Identification of Clubroot (Plasmodiophora brassicae) Resistance Loci in Chinese Cabbage (Brassica rapa ssp. pekinensis) with Recessive Character
Abstract
:1. Introduction
2. Materials and Methods
2.1. Population Construction and Pathogen Isolates
2.2. Pathogen Inoculation and Disease Evaluation
2.3. Analysis of Markers Linked to Identified CR Genes/Loci
2.4. Bulk Segregant Analysis Sequencing (BSA-Seq)
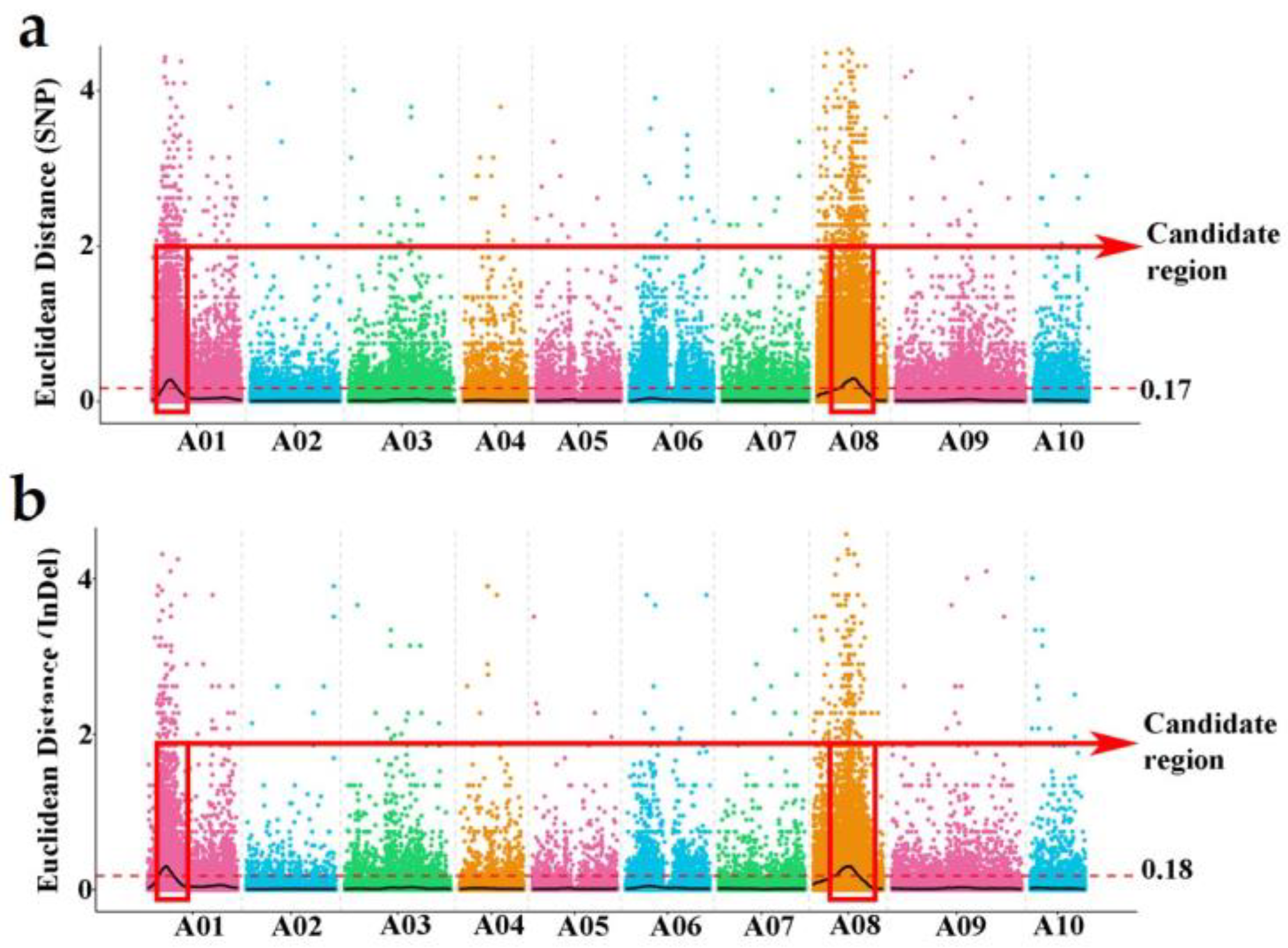
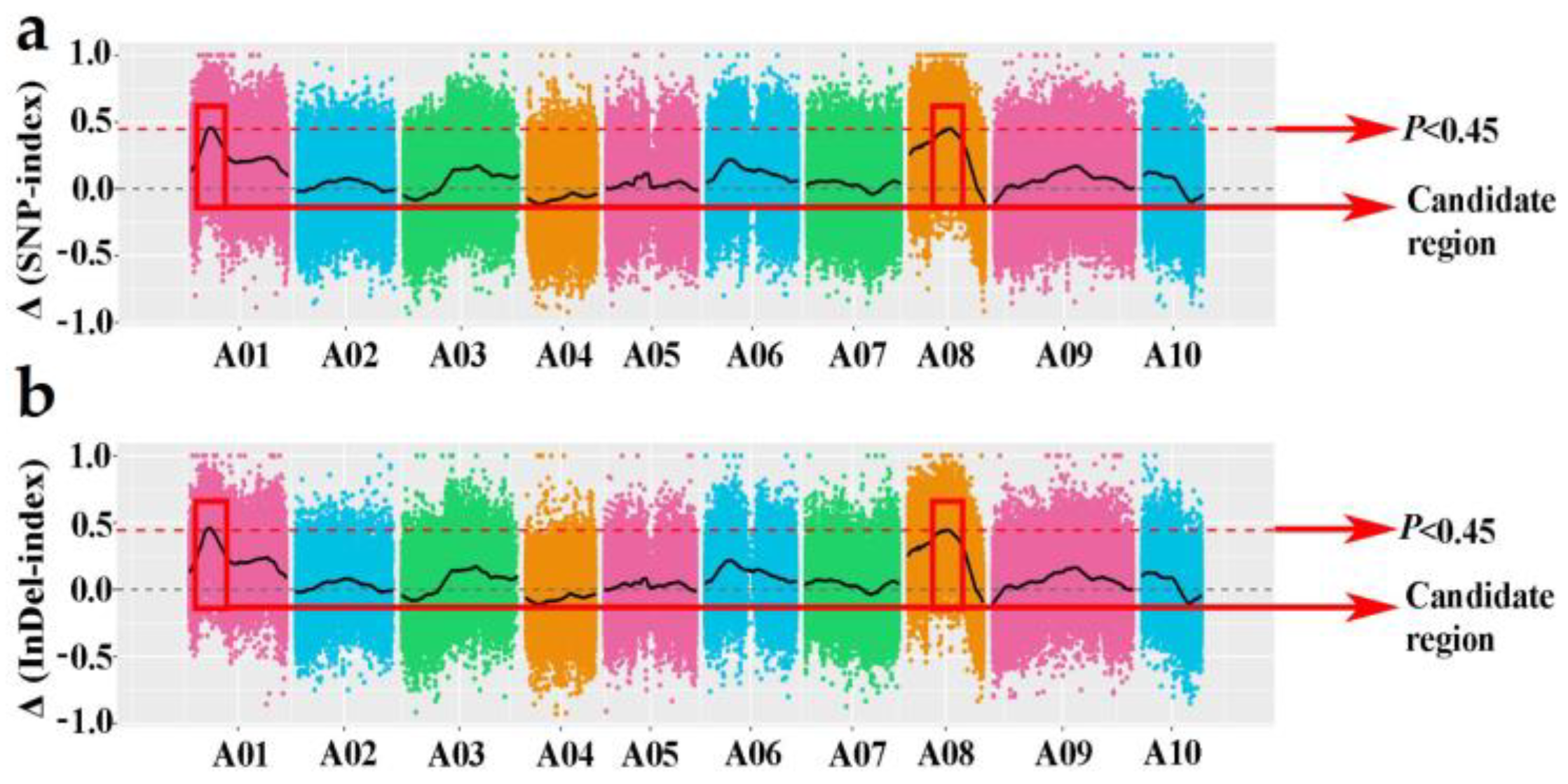
2.5. Association Mapping Analysis and Target Region Annotation
2.6. SNP/InDel Markers Development and Genotyping in Target Region
2.7. Expression Analysis
3. Results
3.1. Phenotype Evaluation of P. brassicae Infection in Different Populations
3.2. Identification of Markers Linked to CR Genes/Loci and Whole Genome Sequencing Analysis
3.3. Association Analysis
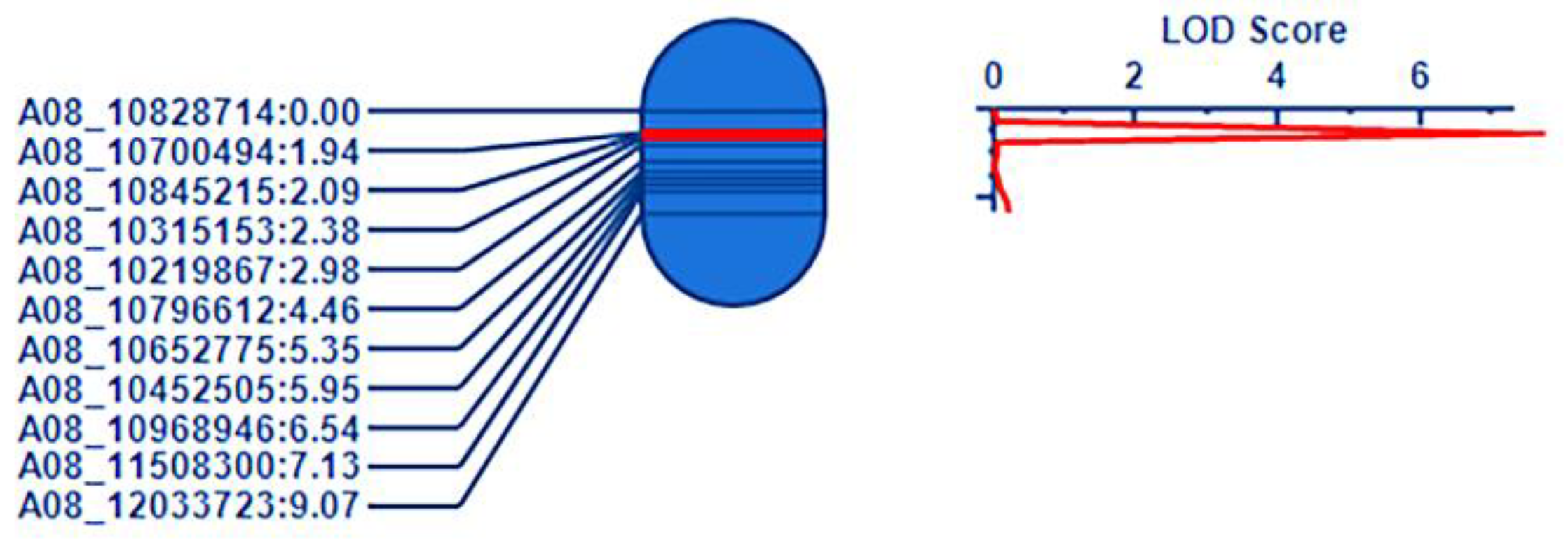
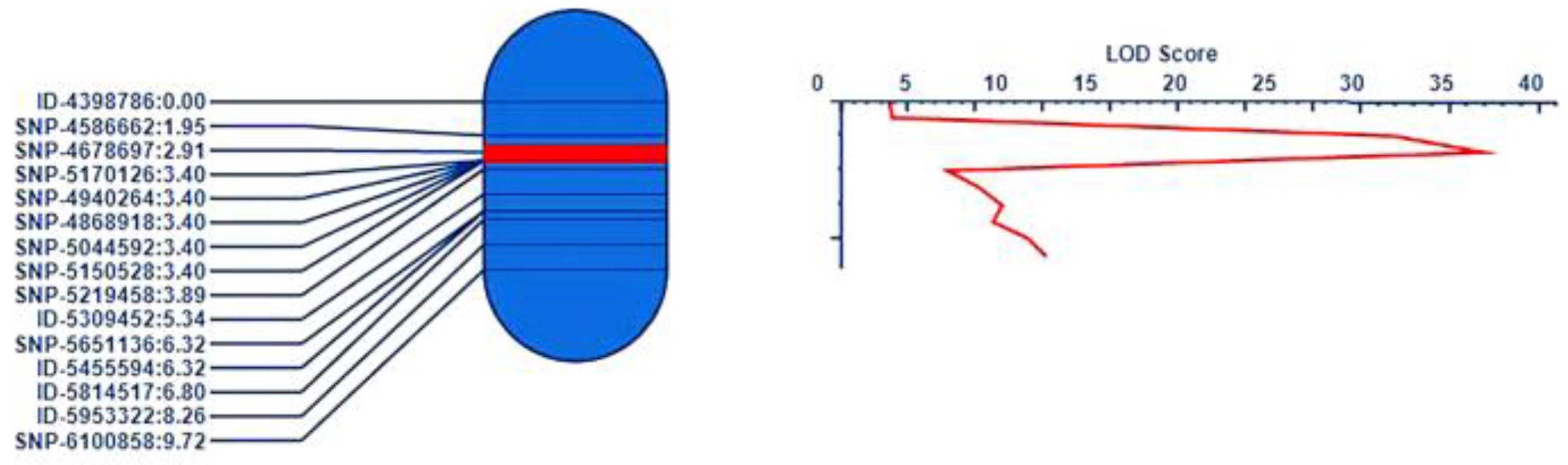
3.4. QTL Mapping of Cr4Ba8.1
3.5. Candidate Gene Prediction of Cr4Ba8.1
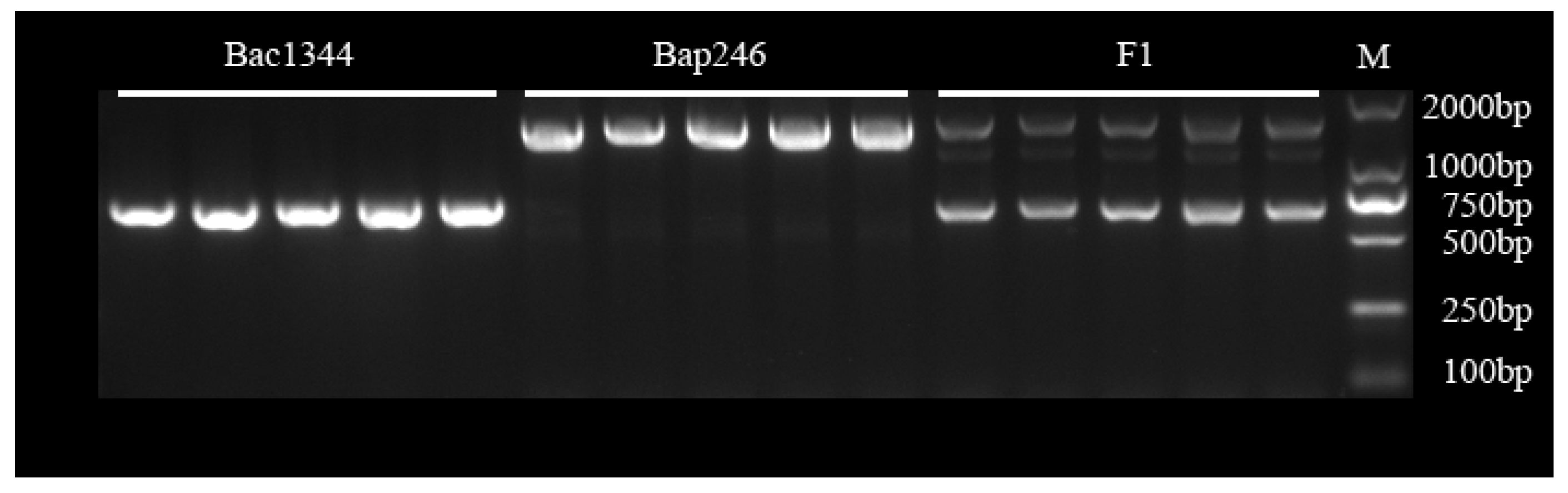
3.6. Molecular Marker of Bra020861
3.7. QTL Mapping of Cr4Ba1.1
3.8. Candidate Gene Prediction of Cr4Ba1.1
3.9. Expression Characteristic Analysis of Cr4Ba1.1 Candidate Genes
4. Discussion
4.1. Rapid Mapping Strategy of CR Loci
4.2. The CR Mechanism of Bap246
4.3. The Interaction between Cr4Ba1.1 and Cr4Ba8.1
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pang, W.; Fu, P.; Li, X.; Zhan, Z.; Yu, S.; Piao, Z. Identification and mapping of the clubroot resistance gene CRd in Chinese cabbage (Brassica rapa ssp. pekinensis). Front. Plant Sci. 2018, 9, 653. [Google Scholar] [CrossRef] [PubMed]
- Chai, A.L.; Xie, X.W.; Shi, Y.X.; Li, B.J. Research status of clubroot (Plasmodiophora brassicae) on cruciferous crops in China. Can. J. Plant Pathol. 2014, 36, 142–153. [Google Scholar] [CrossRef]
- Bhattacharya, I.; Dutta, S.; Mondal, S.; Mondal, B. Clubroot disease on Brassica crops in India. Can. J. Plant Pathol. 2014, 36, 154–160. [Google Scholar] [CrossRef]
- Donald, E.C.; Porter, I. Clubroot in Australia: The history and impact of Plasmodiophora brassicae in Brassica crops and research efforts directed towards its control. Can. J. Plant Pathol. 2014, 36, 66–84. [Google Scholar] [CrossRef]
- Rahman, H.; Peng, G.; Yu, F.; Falk, K.C.; Selvaraj, G. Genetics and breeding for clubroot resistance in Canadian spring canola (Brassica napus L.). Can. J. Plant Pathol. 2014, 36, 122–134. [Google Scholar] [CrossRef]
- Řičařová, V.; Kazda, J.; Singh, K.; Science, P.R. Clubroot caused by Plasmodiophora brassicae Wor.: A review of emerging serious disease of oilseed rape in the Czech Republic. Plant Protect. Sci. 2016, 52, 71–86. [Google Scholar] [CrossRef]
