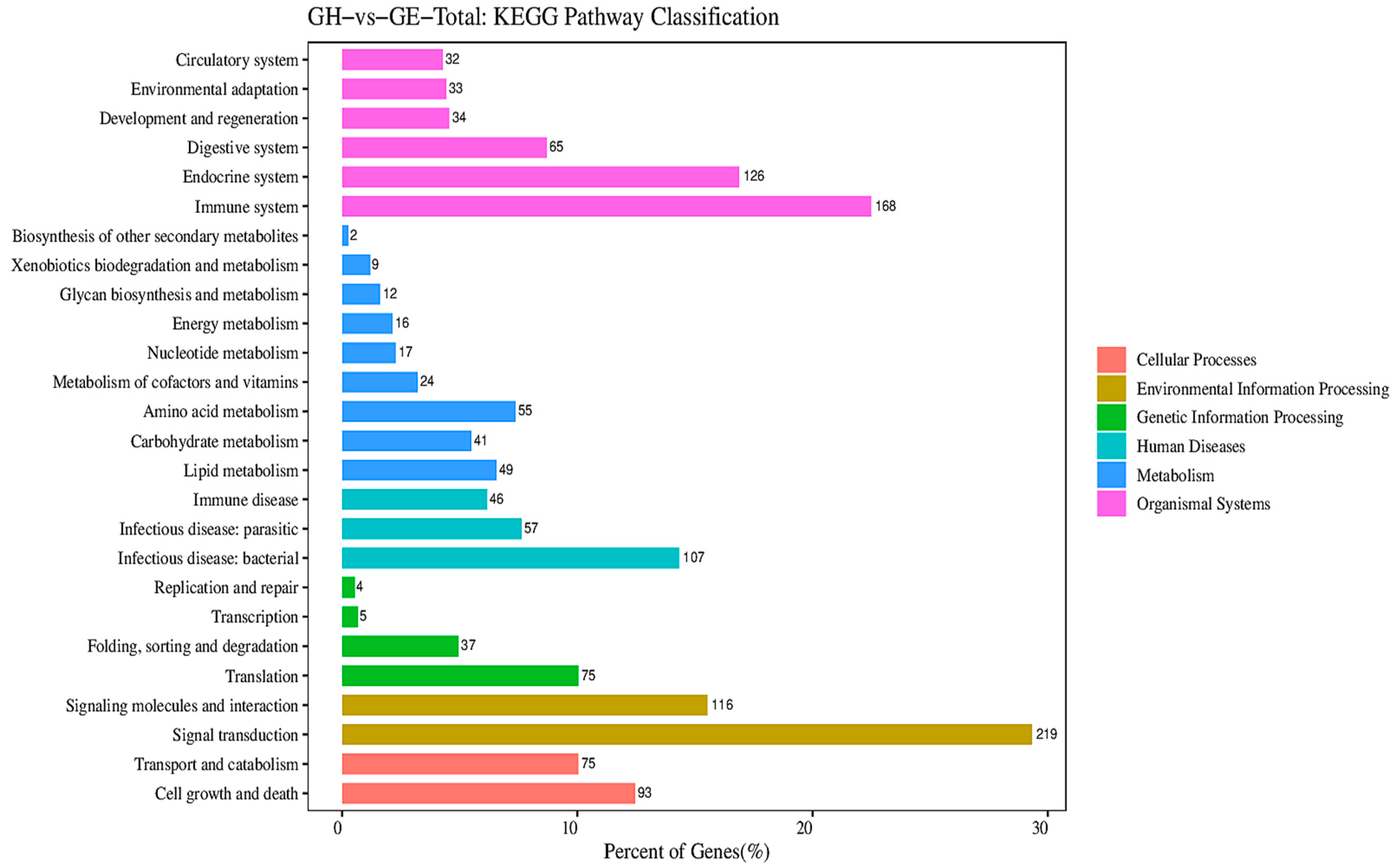
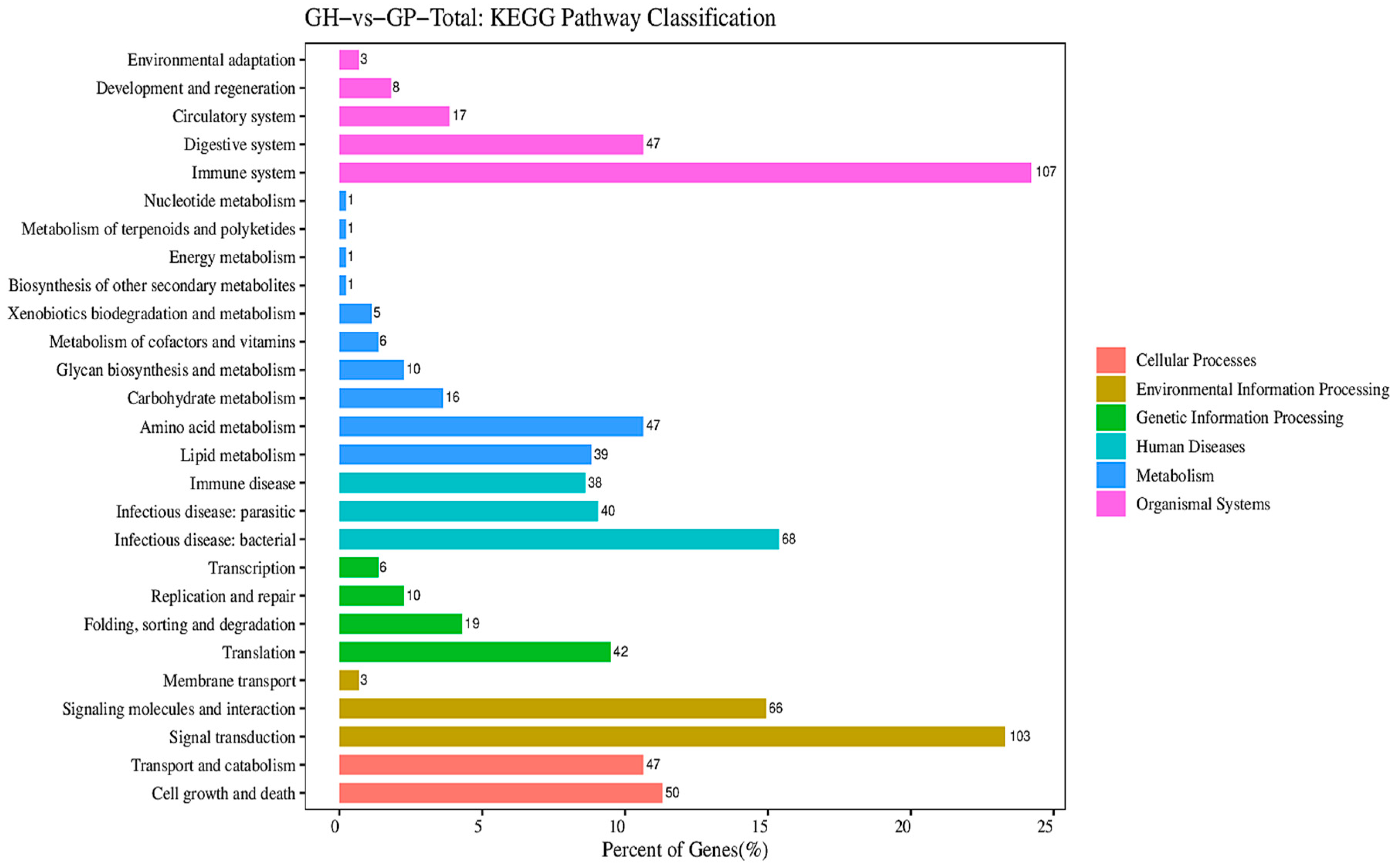
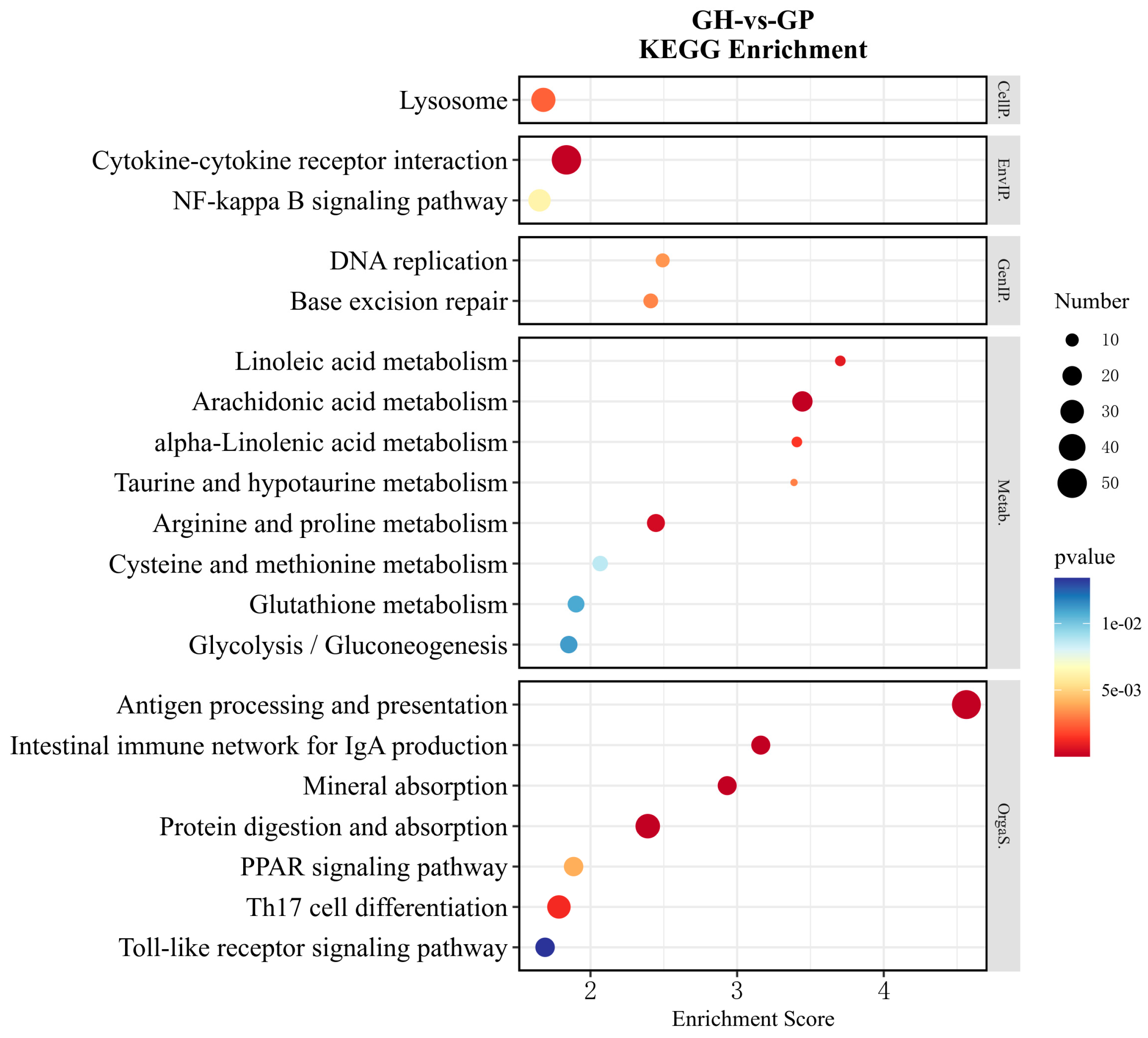
3.1. Metabolism-Related DEG Pathway Analysis
3.2. Validation of DEGs by Quantitative Real-Time PCR (qRT-PCR)
4. Discussion
4.1. Differences in the Metabolism of F1 Hybrid Gymnocypris and Its Parents
4.2. Differences in the Relative Growth Pathways of F1 Hybrid Gymnocypris and Its Parents
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chandhini, S.; Rejish, K.V.J. Transcriptomics in aquaculture: Current status and applications. Rev. Aquac. 2019, 11, 1379–1397. [Google Scholar] [CrossRef]
- Waiho, K.; Ikhwanuddin, M.; Afiqah-Aleng, N.; Shu-Chien, A.C.; Wang, Y.J.; Ma, H.Y.; Fazhan, H. Transcriptomics in advancing portunid aquaculture: A systematic review. Rev. Aquac. 2022, 14, 2064–2088. [Google Scholar] [CrossRef]
- Lowe, R.; Shirley, N.; Bleackley, M.; Dolan, S.; Shafee, T. Transcriptomics technologies. PLoS Comput. Biol. 2017, 13, e1005457. [Google Scholar] [CrossRef]
- Sun, Y.; Guo, C.Y.; Wang, D.D.; Li, X.F.; Shi, Q.; Hu, G.J.; Fang, C.; Lin, H.R.; Zhang, Y. Transcriptome analysis reveals the molecular mechanisms underlying growth superiority in a novel grouper hybrid (Epinephelus fuscogutatus ♀ × Epinephelus lanceolatus ♂). BMC Genet. 2016, 17, 24. [Google Scholar] [CrossRef]
- Gomes, F.; Watanabe, L.; Vianez, J.; Nunes, M.; Cardoso, J.; Lima, C.; Schneider, H.; Sampaio, I. Comparative analysis of the transcriptome of the Amazonian fish species Colossoma macropomum (tambaqui) and hybrid tambacu by next generation sequencing. PLoS ONE 2019, 14, e0212755. [Google Scholar] [CrossRef]
- Wang, Y.D.; Yao, J.J.; Liao, A.M.; Tan, H.F.; Luo, Y.X.; Wu, P.; Wang, S.; Zhang, C.; Qin, Q.B.; Tao, M.; et al. The formation of hybrid fish derived from hybridization of Megalobrama amblycephala (♀) × Siniperca chuatsi (♂). Aquaculture 2022, 548, 737547. [Google Scholar] [CrossRef]
