Diversity, Distribution, and Chromosomal Rearrangements of TRIP1 Repeat Sequences in Escherichia coli
Abstract
:1. Introduction
2. Materials and Methods
2.1. Genome Sequences
2.2. Identification and Localization of the TRIP1 Repeat Sequences
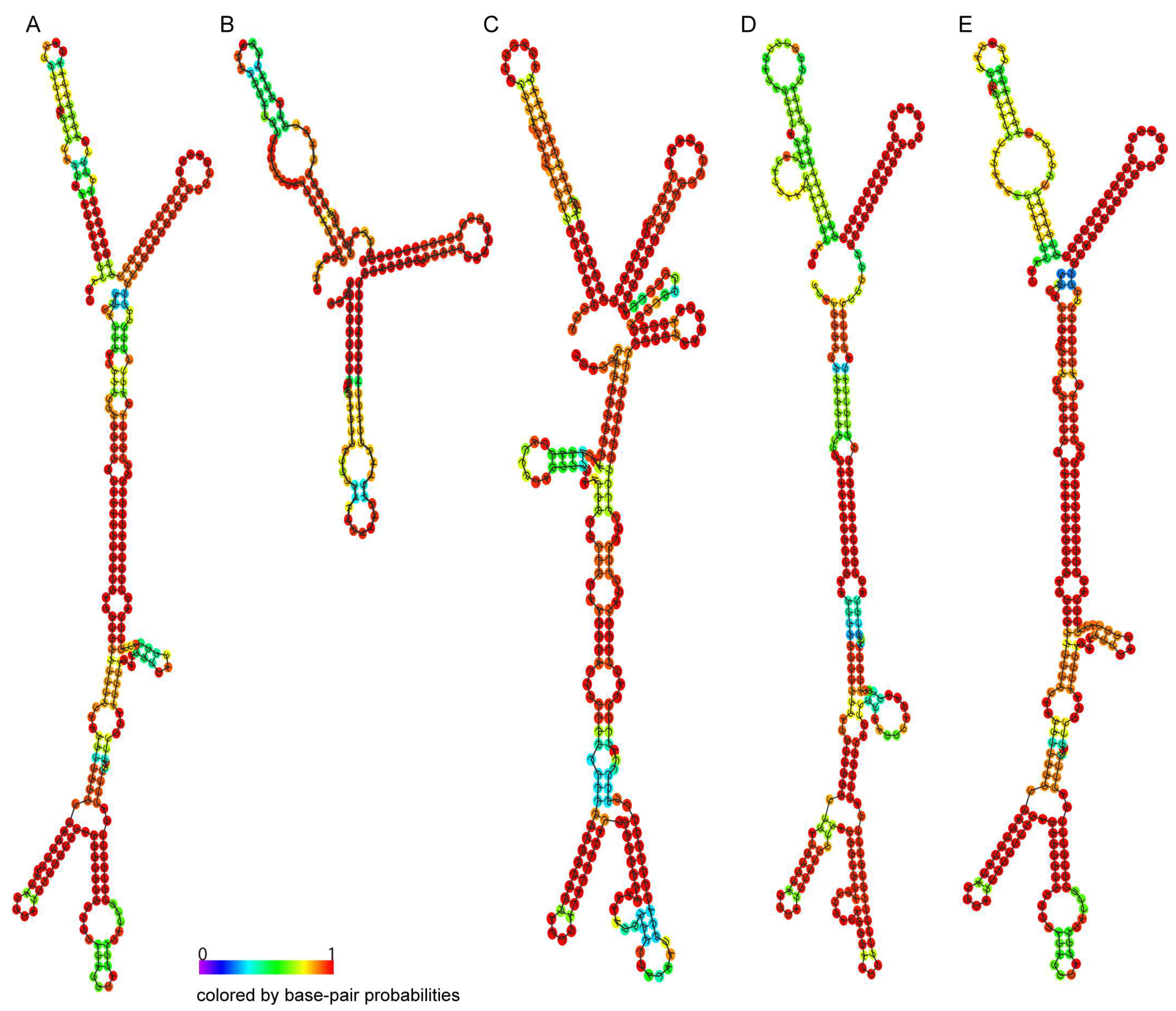
2.3. RNA Structure Prediction of the TRIP1 Repeat Sequences
2.4. Identification of Chromosomal Arrangement Caused by TRIP1 Repeat Sequences
3. Results
3.1. The Distribution and Copy Number of TRIP1 Repeats in E. coli Genomes
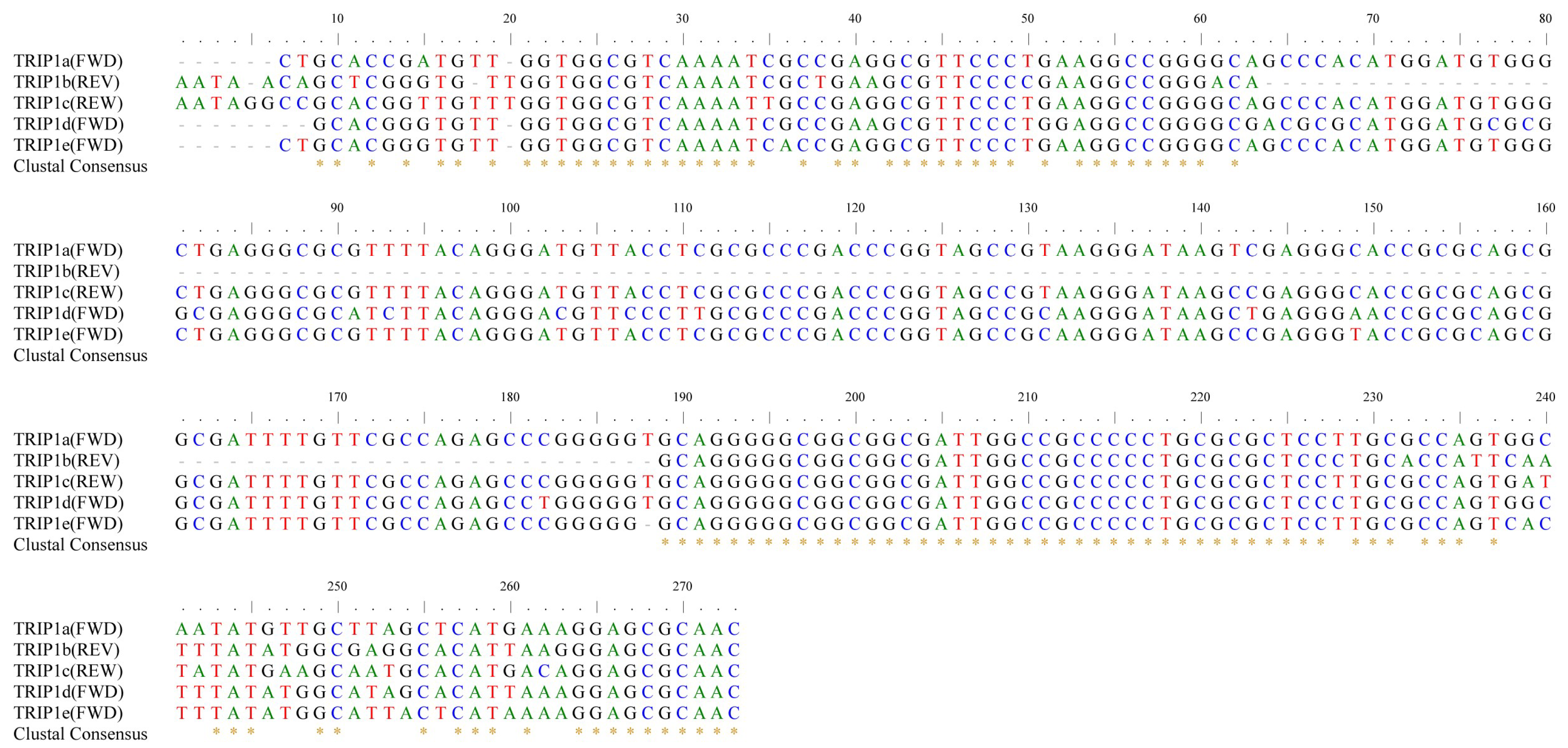
3.2. Distribution and Genome Localization of the Five Types of TRIP1 Repeats in E. coli
3.3. Chromosomal Rearrangements Caused by TRIP1 Repeat Sequences
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Smirnov:, G.B. Repeats in bacterial genomes: Evolutionary considerations. Mol. Genet. Mikrobiol. I Virusol. 2010, 2, 11–20. [Google Scholar] [CrossRef]
- Patel, S. Drivers of bacterial genomes plasticity and roles they play in pathogen virulence, persistence and drug resistance. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2016, 45, 151–164. [Google Scholar] [CrossRef]
- Čutová, M.; Manta, J.; Porubiaková, O.; Kaura, P.; Šťastný, J.; Jagelská, E.B.; Goswami, P.; Bartas, M.; Brázda, V. Divergent distributions of inverted repeats and G-quadruplex forming sequences in Saccharomyces cerevisiae. Genomics 2020, 112, 1897–1901. [Google Scholar] [CrossRef]
- Waters, E.V.; Tucker, L.A.; Ahmed, J.K.; Wain, J.; Langridge, G.C. Impact of Salmonella genome rearrangement on gene expression. Evol. Lett. 2022, 6, 426–437. [Google Scholar] [CrossRef]
- Delihas, N. Impact of small repeat sequences on bacterial genome evolution. Genome Biol. Evol. 2011, 3, 959–973. [Google Scholar] [CrossRef]
- Brazda, V.; Fojta, M.; Bowater, R.P. Structures and stability of simple DNA repeats from bacteria. Biochem. J. 2020, 477, 325–339. [Google Scholar] [CrossRef] [PubMed]
- Bowater, R.P.; Bohálová, N.; Brázda, V. Interaction of Proteins with Inverted Repeats and Cruciform Structures in Nucleic Acids. Int. J. Mol. Sci. 2022, 23, 6171. [Google Scholar] [CrossRef] [PubMed]