- Liu, X.; Strelkov, S.E.; Sun, R.; Hwang, S.F.; Fredua-Agyeman, R.; Li, F.; Zhang, S.; Li, G.; Zhang, S.; Zhang, H. Histopathology of the Plasmodiophora brassicae-Chinese cabbage interaction in hosts carrying different sources of resistance. Front. Plant Sci. 2021, 12, 783550. [Google Scholar] [CrossRef]
- Tso, H.H.; Galindo-González, L.; Strelkov, S.E. Current and future pathotyping platforms for Plasmodiophora brassicae in Canada. Plants 2021, 10, 1446. [Google Scholar] [CrossRef]
- Asano, K.T. Life cycle of Plasmodiophora brassicae. J. Plant Growth Regul. 2009, 28, 203–211. [Google Scholar]
- Rolfe, S.; Fleming, P.J.; Smith, R.; Scholes, D.; Malinowski, P.R.; Biology, M. Gall formation in clubroot-infected arabidopsis results from an increase in existing meristematic activities of the host but is not essential for the completion of the pathogen life cycle. Plant J. 2012, 71, 226–238. [Google Scholar]
- Walerowski, P.; Gündel, A.; Yahaya, N.; Truman, W.; Sobczak, M.; Olszak, M.; Rolfe, S.; Borisjuk, L.; Malinowski, R. Clubroot disease stimulates early steps of phloem differentiation and recruits sweet sucrose transporters within developing galls. Plant Cell 2019, 12, 3058–3073. [Google Scholar] [CrossRef]
- Dixon, G.R. The occurrence and economic impact of Plasmodiophora brassicae and clubroot disease. J. Plant Growth Regul. 2009, 28, 194–202. [Google Scholar] [CrossRef]
- Fredua-Agyeman, R.; Hwang, S.F.; Strelkov, S.E.; Zhou, Q.; Feindel, D. Assessment of resistance to ‘new’ virulent populations of Plasmodiophora brassicae reveals potential loss of clubroot resistance genes from donor parent Brassica rapa L. subsp. rapifera (ECD 04) during doubled haploid production. Plant Pathol. 2017, 67, 892–901. [Google Scholar]
- Peng, G.; Lahlali, R.; Hwang, S.F.; Pageau, D.; Hynes, R.K.; Mcdonald, M.R.; Gossen, B.D.; Strelkov, S.E. Crop rotation, cultivar resistance, and fungicides/biofungicides for managing clubroot (Plasmodiophora brassicae) on canola. Can. J. Plant Pathol. 2014, 36, 99–112. [Google Scholar] [CrossRef]
- Chen, J.; Jing, J.; Zhan, Z.; Zhang, T.; Zhang, C.; Piao, Z. Identification of novel QTLs for isolate-specific partial resistance to Plasmodiophora brassicae in Brassica rapa. PLoS ONE 2013, 8, e85307. [Google Scholar] [CrossRef] [PubMed]
- Hirai, M.; Harada, T.; Kubo, N.; Tsukada, M.; Suwabe, K.; Matsumoto, S. A novel locus for clubroot resistance in Brassica rapa and its linkage markers. Theor. Appl. Genet. 2004, 108, 639–643. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, K.; Saito, A.; Hayashida, N.; Taguchi, G.; Matsumoto, E. Mapping of isolate-specific qtls for clubroot resistance in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Theor. Appl. Genet. 2008, 117, 759–767. [Google Scholar] [CrossRef]
- Matsumoto, E.; Yasui, C.; Ohi, M.; Tsukada, M. Linkage analysis of rflp markers for clubroot resistance and pigmentation in Chinese cabbage (Brassica rapa ssp. pekinensis). (Brassica rapa ssp. pekinensis). Euphytica 1998, 104, 79–86. [Google Scholar] [CrossRef]
