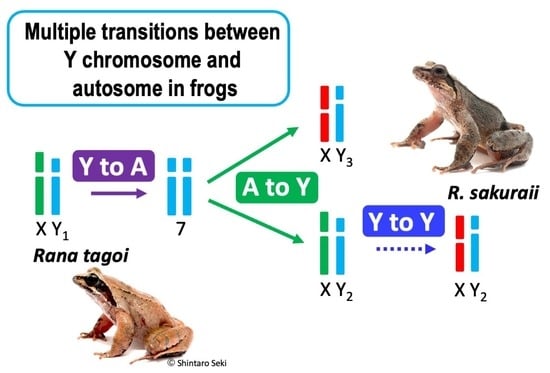
Multiple Transitions between Y Chromosome and Autosome in Tago’s Brown Frog Species Complex
Abstract
:1. Introduction
2. Materials and Methods
2.1. Frogs
2.2. Mitochondrial Sequence Analysis
2.3. Chromosome Preparation and Banding Techniques
2.4. Genotyping by Sequencing Using DArTseq
2.5. Identification of Sex-Linked Markers
2.6. Alignment of Sex-Linked Genes to Rana temporaria Reference Genome
2.7. Principal Component, Structure, and Gene Tree Analysis
3. Results
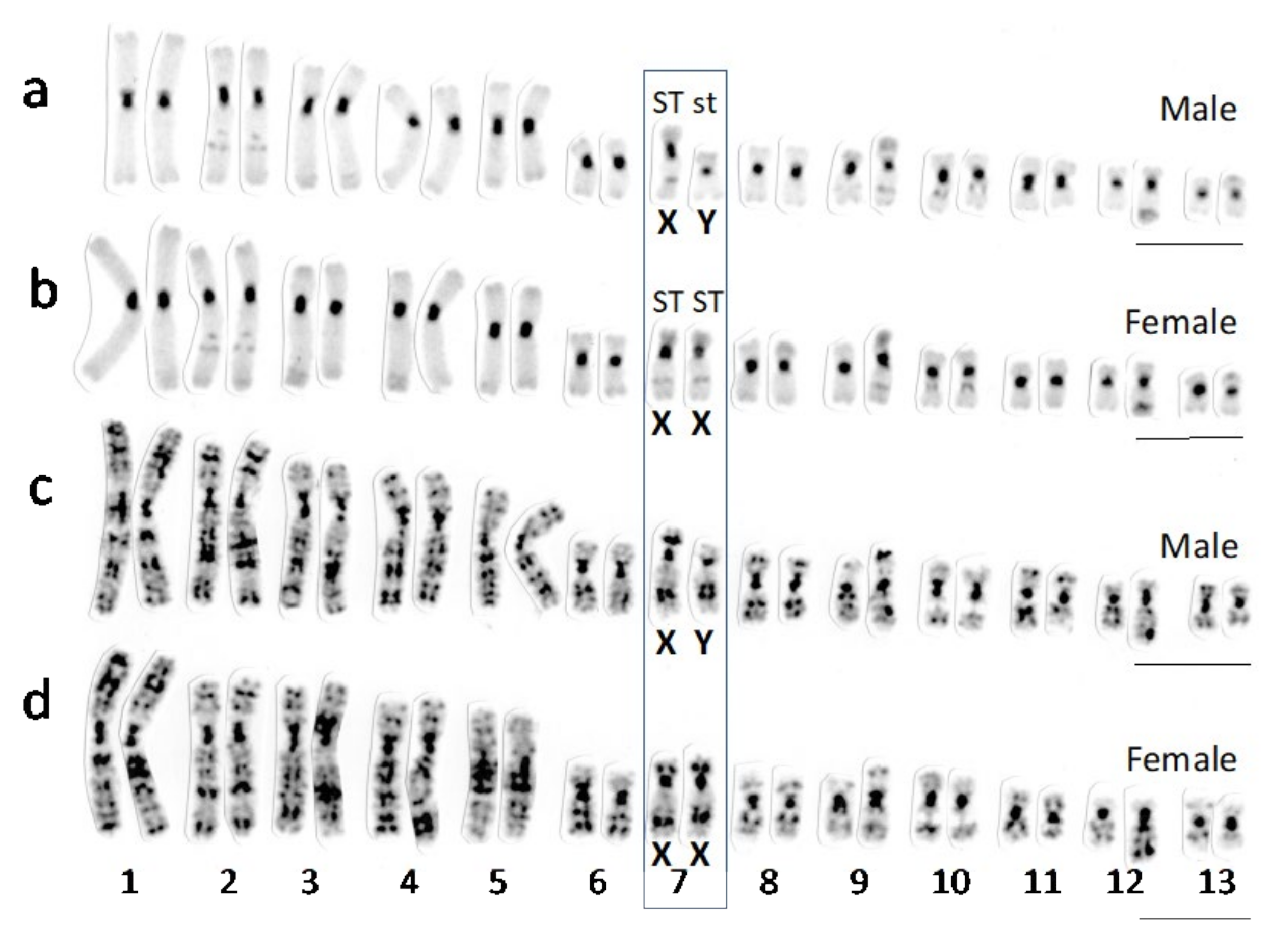
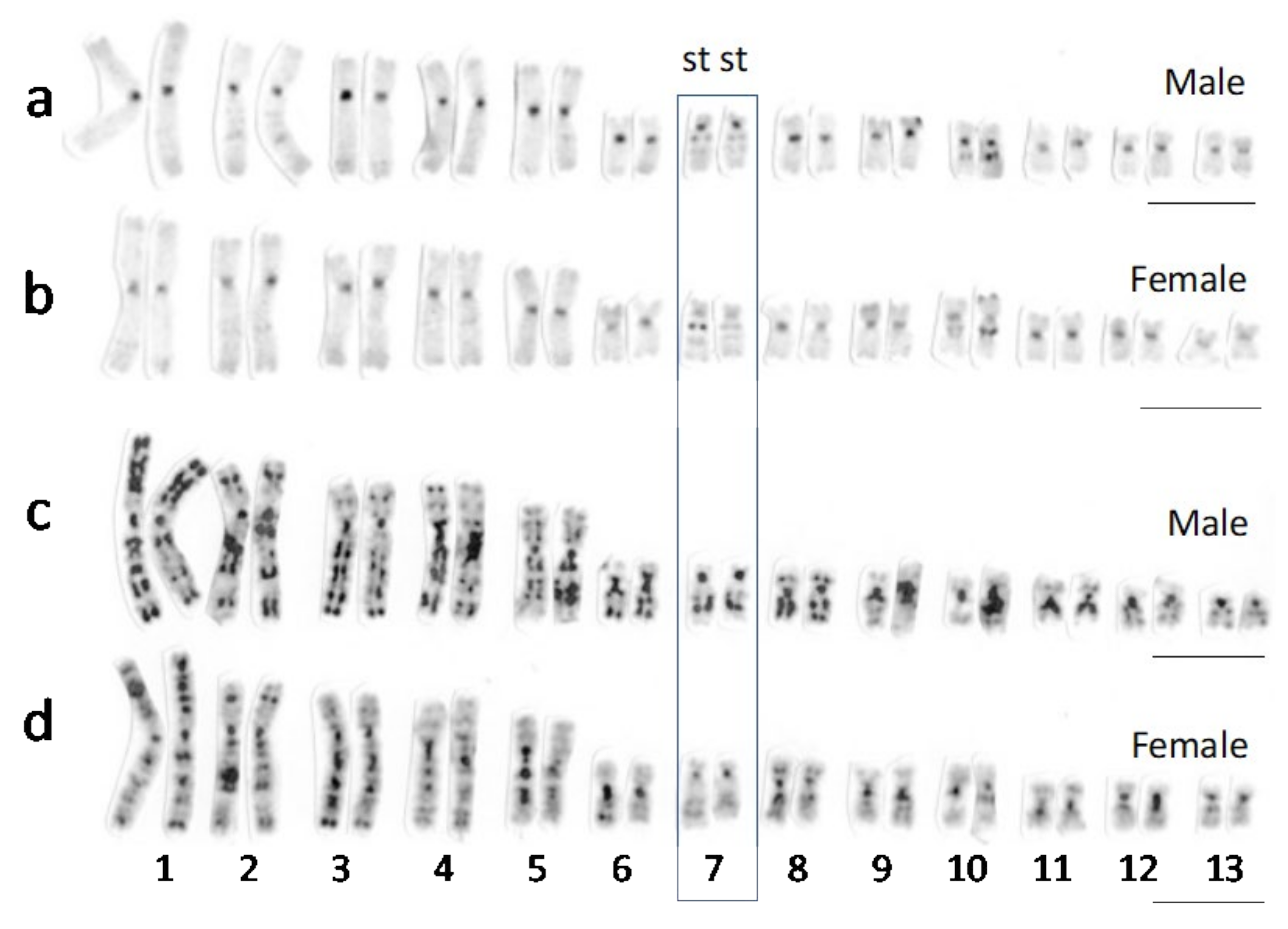
3.1. Sex Chromosomes of R. tagoi from Chiba (East Japan) and Akita (North Japan) Populations
3.2. Identification of Sex Chromosomes Based on the Sex-Linked SNP Markers
3.3. The Origins of Y Chromosomes and Autosome 7
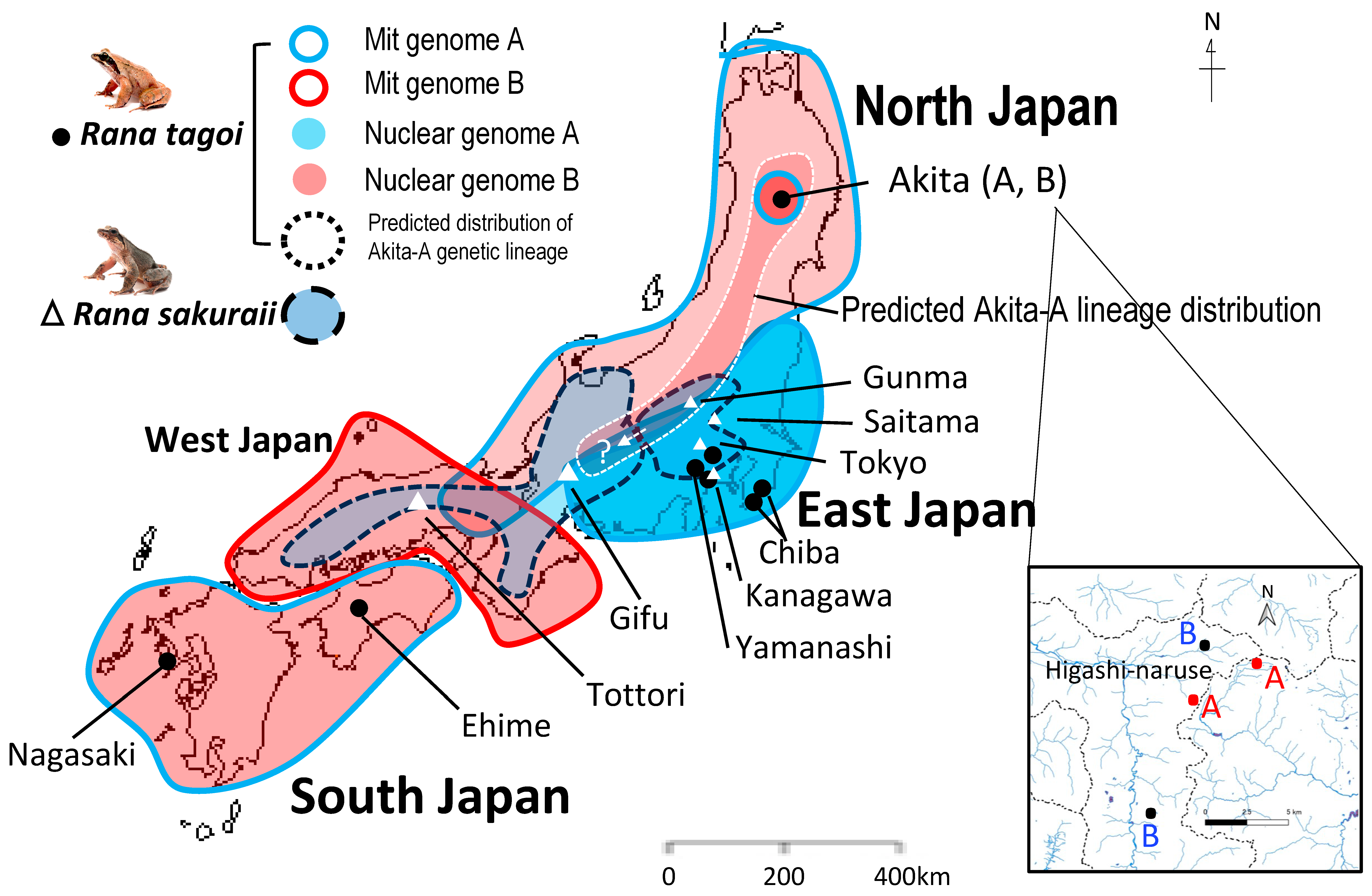
3.4. The Population Dynamics of the Two Species
4. Discussion
4.1. Sex Chromosomes in Tago’s Brown Frogs
4.2. Transitions between Y Chromosome and Autosome
4.3. Hybridization and Sex Chromosome Turnover
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Miura, I. Sex determination and sex-chromosomes in Amphibia. Sex. Dev. 2018, 11, 298–306. [Google Scholar] [CrossRef]
- Jeffries, D.L.; Lavanchy, G.; Sermier, R.; Sredl, M.J.; Miura, I.; Borzée, A.; Barrow, L.N.; Canestrelli, D.; Crochet, P.A.; Dufresnes, C.; et al. A rapid rate of sex-chromosome turnover and non-random transitions in true frogs. Nat. Commun. 2018, 9, 4088. [Google Scholar] [CrossRef] [PubMed]
- Sumida, M.; Nishioka, M. Sex-linked genes and linkage maps in amphibians. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2000, 26, 257–270. [Google Scholar] [CrossRef] [PubMed]
- Keating, S.E.; Blumer, M.; Grismer, L.L.; Lin, A.; Nielsen, S.V.; Thura, M.K.; Wood, P.L., Jr.; Quah, E.S.H.; Gamble, T. Sex Chromosome Turnover in Bent-Toed Geckos (Cyrtodactylus). Genes 2021, 12, 116. [Google Scholar] [CrossRef]
- Kikuchi, K.; Hamaguchi, S. Novel sex-determining genes in fish and sex-chromosome evolution. Dev. Dyn. 2013, 242, 339–353. [Google Scholar] [CrossRef]