- Liu, S.J. Distant hybridization leads to different ploidy fishes. Sci. China Life Sci. 2010, 53, 416–425. [Google Scholar] [CrossRef]
- Yu, J.H.; Xia, D.Q.; Yang, H.; He, Y.H.; Wu, T.T. Morphology of the progenies of Oreochromis aurea (♀) × Sinipperca chuatai (♂). J. Fish China 2003, 27, 431–435. [Google Scholar]
- Liu, S.J.; Qin, Q.B.; Xiao, J.; Lu, W.T.; Liu, Y. The formation of the polyploid hybrids from different subfamily fish crossings and its evolutionary significance. Genetics 2006, 176, 1023–1034. [Google Scholar] [CrossRef]
- Xiao, J.; Kang, X.W.; Xie, L.H.; Qin, Q.B.; He, Z.L.; Hu, F.Z.; Zhang, C.; Zhao, R.R.; Wang, J.; Luo, K.K.; et al. The fertility of the hybrid lineage derived from female Megalobrama amblycephala × male Culter alburnus. Anim. Reprod. Sci. 2014, 151, 61–70. [Google Scholar] [CrossRef]
- Liu, S.J.; Sun, Y.D.; Zhang, C.; Luo, K.K.; Liu, Y. Production of gynogenetic progeny from allotetraploid hybrids red crucian carp × common carp. Aquaculture 2004, 236, 193–200. [Google Scholar] [CrossRef]
- Yang, S.Y.; Liu, Z.; Yang, Z.Z.; Zhao, Z.M.; Zhang, C.Y.; Gong, Q.; Du, X.G.; Wu, J.Y.; Feng, Y.; Du, J.; et al. Improvement of skeletal muscle growth by GH/IGF growth-axis contributes to growth performance in commercial fleshy sturgeon. Aquaculture 2021, 543, 736929. [Google Scholar] [CrossRef]
- Wang, Y.S.; Gonzalez, R.J.; Patrick, M.L.; Grosell, M.; Wood, C.M. Unusual physiology of scale-less carp, Gymnocypris przewalskii, in Lake Qinghai: A high altitude alkaline saline lake. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2003, 134, 409–421. [Google Scholar] [CrossRef]