- Piao, Z.Y.; Deng, Y.Q.; Choi, S.R.; Park, Y.J.; Lim, Y.P. Scar and caps mapping of CRb, a gene conferring resistance to Plasmodiophora brassicae in Chinese cabbage (Brassica rapa ssp. pekinensis). Theor. Appl. Genet. 2004, 108, 1458–1465. [Google Scholar] [CrossRef]
- Zhen, H.; Peng, G.; Liu, X.; Abhinandan, D.; Falk, K.C.; Gossen, B.D.; Mcdonald, M.R.; Yu, F. Fine mapping of a clubroot resistance gene in Chinese cabbage using SNP markers identified from bulked segregant RNA sequencing. Front. Plant Sci. 2017, 8, 1448. [Google Scholar]
- Matsumoto, E.; Hayashida, N.; Sakamoto, K.; Ohi, M. Behavior of DNA markers linked to a clubroot resistance gene in segregating populations of Chinese cabbage (Brassica rapa ssp. pekinensis). J.-Stage 2005, 74, 367–373. [Google Scholar] [CrossRef]
- Diederichsen, E.; Frauen, M.; Linders, E.; Hatakeyama, K.; Hirai, M. Status and perspectives of clubroot resistance breeding in crucifer crops. J. Plant Growth Regul. 2009, 28, 265–281. [Google Scholar] [CrossRef]
- Yu, F.; Zhang, X.; Peng, G.; Falk, K.C.; Strelkov, S.E.; Gossen, B.D. Genotyping-by-sequencing reveals three QTL for clubroot resistance to six pathotypes of Plasmodiophora brassicae in Brassica rapa. Sci. Rep. 2017, 7, 4516. [Google Scholar] [CrossRef] [PubMed]
- Suwabe, K.; Tsukazaki, H.; Iketani, H.; Hatakeyama, K.; Fujimura, M.; Nunome, T.; Fukuoka, H.; Matsumoto, S.; Hirai, M. Identification of two loci for resistance to clubroot (Plasmodiophora brassicae woronin) in Brassica rapa L. Theor. Appl. Genet. 2003, 107, 997–1002. [Google Scholar] [CrossRef] [PubMed]
- Suwabe, K.; Tsukazaki, H.; Iketani, H.; Hatakeyama, K.; Kondo, M.; Fujimura, M.; Nunome, T.; Fukuoka, H.; Hirai, M.; Matsumoto, S. Simple sequence repeat-based comparative genomics between and: The genetic origin of clubroot resistance. Genetics 2006, 173, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Strelkov, S.E.; Hwang, S.F.; Manolii, V.P.; Cao, T.; Feindel, D. Emergence of new virulence phenotypes of Plasmodiophora brassicae on canola (Brassica napus) in Alberta, Canada. Eur. J. Plant Pathol. 2016, 145, 517–529. [Google Scholar] [CrossRef]
- Han, Y.; Lv, P.; Hou, S.; Li, S.; Ji, G.; Ma, X.; Du, R.; Liu, G. Combining next generation sequencing with bulked segregant analysis to fine map a stem moisture locus in sorghum (Sorghum bicolor L. Moench). PLoS ONE 2015, 10, e0127065. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Xu, W.; Song, Q.; Tan, L.; Liu, J.; Zhu, Z.; Fu, Y.; Su, Z.; Sun, C. Microarray-assisted fine-mapping of quantitative trait loci for cold tolerance in rice. Mol. Plant 2013, 6, 757–767. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Zhen, L.; Liu, Z.; Guo, Y.; Qiu, L.J. Next-generation sequencing from bulked-segregant analysis accelerates the simultaneous identification of two qualitative genes in soybean. Front. Plant Sci. 2017, 8, 919. [Google Scholar] [CrossRef]