- Bachtrog, D.; Mank, J.E.; Peichel, C.L.; Kirkpatrick, M.; Otto, S.P.; Ashman, T.L.; Hahn, M.W.; Kitano, J.; Mayrose, I.; Ming, R.; et al. Sex determination: Why so many ways of doing it? PLoS Biol. 2014, 12, e1001899. [Google Scholar] [CrossRef]
- Graves, J.A. Evolution of vertebrate sex-chromosomes and dosage compensation. Nat. Rev. Genet. 2016, 17, 33–46. [Google Scholar] [CrossRef]
- Zhou, Q.; Zhang, J.; Bachtrog, D.; An, N.; Huang, Q.; Jarvis, E.D.; Gilbert, M.T.; Zhang, G. Complex evolutionary trajectories of sex-chromosomes across bird taxa. Science 2014, 346, 1246338. [Google Scholar] [CrossRef]
- Uno, Y.; Nozu, R.; Kiyatake, I.; Higashiguchi, N.; Sodeyama, S.; Murakumo, K.; Sato, K.; Kuraku, S. Cell culture-based karyotyping of orectolobiform sharks for chromosome-scale genome analysis. Commun. Biol. 2020, 3, 652. [Google Scholar] [CrossRef] [PubMed]
- Ogata, M.; Suzuki, K.; Yuasa, Y.; Miura, I. Sex-chromosome evolution from a heteromorphic to a homomorphic system by inter-population hybridization in a frog. Philos. Trans. R. Soc. B 2021, 376, 20200105. [Google Scholar] [CrossRef] [PubMed]
- Pennell, M.W.; Mank, J.E.; Peichel, C.L. Transitions in sex determination and sex chromosomes across vertebrate species. Mol. Ecol. 2018, 27, 3950–3963. [Google Scholar] [CrossRef]
- Perrin, N. Sex-chromosome evolution in frogs: What role for sex-antagonistic genes? Philos. Trans. R. Soc. Lond. B Biol. Sci. 2021, 37, 20200094. [Google Scholar] [CrossRef]
- Graves, J.A. Did sex-chromosome turnover promote divergence of the major mammal groups?: De novo sex-chromosomes and drastic rearrangements may have posed reproductive barriers between monotremes, marsupials and placental mammals. Bioessays 2016, 38, 734–743. [Google Scholar] [CrossRef]
- Dufresnes, C.; Borzée, A.; Horn, A.; Stöck, M.; Ostini, M.; Sermier, R.; Wassef, J.; Litvinchuck, S.N.; Kosch, T.A.; Waldman, B.; et al. Sex-Chromosome Homomorphy in Palearctic Tree Frogs Results from Both Turnovers and X-Y Recombination. Mol. Biol. Evol. 2015, 32, 2328–2337. [Google Scholar] [CrossRef]