- Porubiaková, O.; Havlík, J.; Indu; Šedý, M.; Přepechalová, V.; Bartas, M.; Bidula, S.; Šťastný, J.; Fojta, M.; Brázda, V. Variability of Inverted Repeats in All Available Genomes of Bacteria. Microbiol. Spectr. 2023, 11, e0164823. [Google Scholar] [CrossRef] [PubMed]
- Qian, Z.; Adhya, S. DNA repeat sequences: Diversity and versatility of functions. Curr. Genet. 2017, 63, 411–416. [Google Scholar] [CrossRef] [PubMed]
- Mahfooz, S.; Shankar, G.; Narayan, J.; Singh, P.; Akhter, Y. Simple sequence repeat insertion induced stability and potential ‘gain of function’ in the proteins of extremophilic bacteria. Extrem. Life Under Extrem. Cond. 2022, 26, 17. [Google Scholar] [CrossRef] [PubMed]
- Malhotra, N.; Seshasayee, A.S.N. Replication-Dependent Organization Constrains Positioning of Long DNA Repeats in Bacterial Genomes. Genome Biol. Evol. 2022, 14, evac102. [Google Scholar] [CrossRef]
- Fozo, E.M.; Kawano, M.; Fontaine, F.; Kaya, Y.; Mendieta, K.S.; Jones, K.L.; Ocampo, A.; Rudd, K.E.; Storz, G. Repression of small toxic protein synthesis by the Sib and OhsC small RNAs. Mol. Microbiol. 2008, 70, 1076–1093. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.; Rudd, K.E.; Deutscher, M.P. A role for REP sequences in regulating translation. Mol. Cell 2015, 58, 431–439. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Fozo, E.M. sRNA antitoxins: More than one way to repress a toxin. Toxins 2014, 6, 2310–2335. [Google Scholar] [CrossRef] [PubMed]
- Achaz, G.; Coissac, E.; Netter, P.; Rocha, E.P. Associations between inverted repeats and the structural evolution of bacterial genomes. Genetics 2003, 164, 1279–1289. [Google Scholar] [CrossRef]
- Eisen, J.A.; Heidelberg, J.F.; White, O.; Salzberg, S.L. Evidence for symmetric chromosomal inversions around the replication origin in bacteria. Genome Biol. 2000, 1, Research0011. [Google Scholar] [CrossRef]
- Hughes, D. Evaluating genome dynamics: The constraints on rearrangements within bacterial genomes. Genome Biol. 2000, 1, Reviews0006. [Google Scholar] [CrossRef] [PubMed]
- Tillier, E.R.; Collins, R.A. Genome rearrangement by replication-directed translocation. Nat. Genet. 2000, 26, 195–197. [Google Scholar] [CrossRef]
- Suyama, M.; Bork, P. Evolution of prokaryotic gene order: Genome rearrangements in closely related species. Trends Genet. TIG 2001, 17, 10–13. [Google Scholar] [CrossRef]