- Trick, M.; Adamski, N.M.; Mugford, S.G.; Jiang, C.C.; Febrer, M.; Uauy, C. Combining snp discovery from next-generation sequencing data with bulked segregant analysis (BSA) to fine-map genes in polyploid wheat. BMC Plant Biol. 2012, 12, 14. [Google Scholar] [CrossRef]
- Zhu, J.; Chen, J.; Gao, F.; Xu, C.; Wu, H.; Chen, K.; Si, Z.; Yan, H.; Zhang, T. Rapid mapping and cloning of the virescent-1 gene in cotton by bulked segregant analysis-next generation sequencing and virus-induced gene silencing strategies. J. Exp. Bot. 2017, 68, 4125–4135. [Google Scholar] [CrossRef]
- Liang, T.; Chi, W.; Huang, L.; Qu, M.; Chen, S. Bulked segregant analysis coupled with whole-genome sequencing (bsa-seq) mapping identifies a novel pi21 haplotype conferring basal resistance to rice blast disease. Int. J. Mol. Sci. 2020, 21, 2162. [Google Scholar] [CrossRef]
- Zheng, X.; Chen, X.; Zhang, X.; Lin, Z.; Shang, J.; Xu, J.; Zhai, W.; Zhu, L. Isolation and identification of a gene in response to rice blast disease in rice. Plant Mol. Biol. 2004, 54, 99–109. [Google Scholar] [CrossRef]
- Du, H.; Wen, C.; Zhang, X.; Xu, X.; Yang, J.; Chen, B.; Geng, S. Identification of a Major QTL (qRRs-10.1) That Confers Resistance to Ralstonia solanacearum in Pepper (Capsicum annuum) Using SLAF-BSA and QTL Mapping. Int. J. Mol. Sci. 2019, 20, 5887. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhang, C.; Chen, H.; Yuan, M.; Nipper, R.; Prakash, C.S.; Zhuang, W.; He, G. Qtl mapping for bacterial wilt resistance in peanut (Arachis hypogaea L.). Mol. Breed. 2016, 36, 13. [Google Scholar] [CrossRef] [PubMed]
- She, H.; Qian, W.; Zhang, H.; Liu, Z.; Wang, X.; Wu, J.; Feng, C.; Correll, J.C.; Xu, Z. Fine mapping and candidate gene screening of the downy mildew resistance gene RPF1 in spinach. Theor. Appl. Genet. 2018, 131, 2529–2541. [Google Scholar] [CrossRef] [PubMed]
- Win, K.T.; Vegas, J.; Zhang, C.; Song, K.; Le, S. Qtl mapping for downy mildew resistance in cucumber via bulked segregant analysis using next-generation sequencing and conventional methods. Theor. Appl. Genet. 2017, 130, 199–211. [Google Scholar] [CrossRef] [PubMed]
- Chunthawodtiporn, J.; Hill, T.; Stoffel, K.; Deynze, A.V. Genetic analysis of resistance to multiple isolates of phytophthora capsici and linkage to horticultural traits in bell pepper. Hort. Sci. 2019, 7, 8. [Google Scholar] [CrossRef]
- Vogel, G.; Laplant, K.E.; Mazourek, M.; Gore, M.A.; Smart, C.D. A combined bsa-seq and linkage mapping approach identifies genomic regions associated with phytophthora root and crown rot resistance in squash. Theor. Appl. Genet. 2021, 134, 1015–1031. [Google Scholar] [CrossRef] [PubMed]
- Gan, C.; Deng, X.; Cui, L.; Yu, X.; Piao, Z. Construction of a high-density genetic linkage map and identification of quantitative trait loci associated with clubroot resistance in radish (Raphanus sativus L.). Mol. Breed. 2019, 39, 116. [Google Scholar] [CrossRef]