- Kitano, J.; Ross, J.A.; Mori, S.; Kume, M.; Jones, F.C.; Chan, Y.F.; Absher, D.M.; Grimwood, J.; Schmutz, J.; Myers, R.M.; et al. A role for a neo-sex chromosome in stickleback speciation. Nature 2009, 46, 1079–1083. [Google Scholar] [CrossRef] [PubMed]
- Nishioka, M.; Sumida, M. The Position of sex-determining genes in the chromosomes of Rana nigromaculata and Rana brevipoda. Sci. Rep. Lab. Amphibian Biol. Hiroshima Univ. 1990, 13, 51–97. [Google Scholar]
- Sumida, M.; Nishioka, M. Geographic variability of sex-linked loci in the Japanese brown frog, Rana japonica. Sci. Rep. Lab. Amphibian Biol. Hiroshima Univ. 1990, 13, 173–195. [Google Scholar]
- Eto, K.; Matsui, M.; Sugahara, T.; Tanaka-Ueno, T. Highly complex mitochondrial DNA genealogy in an endemic Japanese subterranean breeding brown frog Rana tagoi (Amphibia, Anura, Ranidae). Zool. Sci. 2012, 29, 662–671. [Google Scholar] [CrossRef]
- Eto, K.; Matsui, M. Cytonuclear discordance and historical demography of two brown frogs, Rana tagoi and R. sakurai (Amphibia: Ranidae). Mol. Phylogenet. Evol. 2014, 79C, 231–239. [Google Scholar] [CrossRef]
- Okada, Y. Notes on Japanese frogs. Annot. Zool. Jpn./Nihon dōbutsugaku ihō Tokyo 1928, 11, 269–277. [Google Scholar]
- Matsui, T.; Matsui, M. A new brown frog (genus Rana) from Honshu, Japan. Herpetologica 1990, 46, 78–85. [Google Scholar]
- Ryuzaki, M.; Hasegawa, Y.; Kuramoto, M. A new brown frog of the genus Rana from Japan (Anura: Ranidae) revealed by cytological and bioacoustic studies. Alytes 2014, 31, 49–58. [Google Scholar]
- Eto, K.; Matsui, M.; Sugahara, T. Description of a new subterranean breeding brown frog (Ranidae: Rana) from Japan. Zootaxa 2022, 5209, 401–425. [Google Scholar] [CrossRef]
- Eto, K.; Matsui, M. A New Brown Frog from the Goto Islands, Japan with Taxonomic Revision on the Subspecific Relationships of Rana tagoi (Amphibia: Anura: Ranidae). Curr. Herpetol. 2023, 42, 191–209. [Google Scholar] [CrossRef]