- Xiong, F.; Chen, D.Q.; Duan, X.B. Threatened fishes of the world: Gymnocypris przewalskii (Kessler, 1876) (Cyprinidae: Schizothoracinae). Environ. Biol. Fishes 2010, 87, 351–352. [Google Scholar] [CrossRef]
- Zhang, R.Y.; Ludwig, A.; Zhang, C.F.; Tong, C.; Li, G.G.; Tang, Y.T.; Peng, Z.G.; Zhao, K. Local adaptation of Gymnocypris przewalskii (Cyprinidae) on the Tibetan Plateau. Sci. Rep. 2015, 5, 9780. [Google Scholar] [CrossRef]
- Wood, C.M.; Du, J.Z.; Rogers, J.; Brauner, C.J.; Richards, J.G.; Semple, J.W.; Murray, B.W.; Chen, X.Q.; Wang, Y.X. Przewalski’s Naked Carp (Gymnocypris przewalskii): An Endangered Species Taking a Metabolic Holiday in Lake Qinghai, China. Physiol. Biochem. Zool 2007, 80, 59–77. [Google Scholar] [CrossRef]
- Zhao, K.; Li, J.B.; Yang, G.S.; Duan, Z.Y.; He, S.P.; Chen, Y.Y. Molecular phylogeny of endemic Gymnocyprinus in Qinghai lake and its adjacent water systems. Chin. Sci. Bull. 2005, 50, 1348–1355. (In Chinese) [Google Scholar] [CrossRef]
- Wang, F.Y.; Wang, L.H.; Liu, D.; Gao, Q.; Nie, M.M.; Zhu, S.H.; Chao, Y.; Yang, C.J.; Zhang, C.F.; Yi, R.G.; et al. Chromosome-level assembly of Gymnocypris eckloni genome. Sci. Data 2022, 9, 464. [Google Scholar] [CrossRef]
- Zhang, H.C.; Xu, B.K.; A, L.L.; Ma, Q.H.; Gao, Q.; Tian, W.G.; Yu, L.X.; Liang, J. Cloning, tissue expression and polymorphism of MHCⅡ gene of Gymnocypris. J. Fish. China 2023, 1–18. [Google Scholar]
- Bartley, D.M.; Rana, K.; Immink, A.J. The use of inter-specific hybrids in aquaculture and fisheries. Rev. Fish Biol. Fisher 2000, 10, 325–337. [Google Scholar] [CrossRef]
- Zhou, H. Genetic Analysis for Two Grouper Hybrids and Their Parents. Master’s Thesis, Sun Yat-sen University, Guangzhou, China, 2011. [Google Scholar]
- Zhou, Y.H.; Li, J.Y.; Qi, H.F.; Ban, X.; Yang, J.X.; Du, H. Riverbed substrate requirements for natural reproduction of Gymnocypris przewalskii. Animals 2021, 11, 3246. [Google Scholar] [CrossRef]