- Zhu, H.; Zhai, W.; Li, X.; Zhu, Y. Two QTLs controlling Clubroot resistance identified from Bulked Segregant Sequencing in Pakchoi (Brassica campestris ssp. chinensis Makino). Sci. Rep. 2019, 9, 9228. [Google Scholar] [CrossRef] [PubMed]
- Abe, A.; Kosugi, S.; Yoshida, K.; Natsume, S.; Takagi, H.; Kanzaki, H.; Matsumura, H.; Yoshida, K.; Mitsuoka, C.; Tamiru, M.; et al. Genome sequencing reveals agronomically important loci in rice using mutmap. Nat. Biotechnol. 2012, 30, 174–178. [Google Scholar] [CrossRef] [PubMed]
- Magwene, P.M.; Willis, J.H.; Kelly, J.K. The statistics of bulk segregant analysis using next generation sequencing. PLoS Comput. Biol. 2011, 7, e1002255. [Google Scholar] [CrossRef] [PubMed]
- Hill, J.T.; Demarest, B.L.; Bisgrove, B.W.; Gorsi, B.; Su, Y.C.; Yost, H.J. Mmappr: Mutation mapping analysis pipeline for pooled RNA-seq. Genome Res. 2013, 23, 687–697. [Google Scholar] [CrossRef] [PubMed]
- Ertiro, B.T.; Ogugo, V.; Worku, M.; Das, B.; Olsen, M.; Labuschagne, M.; Semagn, K. Comparison of kompetitive allele specific PCR (KASP) and genotyping by sequencing (GBS) for quality control analysis in maize. BMC Genom. 2015, 16, 908. [Google Scholar] [CrossRef] [PubMed]
- Bourras, S.; Mcnally, K.E.; Ben-David, R.; Parlange, F.; Keller, B. Multiple avirulence loci and allele-specific effector recognition control the pm3 race-specific resistance of wheat to powdery mildew. Plant Cell 2015, 27, 2991–3012. [Google Scholar] [PubMed]
- Semagn, K.; Babu, R.; Hearne, S.; Olsen, M. Single nucleotide polymorphism genotyping using kompetitive allele specific PCR (KASP): Overview of the technology and its application in crop improvement. Mol. Breed. 2014, 33, 1–14. [Google Scholar] [CrossRef]
- Williams, P. A system for the determination of races of Plasmodiophora brassicae that infect cabbage and rutabaga. Phytopathology 1966, 6, 624–626. [Google Scholar]
- Johnston, T.D. Clubroot in Brassica a standard inoculation technique and the specification of races. Plant Pathol. 1968, 17, 184–187. [Google Scholar] [CrossRef]
- Yang, X.X.; Huang, X.Q.; Wen-Xian, W.U.; Xiang, Y.J.; Liu, Y. Effects of different rotation patterns on the occurrence of clubroot disease and diversity of rhizosphere microbes. J. Integr. Agric. 2020, 19, 2265–2273. [Google Scholar] [CrossRef]
- Ueno, H.; Matsumoto, E.; Aruga, D.; Kitagawa, S.; Matsumura, H.; Hayashida, N. Molecular characterization of the cra gene conferring clubroot resistance in Brassica rapa. Plant Mol. Biol. 2012, 80, 621–629. [Google Scholar] [CrossRef]