- Kuwana, C.; Fujita, H.; Tagami, M.; Matsuo, T.; Miura, I. Evolution of Sex Chromosome Heteromorphy in Geographic Populations of the Japanese Tago’s Brown Frog Complex. Cytogenet. Genome Res. 2021, 161, 23–31. [Google Scholar] [CrossRef]
- Ryuzaki, M.; Hanada, H.; Okumoto, H.; Takizawa, N.; Nishioka, M. Evidence for heteromorphic sex chromosomes in males of Rana tagoi and Rana sakuraii in Nishitama district of Tokyo (Anura: Ranidae). Chromosome Res. 1999, 7, 31–42. [Google Scholar] [CrossRef]
- Mawaribuchi, S.; Ito, M.; Ogata, M.; Yoshimura, Y.; Miura, I. Parallel Evolution of Sex-Linked Genes across XX/XY and ZZ/ZW Sex-chromosome Systems in the Frog Glandirana rugosa. Genes 2023, 14, 257. [Google Scholar] [CrossRef]
- Edgar, R.C.M. Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Darriba, D.; Posada, D.; Stamatakis, A.M.K.A.; Morel, B.; Flouri, T. ModelTest-NG: A New and Scalable Tool for the Selection of DNA and Protein Evolutionary Models. Mol. Biol. Evol. 2020, 37, 291–294. [Google Scholar] [CrossRef]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef]
- Sumner, A.T. A simple technique for demonstrating centromeric heterochromatin. Exp. Cell Res. 1972, 75, 304–306. [Google Scholar] [CrossRef]
- Takayama, S.; Taniguchi, T.; Iwashita, Y. Application of the 4Na-EDTA Giemsa staining method for analysis of DNA replication pattern. Chromosome Inf. Serv. 1981, 31, 36–38. [Google Scholar]
- Kilian, A.; Wenzl, P.; Huttner, E.; Carling, J.; Xia, L.; Blois, H.; Caig, V.; Heller-Uszynska, K.; Jaccoud, D.; Hopper, C. Diversity arrays technology: A generic genome profiling technology on open platforms. In Data Production and Analysis in Population Genomics: Methods and Protocols; Huwana Press: Totowa, NJ, USA, 2012; pp. 67–89. [Google Scholar]
- Nguyen, D.H.; Ponjarat, M.J.; Laopichienpong, N.; Kraichak, E.; Panthum, T.; Singchat, W.; Ahmad, S.F.; Muangmai, N.; Duengkae, P.; Peyachoknagul, S. Genome-wide SNP analysis suggests male heterogamety in bighead catfish (Clarias macrocephalus). Aquaculture 2021, 543, 737005. [Google Scholar] [CrossRef]