- De-Assis, L.A.; Rezende, T.T.; Dias, M.A.D.; De Freitas, M.A.F.; Hilsdorf, R.T.S.; Wagner, A. The development of genetically improved red tilapia lines through the backcross breeding of two Oreochromis niloticus strains. Aquaculture 2017, 472, 17–22. [Google Scholar] [CrossRef]
- Hedgecock, D.; Lin, J.Z.; de-Cola, S.; Haudenschild, C.D.; Meyer, E.; Manahan, D.T.; Bowen, B. Transcriptomic analysis of growth heterosis in larval Pacific oysters (Crassostrea gigas). Proc. Natl. Acad. Sci. USA 2007, 104, 2313–2318. [Google Scholar] [CrossRef]
- He, S.; Liang, X.F.; Sun, J.; Ling Li, L.; Yu, Y.; Huang, W.; Qu, C.M.; Cao, L.; Bai, X.L.; Tao, Y.X. Insights into food preference in hybrid F1 of Siniperca chuatsi (♀) × Siniperca scherzeri (♂) mandarin fish through transcriptome analysis. BMC Genom. 2013, 14, 601. [Google Scholar] [CrossRef]
- Tixier, V.; Bataillé, L.; Etard, C.; Jaglaet, K. Glycolysis supports embryonic muscle growth by promoting myoblast fusion. Proc. Natl. Acad. Sci. USA 2013, 110, 18982–18987. [Google Scholar] [CrossRef]
- Wang, N.; Tian, Y.S.; Zhang, J.J.; Li, Z.T.; Cheng, M.L.; Wu, Y.P. Involvement of glycolysis activation in flatfish sexual size dimorphism: Insights from transcriptomic analyses of Platichthys stellatus and Cynoglossus semilaevis. Comp. Biochem. Phys. D Genom. Proteom. 2021, 39, 100832. [Google Scholar] [CrossRef]
- Gao, X.L.; Wang, H.Z.; Yang, J.J.; Chen, J.M.; Jie, J.; Li, L.W.; Liu, Z.R. Reciprocal regulation of protein kinase and pyruvate kinase activities of pyruvate kinase M2 by growth signals. J. Biol. Chem. 2013, 288, 15971–15979. [Google Scholar] [CrossRef]
- Peterside, I.E.; Selak, M.A.; Simmons, R.A. Impaired oxidative phosphorylation in hepatic mitochondria in growth-retarded rats. Am. J. Physiol. Endocrinol. Metab. 2003, 285, E1258. [Google Scholar] [CrossRef]