- Zhang, T.; Zhao, Z.; Zhang, C.; Pang, W.; Choi, S.R.; Lim, Y.P.; Piao, Z. Fine genetic and physical mapping of the CRb gene conferring resistance to clubroot disease in Brassica rapa. Mol. Breed. 2014, 34, 1173–1183. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, X.; Xu, H.; Song, B.; Fan, X.; Snowdon, R. Introgression of clubroot resistance into an elite pak choi inbred line through marker-assisted introgression breeding. Plant Breed. 2016, 135, 471–475. [Google Scholar] [CrossRef]
- Saito, M.; Kubo, N.; Matsumoto, S.; Suwabe, K.; Tsukada, M.; Hirai, M. Fine mapping of the clubroot resistance gene, Crr3, in Brassica rapa. Theor. Appl. Genet. 2006, 114, 81–91. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map (sam) format and samtools. Bioinformatics 2009, 25, 2078. [Google Scholar] [CrossRef]
- Mckenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K. The genome analysis toolkit: A mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. Annovar: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 28, e164. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Zhou, L.; Li, Q.; Wei, D.; Ren, X.; Song, H.; Mei, J.; Si, J.; Qian, W. Identification of Quantitative Trait Loci for Clubroot Resistance in Brassica oleracea with the Use of Brassica SNP Microarray. Front. Plant Sci. 2018, 9, 822. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2011, 25, 402. [Google Scholar] [CrossRef] [PubMed]
- Jahn, L.; Mucha, S.; Bergmann, S.; Horn, C.; Staswick, P.; Steffens, B.; Siemens, J.; Ludwig-Müller, J. The clubroot pathogen (Plasmodiophora brassicae) influences auxin signaling to regulate auxin homeostasis in Arabidopsis. Plants 2013, 2, 726–749. [Google Scholar] [CrossRef] [PubMed]
- Hatakeyama, K.; Suwabe, K.; Tomita, R.N.; Kato, T.; Nunome, T.; Fukuoka, H.; Matsumoto, S. Identification and characterization of Crr1a, a gene for resistance to clubroot disease (Plasmodiophora brassicae Woronin) in Brassica rapa L. PLoS ONE 2013, 8, e54745. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Xie, J.; Wang, H.; Zhong, X.; Li, H.; Yu, J.; Kang, J. Identification and expression profiling analysis of NBS-LRR genes involved in Fusarium oxysporum f.Sp. Conglutinans resistance in cabbage. 3 Biotech 2019, 9, 202. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Jia, Z.H.; Zhang, J.Y.; Liu, M.; Guo, Z.R.; Wang, G. Identification and analysis of NBS-LRR Genes in Actinidia chinensis genome. Plants 2020, 9, 1350. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Galindo-González, L.; Manolii, V.; Hwang, S.F.; Strelkov, S.E. Comparative transcriptome analysis of rutabaga (Brassica napus) cultivars indicates activation of salicylic acid and ethylene-mediated defenses in response to Plasmodiophora brassicae. Int. J. Mol. Sci. 2020, 21, 8381. [Google Scholar] [CrossRef]