- Choi, S.; Ohta, S.; Ohtsubo, E. A novel IS element, IS621, of the IS110/IS492 family transposes to a specific site in repetitive extragenic palindromic sequences in Escherichia coli. J. Bacteriol. 2003, 185, 4891–4900. [Google Scholar] [CrossRef]
- Partridge, S.R.; Hall, R.M. The IS1111 family members IS4321 and IS5075 have subterminal inverted repeats and target the terminal inverted repeats of Tn21 family transposons. J. Bacteriol. 2003, 185, 6371–6384. [Google Scholar] [CrossRef] [PubMed]
- Tobes, R.; Pareja, E. Bacterial repetitive extragenic palindromic sequences are DNA targets for Insertion Sequence elements. BMC Genom. 2006, 7, 62. [Google Scholar] [CrossRef]
- van Belkum, A. Short sequence repeats in microbial pathogenesis and evolution. Cell. Mol. Life Sci. CMLS 1999, 56, 729–734. [Google Scholar] [CrossRef] [PubMed]
- Dame, R.T.; Rashid, F.M.; Grainger, D.C. Chromosome organization in bacteria: Mechanistic insights into genome structure and function. Nat. Rev. Genet. 2020, 21, 227–242. [Google Scholar] [CrossRef] [PubMed]
- Lioy, V.S.; Junier, I.; Boccard, F. Multiscale Dynamic Structuring of Bacterial Chromosomes. Annu. Rev. Microbiol. 2021, 75, 541–561. [Google Scholar] [CrossRef]
- Rudd, K.E. Novel intergenic repeats of Escherichia coli K-12. Res. Microbiol. 1999, 150, 653–664. [Google Scholar] [CrossRef]
- Edgar, R. Taxonomy annotation and guide tree errors in 16S rRNA databases. PeerJ 2018, 6, e5030. [Google Scholar] [CrossRef]
- Alzohairy, A.M. BioEdit: An important software for molecular biology. GERF Bull Biosci 2011, 2, 60–61. [Google Scholar]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Loman, N.J.; Pallen, M.J. Twenty years of bacterial genome sequencing. Nat. Rev. Microbiol. 2015, 13, 787–794. [Google Scholar] [CrossRef]
- Dubey, S.; Majumder, P.; Penmatsa, A.; Sardesai, A.A. Topological analyses of the L-lysine exporter LysO reveal a critical role for a conserved pair of intramembrane solvent-exposed acidic residues. J. Biol. Chem. 2021, 297, 101168. [Google Scholar] [CrossRef] [PubMed]