- Yang, H.C.; Yu, H.; Liu, Y.C.; Chen, T.-L.; Stern, A.; Szecheng, J.L.; Tsun-Yee Chiu, D. IDH-1 deficiency induces growth defects and metabolic alterations in GSPD-1-deficient Caenorhabditis elegans. J. Mol. Med. 2019, 97, 385–396. [Google Scholar] [CrossRef]
- Wang, Q.C.; Xu, Z.; Ai, Q.H. Arginine metabolism and its functions in growth, nutrient utilization, and immunonutrition of fish. Anim. Nutr. 2021, 7, 716–727. [Google Scholar] [CrossRef]
- Wang, B.; Wu, L.J.; Chen, J.; Dong, L.L.; Chen, C.; Wen, Z.; Hu, J.; Fleming, I.; Wang, D.W. Metabolism pathways of arachidonic acids: Mechanisms and potential therapeutic targets. Signal Transduct. Target. Ther. 2021, 6, 94. [Google Scholar] [CrossRef]
- Rubin, P.; Mollison, K.W. Pharmacotherapy of diseases mediated by 5-lipoxygenase pathway eicosanoids. Prostaglandins Other Lipid Mediat. 2007, 83, 188–197. [Google Scholar] [CrossRef]
- Jiang, W.G.; Douglas-Jones, A.; Mansel, R.E. Levels of expression of lipoxygenases and cyclooxygenase-2 in human breast cancer. Prostaglandins Leukot. Essent. Fat. Acids. 2003, 69, 275–281. [Google Scholar] [CrossRef]
- Fuentes, E.N.; Valdés, J.A.; Molina, A.; Björnsson, B.T. Regulation of skeletal muscle growth in fish by the growth hormone–insulin-like growth factor system. Gen. Comp. Endocrinol. 2013, 192, 136–148. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.J.; Chen, F.Y.; Cao, L.F.; Zhang, Y.; Zhong, H. Functional Enrichment Analysis by David in Transgenic MYCN Zebrafish Model. Blood 2012, 120, 5114. [Google Scholar] [CrossRef]
- Fuhrmann, V.; Kinkl, N.; Leveillard, T.; Sahel, J.; Hicks, D. Fibroblast growth factor receptor 4 (FGFR4) is expressed in adult rat and human retinal photoreceptors and neurons. J. Mol. Neurosci. 1999, 13, 187–197. [Google Scholar] [CrossRef]