- Shams, F.; Dyer, F.; Thompson, R.; Duncan, P.; Thiem, J.D.; Kilian, A.; Ezaz, T. Application of DArT seq derived SNP tags for comparative genome analysis in fishes: An alternative pipeline using sequence data from a non-traditional model species, Macquaria ambigua. PLoS ONE 2019, 14, e0226365. [Google Scholar] [CrossRef] [PubMed]
- Hill, P.L.; Burridge, C.P.; Ezaz, T.; Wapstra, E. Conservation of sex-linked markers among conspecific populations of a viviparous skink, Niveoscincus ocellatus, exhibiting genetic and temperature-dependent sex determination. Genome Biol. Evol. 2018, 10, 1079–1087. [Google Scholar] [CrossRef] [PubMed]
- Charlesworth, D. Evolution of recombination rates between sex chromosomes. Philos. Trans. R. Soc. B 2017, 372, 20160456. [Google Scholar] [CrossRef] [PubMed]
- Gruber, B.; Unmack, P.J.; Berry, O.F.; Georges, A. dartr: An r package to facilitate analysis of SNP data generated from reduced representation genome sequencing. Mol. Ecol. Resour. 2018, 18, 691–699. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing. 2020. [Google Scholar]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; Von Holdt, B.M. TRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Miura, I.; Shams, F.; Jeffries, D.L.; Katsura, Y.; Mawaribuchi, S.; Perrin, N.; Ito, M.; Ogata, M.; Ezaz, T. Identification of ancestral sex chromosomes in the frog Glandirana rugosa bearing XX-XY and ZZ-ZW sex-determining systems. Mol. Ecol. 2022, 1, 859–3870. [Google Scholar] [CrossRef]
- Vicoso, B.; Bachtrog, D. Reversal of an ancient sex-chromosome to an autosome in Drosophila. Nature 2013, 499, 332–335. [Google Scholar] [CrossRef] [PubMed]
- Dixon, G.; Kitano, J.; Kirkpatrick, M. The origin of a new sex-chromosome by introgression between two stickleback fishes. Mol. Biol. Evol. 2018, 36, 28–38. [Google Scholar] [CrossRef]
- Tuttle, E.M.; Bergland, A.O.; Korody, M.L.; Brewer, M.S.; Newhouse, D.J.; Minx, P.; Stager, M.; Betuel, A.; Cheviron, Z.A.; Warren, W.C.; et al. 2Divergence and Functional Degradation of a Sex-chromosome-like Supergene. Curr. Biol. 2016, 26, 344–350. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.J.; Veltsos, P. The Diversity and Evolution of Sex-chromosomes in Frogs. Genes 2021, 12, 483. [Google Scholar] [CrossRef] [PubMed]