- Pathania, A.; Sardesai, A.A. Distinct Paths for Basic Amino Acid Export in Escherichia coli: YbjE (LysO) Mediates Export of L-Lysine. J. Bacteriol. 2015, 197, 2036–2047. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.; Zhao, Y.; Zhao, W.; Xie, H.; Chen, Y.; Tong, Q.; Yang, J. Dynamics properties of membrane proteins in native cell membranes revealed by solid-state NMR spectroscopy. Biochim. Biophys. Acta Biomembr. 2022, 1864, 183791. [Google Scholar] [CrossRef]
- Bracher, S.; Schmidt, C.C.; Dittmer, S.I.; Jung, H. Core Transmembrane Domain 6 Plays a Pivotal Role in the Transport Cycle of the Sodium/Proline Symporter PutP. J. Biol. Chem. 2016, 291, 26208–26215. [Google Scholar] [CrossRef] [PubMed]
- Grosse, C.; Scherer, J.; Koch, D.; Otto, M.; Taudte, N.; Grass, G. A new ferrous iron-uptake transporter, EfeU (YcdN), from Escherichia coli. Mol. Microbiol. 2006, 62, 120–131. [Google Scholar] [CrossRef]
- Sturm, A.; Schierhorn, A.; Lindenstrauss, U.; Lilie, H.; Brüser, T. YcdB from Escherichia coli reveals a novel class of Tat-dependently translocated hemoproteins. J. Biol. Chem. 2006, 281, 13972–13978. [Google Scholar] [CrossRef]
- Sulavik, M.C.; Houseweart, C.; Cramer, C.; Jiwani, N.; Murgolo, N.; Greene, J.; DiDomenico, B.; Shaw, K.J.; Miller, G.H.; Hare, R.; et al. Antibiotic susceptibility profiles of Escherichia coli strains lacking multidrug efflux pump genes. Antimicrob. Agents Chemother. 2001, 45, 1126–1136. [Google Scholar] [CrossRef]
- Byrne, R.T.; Jenkins, H.T.; Peters, D.T.; Whelan, F.; Stowell, J.; Aziz, N.; Kasatsky, P.; Rodnina, M.V.; Koonin, E.V.; Konevega, A.L.; et al. Major reorientation of tRNA substrates defines specificity of dihydrouridine synthases. Proc. Natl. Acad. Sci. USA 2015, 112, 6033–6037. [Google Scholar] [CrossRef]
- Nechooshtan, G.; Elgrably-Weiss, M.; Sheaffer, A.; Westhof, E.; Altuvia, S. A pH-responsive riboregulator. Genes Dev. 2009, 23, 2650–2662. [Google Scholar] [CrossRef]
- Kim, Y.M.; Ogawa, W.; Tamai, E.; Kuroda, T.; Mizushima, T.; Tsuchiya, T. Purification, reconstitution, and characterization of Na(+)/serine symporter, SstT, of Escherichia coli. J. Biochem. 2002, 132, 71–76. [Google Scholar] [CrossRef]