- Montserrat, N.; Capilla, E.; Navarro, I.; Gutiérrez, J. Metabolic effects of insulin and IGFs on Gilthead sea bream (Sparus aurata) muscle cells. Front. Endocrinol. 2012, 3, 55. [Google Scholar] [CrossRef]
- Codina, M.; García de la Serrana, D.; Sánchez-Gurmaches, J.; Montserrat, N.; Chistyakova, O.; Navarro, I.; Gutiérrez, J. Metabolic and mitogenic effects of IGF-II in rainbow trout (Oncorhynchus mykiss) myocytes in culture and the role of IGF-II in the PI3K/Akt and MAPK signalling pathways. Gen. Comp. Endocrinol. 2008, 157, 116–124. [Google Scholar] [CrossRef]
- Rius-Francino, M.; Acerete, L.; Jiménez-Amilburu, V.; Capilla, E.; Navarro, I.; Gutiérrez, J. Differential effects on proliferation of GH and IGFs in sea bream (Sparus aurata) cultured myocytes. Gen. Comp. Endocrinol. 2011, 172, 44–49. [Google Scholar] [CrossRef]
- Lee, J.T., Jr.; Mccubrey, J.A. The Raf/MEK/ERK signal transduction cascade as a target for chemotherapeutic intervention in leukemia. Leukemia 2002, 16, 486–507. [Google Scholar] [CrossRef] [PubMed]
- Sabatelli, P.; Gualandi, F.; Gara, S.K.; Grumati, P.; Zamparelli, A.; Martoni, E.; Pellegrini, C.; Merlini, L.; Ferlini, A.; Bonaldo, P.; et al. Expression of collagen VI α5 and α6 chains in human muscle and in Duchenne muscular dystrophy-related muscle fibrosis. Matrix Biol. 2012, 31, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Siafakas, A.R.; Richardson, D.R. Growth arrest and DNA damage-45 alpha (GADD45α). Int. J. Biochem. Cell B 2009, 41, 986–989. [Google Scholar] [CrossRef]
- Tang, Y. The Study of mTOR Signal Pathway in the Regulation of the Rapid Growth of Fish Species. Master’s Thesis, Hunan Normal University, Changsha, China, 2020. [Google Scholar]