- Dong, J.; Chen, C.; Chen, Z. Expression profiles of the arabidopsis WRKY gene superfamily during plant defense response. Plant Mol. Biol. 2003, 51, 21–37. [Google Scholar] [CrossRef]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Cao, Y.; Yang, Y.; Zhang, H.; Li, D.; Zheng, Z.; Song, F. Overexpression of a rice defense-related F-box protein gene OsDRF1 in tobacco improves disease resistance through potentiation of defense gene expression. Physiol. Plant 2008, 134, 440–452. [Google Scholar] [CrossRef]
- Van den Burg, H.A.; Tsitsigiannis, D.I.; Rowland, O.; Lo, J.; Rallapalli, G.; Maclean, D.; Takken, F.L.; Jones, J.D. The F-box protein ACRE189/ACIF1 regulates cell death and defense responses activated during pathogen recognition in tobacco and tomato. Plant Cell. 2008, 20, 697–719. [Google Scholar] [CrossRef]
- Fu, J.; Wang, S. Insights into auxin signaling in plant-pathogen interactions. Front. Plant Sci. 2011, 2, 74. [Google Scholar] [CrossRef]
- Ludwig-Müller, J. Auxin homeostasis, signaling, and interaction with other growth hormones during the clubroot disease of Brassicaceae. Plant Signal. Behav. 2014, 9, e28593. [Google Scholar] [CrossRef]
- Li, N.; Han, X.; Feng, D.; Yuan, D.; Huang, L.J. Signaling crosstalk between salicylic acid and ethylene/jasmonate in plant defense: Do we understand what they are whispering? Int. J. Mol. Sci. 2019, 20, 671. [Google Scholar] [CrossRef] [PubMed]


| Materials | F1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| P1 | P2 | Total | Disease Grades | Disease Index (DI) | Resistance Level | |||||
| 0 | 1 | 3 | 5 | 7 | 9 | |||||
| Bap246 | Taqing | 50 | 0 | 0 | 0 | 3 | 5 | 42 | 95.11 | HS |
| Quanmei | 50 | 0 | 0 | 0 | 0 | 10 | 40 | 95.56 | HS | |
| Heatwave | 53 | 0 | 0 | 15 | 23 | 13 | 2 | 56.39 | S | |
| Bak10 | 48 | 2 | 0 | 0 | 0 | 4 | 42 | 93.98 | HS | |
| Bak11 | 50 | 0 | 0 | 0 | 0 | 4 | 46 | 98.22 | HS | |
| Bak12 | 50 | 0 | 0 | 0 | 0 | 0 | 50 | 100.00 | HS | |
| Bac1343 | 50 | 0 | 0 | 22 | 11 | 15 | 2 | 54.22 | S | |
| Bac1344 | 30 | 0 | 0 | 2 | 0 | 13 | 15 | 90.48 | HS | |
| Bae090 | 43 | 0 | 0 | 0 | 0 | 11 | 32 | 85.93 | HS | |
| Bae091 | 50 | 0 | 0 | 0 | 1 | 20 | 29 | 90.22 | HS | |
| Bae092 | 52 | 0 | 0 | 0 | 2 | 4 | 46 | 96.58 | HS | |
| Generation | Total | Disease Grades | DI | |||||
|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 3 | 5 | 7 | 9 | |||
| Bap246 (P1) | 48 | 39 | 9 | 0 | 0 | 0 | 0 | 0.02 |
| Bac1344 (P2) | 33 | 0 | 0 | 0 | 3 | 7 | 23 | 91.25 |
| F1 | 35 | 0 | 0 | 0 | 5 | 5 | 25 | 90.48 |
| F1′ | 37 | 0 | 0 | 0 | 7 | 7 | 23 | 87.39 |
| BC1P1 | 52 | 14 | 15 | 1 | 3 | 7 | 11 | 38.68 |
| BC1P2 | 36 | 0 | 0 | 0 | 2 | 2 | 32 | 96.30 |
| F2 | 849 | 130 | 76 | 85 | 136 | 203 | 219 | 57.62 |
| Generation | Total | Disease Grades | DI | |||||
|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 3 | 5 | 7 | 9 | |||
| Bap246 | 48 | 39 | 9 | 0 | 0 | 0 | 0 | 0.02 |
| Bae090 | 35 | 0 | 0 | 0 | 4 | 7 | 24 | 90.48 |
| F1 | 30 | 0 | 0 | 0 | 2 | 13 | 15 | 87.41 |
| F1′ | 34 | 0 | 0 | 0 | 0 | 13 | 21 | 91.50 |
| BC1P1 | 44 | 15 | 5 | 0 | 2 | 12 | 10 | 47.73 |
| BC1P2 | 36 | 0 | 0 | 0 | 2 | 2 | 32 | 96.30 |