- Eggert, C. Sex determination: The amphibian model. Reprod. Nutr. Dev. 2004, 44, 539–549. [Google Scholar] [CrossRef]
- Ogata, M.; Ohtani, H.; Igarashi, T.; Hasegawa, Y.; Ichikawa, Y.; Miura, I. Change of the heterogametic sex from male to female in the frog. Genetics 2003, 164, 613–620. [Google Scholar] [CrossRef]
- Kozielska, M.; Weissing, F.J.; Beukeboom, L.W.; Pen, I. Segregation distortion and the evolution of sex-determining mechanisms. Heredity 2010, 104, 100–112. [Google Scholar] [CrossRef]
- Saunders, P.A.; Neuenschwander, S.; Perrin, N. Sex chromosome turnovers and genetic drift: A simulation study. J. Evol. Biol. 2018, 31, 1413–1419. [Google Scholar] [CrossRef]
- Van Doorn, G.S.; Kirkpatrick, M. Turnover of sex chromosomes induced by sexual conflict. Nature 2007, 449, 909–912. [Google Scholar] [CrossRef]
- Van Doorn, G.S.; Kirkpatrick, M. Transitions between male and female heterogamety caused by sex-antagonistic selection. Genetics 2010, 186, 629–645. [Google Scholar] [CrossRef] [PubMed]
- Blaser, O.; Grossen, C.; Neuenschwander, S.; Perrin, N. Sex-chromosome turnovers induced by deleterious mutation load. Evolution 2013, 67, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, N.; Studer, T.; Dufresnes, C.; Perrin, N. Sex-chromosome recombination in common frogs brings water to the fountain-of-youth. Mol. Biol. Evol. 2018, 35, 942–948. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.-J.; Veltsos, P.; Toups, M.A.; Rodrigues, N.; Sermier, R.; Jeffries, D.L.; Perrin, N. Tissue specificity and dynamics of sex-biased gene expression in a common frog population with differentiated, yet homomorphic, sex chromosomes. Genes 2018, 9, 294. [Google Scholar] [CrossRef]
- Ma, W.-J.; Veltsos, P.; Sermier, R.; Parker, D.J.; Perrin, N. Evolutionary and developmental dynamics of sex-biased gene expression in common frogs with proto-Y chromosomes. Genome Biol. 2018, 19, 156. [Google Scholar] [CrossRef]