- Lu, H.; Patil, P.; Van Sluys, M.A.; White, F.F.; Ryan, R.P.; Dow, J.M.; Rabinowicz, P.; Salzberg, S.L.; Leach, J.E.; Sonti, R.; et al. Acquisition and evolution of plant pathogenesis-associated gene clusters and candidate determinants of tissue-specificity in xanthomonas. PLoS ONE 2008, 3, e3828. [Google Scholar] [CrossRef] [PubMed]
- Meydan, S.; Klepacki, D.; Karthikeyan, S.; Margus, T.; Thomas, P.; Jones, J.E.; Khan, Y.; Briggs, J.; Dinman, J.D.; Vázquez-Laslop, N.; et al. Programmed Ribosomal Frameshifting Generates a Copper Transporter and a Copper Chaperone from the Same Gene. Mol. Cell 2017, 65, 207–219. [Google Scholar] [CrossRef]
- Fu, Y.; Zepeda-Gurrola, R.C.; Aguilar-Gutiérrez, G.R.; Lara-Ramírez, E.E.; De Luna-Santillana, E.J.; Rodríguez-Luna, I.C.; Sánchez-Varela, A.; Carreño-López, R.; Moreno-Medina, V.R.; Rodríguez-Pérez, M.A.; et al. The detection of inherent homologous recombination between repeat sequences in H. pylori 26695 by the PCR-based method. Curr. Microbiol. 2014, 68, 211–219. [Google Scholar] [CrossRef]
- Shikov, A.E.; Savina, I.A.; Nizhnikov, A.A.; Antonets, K.S. Recombination in Bacterial Genomes: Evolutionary Trends. Toxins 2023, 15, 568. [Google Scholar] [CrossRef] [PubMed]
- Fricke, W.F.; Wright, M.S.; Lindell, A.H.; Harkins, D.M.; Baker-Austin, C.; Ravel, J.; Stepanauskas, R. Insights into the environmental resistance gene pool from the genome sequence of the multidrug-resistant environmental isolate Escherichia coli SMS-3-5. J. Bacteriol. 2008, 190, 6779–6794. [Google Scholar] [CrossRef] [PubMed]
- Touchon, M.; Hoede, C.; Tenaillon, O.; Barbe, V.; Baeriswyl, S.; Bidet, P.; Bingen, E.; Bonacorsi, S.; Bouchier, C.; Bouvet, O.; et al. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 2009, 5, e1000344. [Google Scholar] [CrossRef]
- Treangen, T.J.; Abraham, A.L.; Touchon, M.; Rocha, E.P. Genesis, effects and fates of repeats in prokaryotic genomes. FEMS Microbiol. Rev. 2009, 33, 539–571. [Google Scholar] [CrossRef]
- MacAlasdair, N.; Pesonen, M.; Brynildsrud, O.; Eldholm, V.; Kristiansen, P.A.; Corander, J.; Caugant, D.A.; Bentley, S.D. The effect of recombination on the evolution of a population of Neisseria meningitidis. Genome Res. 2021, 31, 1258–1268. [Google Scholar] [CrossRef]