| F2 | 664 | 87 | 68 | 66 | 85 | 135 | 223 | 60.96 |
| Sample Name | Clean Reads | Clean Bases (G) | CleanQ30 (%) | Clean GC Content (%) | Map Reads Rate (%) | Coverage Rate (%) | Average Depth |
|---|---|---|---|---|---|---|---|
| Bap246 | 34,184,318 | 10.23 | 93.21 | 36.59 | 97.35 | 93.53 | 21 |
| Bac1344 | 36,363,047 | 10.89 | 93.29 | 37.47 | 96.44 | 91.29 | 22 |
| R-pool | 51,366,194 | 15.38 | 93.66 | 37.29 | 96.87 | 96.03 | 29 |
| S-pool | 50,591,219 | 15.14 | 92.86 | 37.53 | 97.14 | 95.95 | 30 |
| Gene ID | Homologous Gene | Annotation | A. thaliana Highest Expression |
|---|---|---|---|
| Bra020860 | AT4G22060 | F-box domain | Silique |
| Bra020861 | AT3G25510 | TIR domain; leucine-rich repeats (2/6 copies); NB-ARC domain | Root |
| Bra020876 | AT4G20940 | Leucine-rich repeats (2 copies) | Flower |
| Gene ID | SNP | InDel | ||||
|---|---|---|---|---|---|---|
| Synonymous Coding | Non-Synonymous Coding | Frameshift Mutation | Codon Deletion | Codon Insertion | Codon Change + Deletion | |
| Bra020860 | 2 | 5 | — | — | — | — |
| Bra020861 | 22 | 49 | 14 | 2 | 2 | 1 |
| Bra020876 | 40 | 12 | — | — | — | — |
| Gene ID | SNP | InDel | ||||
|---|---|---|---|---|---|---|
| Synonymous Coding | Non-Synonymous Coding | Frameshift Mutation | Codon Deletion | Codon Change + Deletion | Codon Change + Insertion | |
| Bra013275 | 3 | 4 | — | — | — | — |
| Bra013281 | 5 | 12 | 3 | — | — | — |
| Bra013289 | 1 | 3 | — | 1 | — | 1 |
| Bra013299 | 12 | 6 | — | — | — | — |
| Bra013310 | — | 1 | — | — | — | — |
| Bra013336 | 7 | 3 | — | — | — | — |
| Bra013339 | 6 | 18 | — | 3 | — | — |
| Bra013341 | 10 | 1 | 1 | — | — | — |
| Bra013345 | 6 | 1 | — | — | — | — |
| Bra013347 | — | — | — | — | — | — |
| Bra013348 | — | — | — | — | — | — |
| Bra013357 | 5 | 6 | 1 | — | 1 | — |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Liu, X.; Zhou, J.; Strelkov, S.E.; Fredua-Agyeman, R.; Zhang, S.; Li, F.; Li, G.; Wu, J.; Sun, R.; et al. Identification of Clubroot (Plasmodiophora brassicae) Resistance Loci in Chinese Cabbage (Brassica rapa ssp. pekinensis) with Recessive Character. Genes 2024, 15, 274. https://doi.org/10.3390/genes15030274
Zhang H, Liu X, Zhou J, Strelkov SE, Fredua-Agyeman R, Zhang S, Li F, Li G, Wu J, Sun R, et al. Identification of Clubroot (Plasmodiophora brassicae) Resistance Loci in Chinese Cabbage (Brassica rapa ssp. pekinensis) with Recessive Character. Genes. 2024; 15(3):274. https://doi.org/10.3390/genes15030274
Chicago/Turabian StyleZhang, Hui, Xitong Liu, Jinyan Zhou, Stephen E. Strelkov, Rudolph Fredua-Agyeman, Shifan Zhang, Fei Li, Guoliang Li, Jian Wu, Rifei Sun, and et al. 2024. "Identification of Clubroot (Plasmodiophora brassicae) Resistance Loci in Chinese Cabbage (Brassica rapa ssp. pekinensis) with Recessive Character" Genes 15, no. 3: 274. https://doi.org/10.3390/genes15030274