- Veltos, P.; Rodrigues, N.; Studer, T.; Ma, W.-J.; Sermier, R.; Leuenberger, J.; Perrin, N. No evidence that Y chromosome differentiation affects male fitness in a Swiss population of common frogs. J. Evol. Biol. 2020, 33, 401–409. [Google Scholar] [CrossRef]
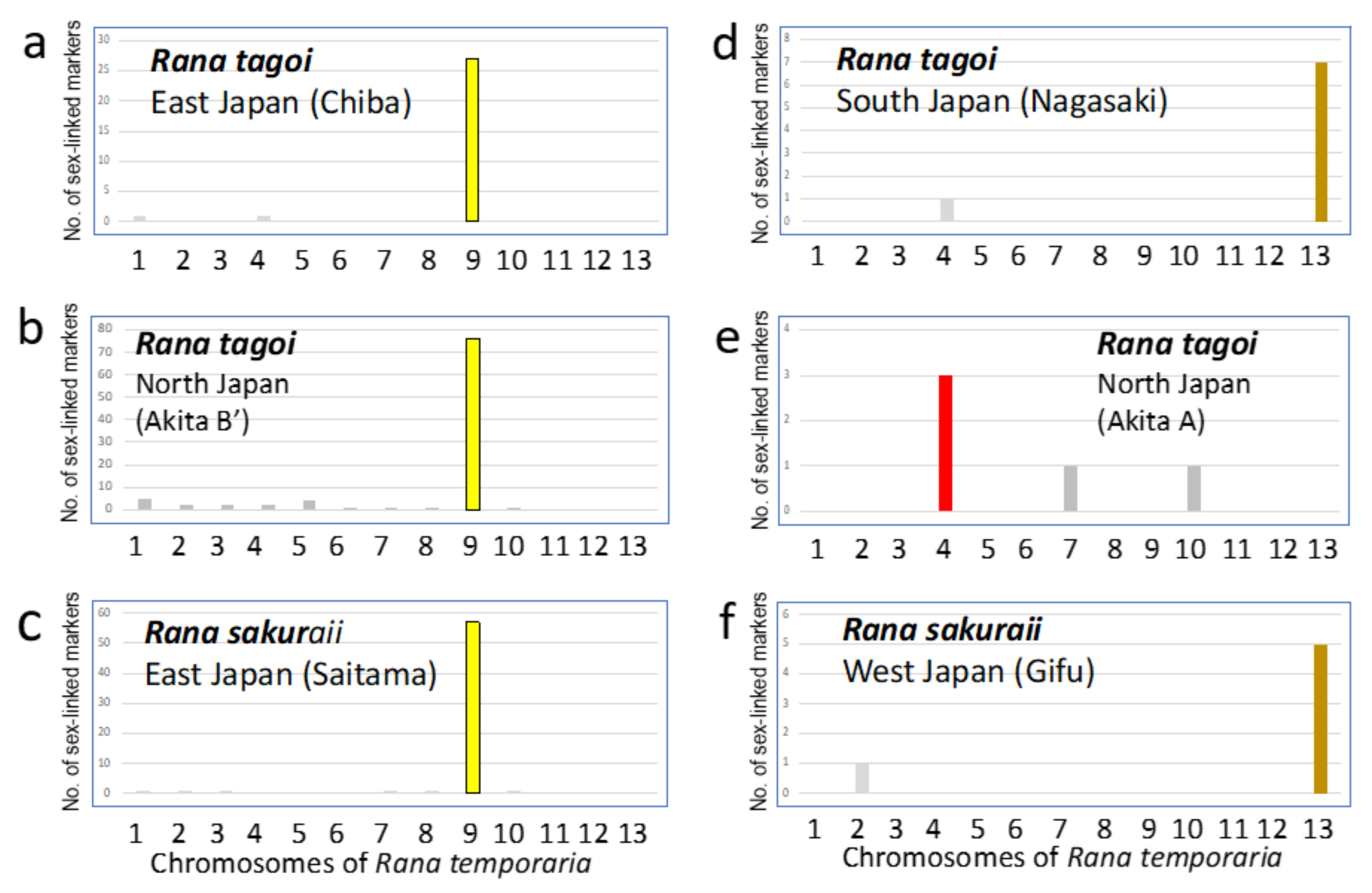
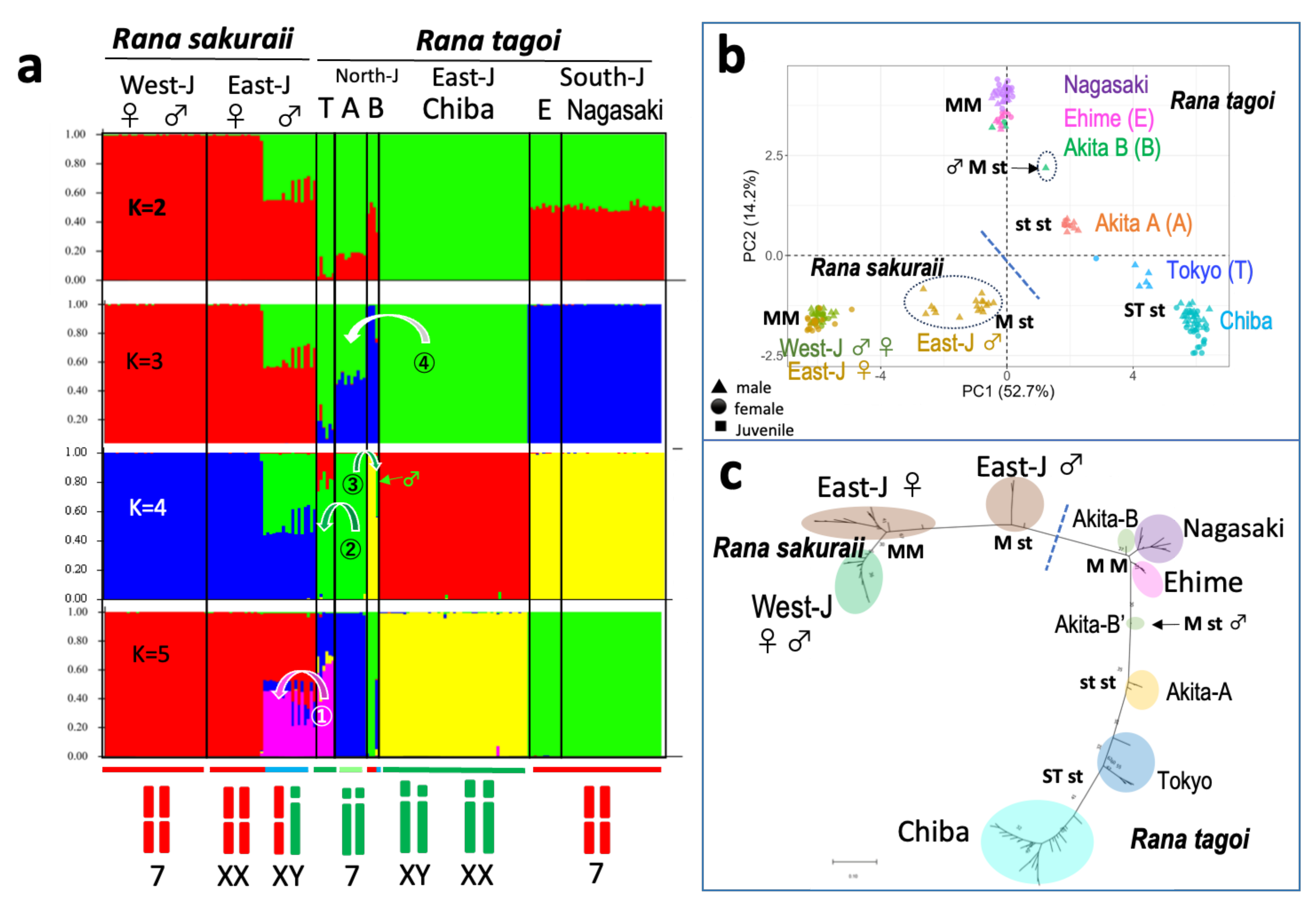
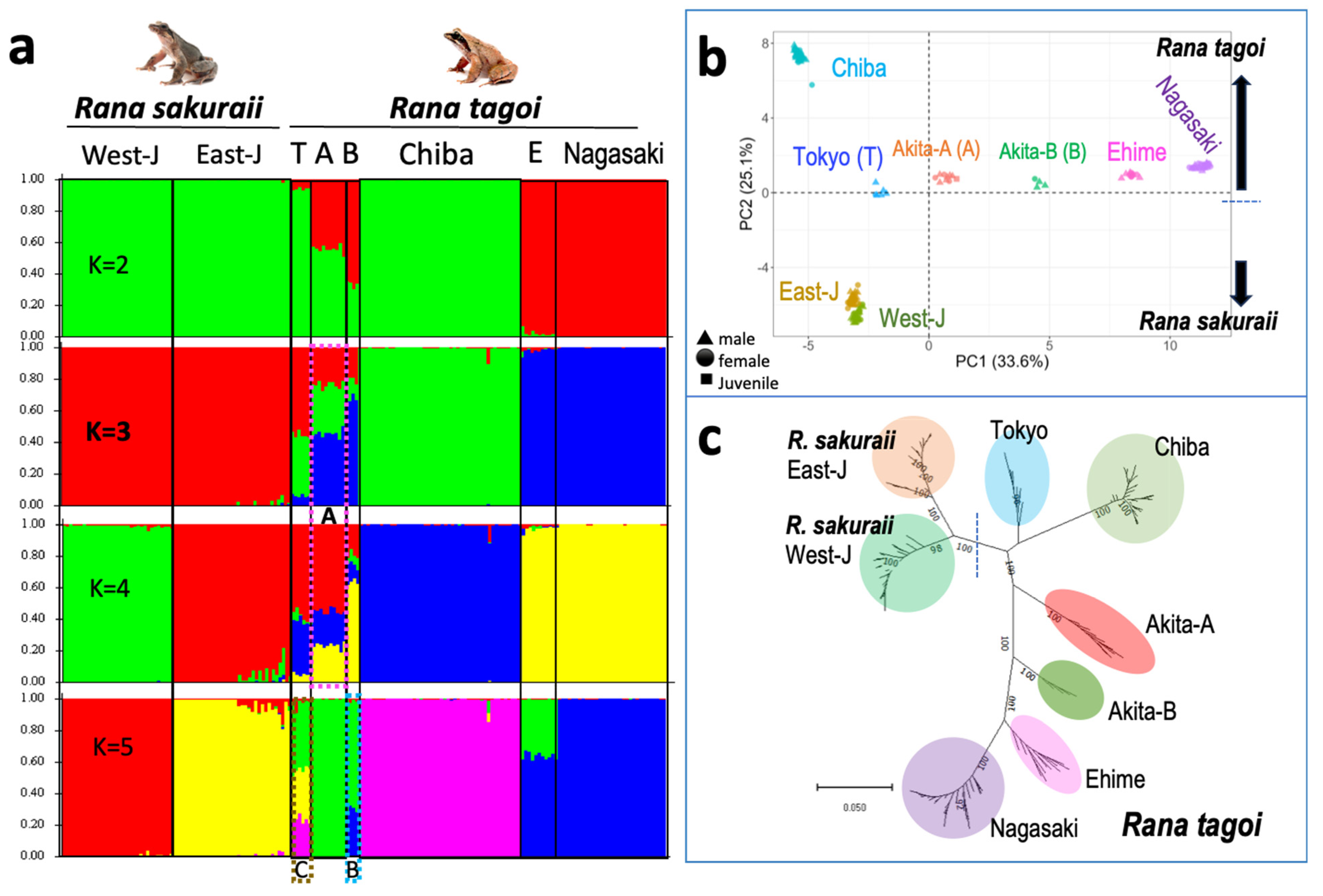

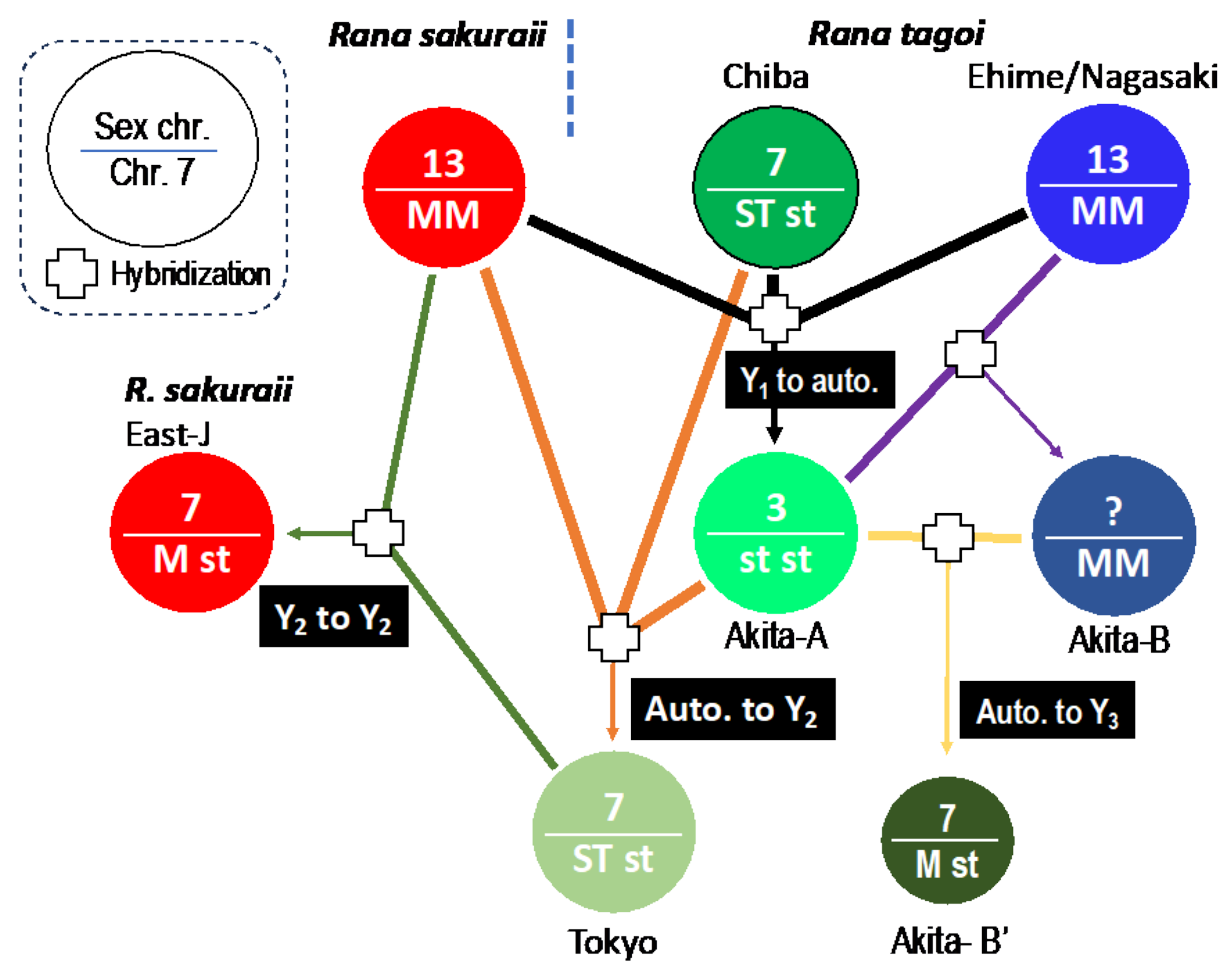
| Population (City) | No. of frogs analyzed | No. of markers heterozygous or present in | |||||||
| Geographic group | Male | Female | Total | ||||||
| Species | Sex chromosome | SNP | SilicoDArT | SNP | SilicoDArT | ||||
| R. sakuraii | Saitama (Hanno) | Male | 9 | Initial sex-linked markers | 197 | 17 | 13 | 3 | |
| East Japan | Female | 10 | False positive | 8 | 0 | 0 | 0 | ||
| Hetero (No. 7 M/st) | True sex-linked markers | 189 | 17 | 13 | 3 | 222 | |||
| Gifu (Gifu) | Male | 10 | Initial sex-linked markers | 12 | 3 | 17 | 3 | ||
| West Japan | Female | 10 | False positive | 3 | 0 | 6 | 0 | ||
| Homo | True sex-linked markers | 9 | 3 | 11 | 3 | 26 | |||
| R. tagoi | Chiba (Ichihara) | Male | 10 | Initial sex-linked markers | 72 | 15 | 36 | 4 | |
| East Japan | Female | 10 | False positive | 7 | 0 | 4 | 0 | ||
| Hetero (No. 7 St/st) | True sex-linked markers | 65 | 15 | 32 | 4 | 116 | |||
| Nagasaki (Nagasaki) | Male | 9 | Initial sex-linked markers | 16 | 5 | 3 | 3 | ||
| South Japan | Female | 10 | False positive | 4 | 0 | 0 | 0 | ||
| Hetero (No. 13) | True sex-linked markers | 12 | 5 | 3 | 3 | 23 | |||
| Akita-A (Higashi-naruse) | Male | 8 | Initial sex-linked markers | 21 | 6 | 19 | 2 | ||
| North Japan | Female | 8 | False positive | 14 | 0 | 15 | 0 | ||
| Homo | True sex-linked markers | 7 | 6 | 4 | 2 | 19 | |||
| Akita-B’ (Higashi-naruse) | Male | 7 | Initial sex-linked markers | 421 | 253 | 106 | 42 | ||
| North Japan | Female | 7 | False positive | 22 | 0 | 33 | 0 | ||
| Hetero (No. 7 M/st) | True sex-linked markers | 399 | 253 | 73 | 42 | 767 | |||
| Species | Population | City | Geographic Group | Sex chr. | Morphology (1) | Chromosome 7 (2) | Analysis | Reference (3) |