- Raghavan, R.; Groisman, E.A.; Ochman, H. Genome-wide detection of novel regulatory RNAs in E. coli. Genome Res. 2011, 21, 1487–1497. [Google Scholar] [CrossRef]
- Qian, Z.; Macvanin, M.; Dimitriadis, E.K.; He, X.; Zhurkin, V.; Adhya, S. A New Noncoding RNA Arranges Bacterial Chromosome Organization. mBio 2015, 6, e00998-15. [Google Scholar] [CrossRef]
- Cho, B.K.; Zengler, K.; Qiu, Y.; Park, Y.S.; Knight, E.M.; Barrett, C.L.; Gao, Y.; Palsson, B. The transcription unit architecture of the Escherichia coli genome. Nat. Biotechnol. 2009, 27, 1043–1049. [Google Scholar] [CrossRef] [PubMed]
- Shaskolskiy, B.; Kravtsov, D.; Kandinov, I.; Dementieva, E.; Gryadunov, D. Genomic Diversity and Chromosomal Rearrangements in Neisseria gonorrhoeae and Neisseria meningitidis. Int. J. Mol. Sci. 2022, 23, 15644. [Google Scholar] [CrossRef] [PubMed]
- González-Torres, P.; Rodríguez-Mateos, F.; Antón, J.; Gabaldón, T. Impact of Homologous Recombination on the Evolution of Prokaryotic Core Genomes. mBio 2019, 10, e02494-18. [Google Scholar] [CrossRef] [PubMed]
- Raeside, C.; Gaffé, J.; Deatherage, D.E.; Tenaillon, O.; Briska, A.M.; Ptashkin, R.N.; Cruveiller, S.; Médigue, C.; Lenski, R.E.; Barrick, J.E.; et al. Large chromosomal rearrangements during a long-term evolution experiment with Escherichia coli. mBio 2014, 5, e01377-14. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Liu, X.; Ning, N.; Li, T.; Wang, H. Diversity, Distribution, and Chromosomal Rearrangements of TRIP1 Repeat Sequences in Escherichia coli. Genes 2024, 15, 236. https://doi.org/10.3390/genes15020236
Li Z, Liu X, Ning N, Li T, Wang H. Diversity, Distribution, and Chromosomal Rearrangements of TRIP1 Repeat Sequences in Escherichia coli. Genes. 2024; 15(2):236. https://doi.org/10.3390/genes15020236
Chicago/Turabian StyleLi, Zhan, Xiong Liu, Nianzhi Ning, Tao Li, and Hui Wang. 2024. "Diversity, Distribution, and Chromosomal Rearrangements of TRIP1 Repeat Sequences in Escherichia coli" Genes 15, no. 2: 236. https://doi.org/10.3390/genes15020236
APA StyleLi, Z., Liu, X., Ning, N., Li, T., & Wang, H. (2024). Diversity, Distribution, and Chromosomal Rearrangements of TRIP1 Repeat Sequences in Escherichia coli. Genes, 15(2), 236. https://doi.org/10.3390/genes15020236