- Chauvin, C.; Koka, V.; Nouschi, A.; Mieulet, V.; Hoareau-Aveilla, C.; Dreazen, A.; Cagnard, N.; Carpentier, W.; Kiss, T.; Meyuhas, O.; et al. Ribosomal protein S6 kinase activity controls the ribosome biogenesis transcriptional program. Oncogene 2014, 33, 474–483. [Google Scholar] [CrossRef] [PubMed]
- Stephens, L.R.; Hughes, K.T.; Irvine, R.F. Pathway of phosphatidylinositol (3, 4, 5)-trisphosphate synthesis in activated neutrophils. Nature 1991, 351, 33–39. [Google Scholar] [CrossRef] [PubMed]



| GH | GE | GP | ||
|---|---|---|---|---|
| Length | initial | 1.52 ± 0.08 | 1.66 ± 0.12 | 1.49 ± 0.05 |
| terminal | 8.49 ± 1.21 b | 11.73 ± 1.00 a | 7.23 ± 0.40 c | |
| Weight | initial | 1.78 ± 0.13 | 1.82 ± 0.19 | 1.64 ± 0.07 |
| terminal | 9.41 ± 2.18 b | 20.69 ± 5.12 a | 5.54 ± 0.71 c | |
| Weight rate (%) | 428.65 ± 52.28 b | 977.60 ± 87.62 a | 259.74 ± 31.45 c | |
| Survival rate (%) | 80.50 ± 1.00 a | 72.33 ± 1.53 b | 68.17 ± 2.93 c | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Y.; Zhou, J.; Dong, Y.; Xu, D.; Qi, D. Transcriptome Analysis Reveals the Molecular Mechanisms Underlying Growth Superiority in a Novel Gymnocypris Hybrid, Gymnocypris przewalskii ♀ × Gymnocypris eckloni ♂. Genes 2024, 15, 182. https://doi.org/10.3390/genes15020182
Zhao Y, Zhou J, Dong Y, Xu D, Qi D. Transcriptome Analysis Reveals the Molecular Mechanisms Underlying Growth Superiority in a Novel Gymnocypris Hybrid, Gymnocypris przewalskii ♀ × Gymnocypris eckloni ♂. Genes. 2024; 15(2):182. https://doi.org/10.3390/genes15020182
Chicago/Turabian StyleZhao, Yun, Junming Zhou, Yanzhen Dong, Dayong Xu, and Dongming Qi. 2024. "Transcriptome Analysis Reveals the Molecular Mechanisms Underlying Growth Superiority in a Novel Gymnocypris Hybrid, Gymnocypris przewalskii ♀ × Gymnocypris eckloni ♂" Genes 15, no. 2: 182. https://doi.org/10.3390/genes15020182