|---|---|---|---|---|---|---|---|---|
| R. tagoi | Akita A | Higashi-naruse | North Japan | 3 | Homo | st/st | SNP | This study |
| Akita-B | Higashi-naruse | ? | Homo | M/M | Chromosome | This study | ||
| Akita-B’ | Higashi-naruse | 7 | Hetero | M/st | SNP/Chromosome | This study | ||
| Chiba | Ichihara | East Japan | 7 | Hetero | ST/st | SNP/Chromosome | This study | |
| Tokyo | Akiruno | 7 | Hetero | ST/st | Chromosome | 1 | ||
| Ehime | Kumankogen | South Japan | 13 | Hetero | M/M | C-banding | 2 | |
| Nagasaki | Nagasaki | 13 | Hetero | M/M | SNP/C-banding | 2, this study | ||
| R. sakuraii | Tokyo, Saitama | Akiruno, Hanno | East Japan | 7 | Hetero | M/st | SNP/Chromosome | This study, 1, 2 |
| Gifu | Gifu | West Japan | 13 | Homo | M/M | SNP | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miura, I.; Shams, F.; Ohki, J.; Tagami, M.; Fujita, H.; Kuwana, C.; Nanba, C.; Matsuo, T.; Ogata, M.; Mawaribuchi, S.; et al. Multiple Transitions between Y Chromosome and Autosome in Tago’s Brown Frog Species Complex. Genes 2024, 15, 300. https://doi.org/10.3390/genes15030300
Miura I, Shams F, Ohki J, Tagami M, Fujita H, Kuwana C, Nanba C, Matsuo T, Ogata M, Mawaribuchi S, et al. Multiple Transitions between Y Chromosome and Autosome in Tago’s Brown Frog Species Complex. Genes. 2024; 15(3):300. https://doi.org/10.3390/genes15030300
Chicago/Turabian StyleMiura, Ikuo, Foyez Shams, Jun’ichi Ohki, Masataka Tagami, Hiroyuki Fujita, Chiao Kuwana, Chiyo Nanba, Takanori Matsuo, Mitsuaki Ogata, Shuuji Mawaribuchi, and et al. 2024. "Multiple Transitions between Y Chromosome and Autosome in Tago’s Brown Frog Species Complex" Genes 15, no. 3: 300. https://doi.org/10.3390/genes15030300
APA StyleMiura, I., Shams, F., Ohki, J., Tagami, M., Fujita, H., Kuwana, C., Nanba, C., Matsuo, T., Ogata, M., Mawaribuchi, S., Shimizu, N., & Ezaz, T. (2024). Multiple Transitions between Y Chromosome and Autosome in Tago’s Brown Frog Species Complex. Genes, 15(3), 300. https://doi.org/10.3390/genes15030300