Population Structure and Genetic Diversity of the Toona ciliata (Meliaceae) Complex Assayed with Chloroplast DNA Markers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling and DNA Extraction
2.2. Primer Screening, Amplification, and Sequencing
2.3. Analysis of Genetic Diversity
2.4. Population Genetic Structure
2.5. Population Demography
3. Results
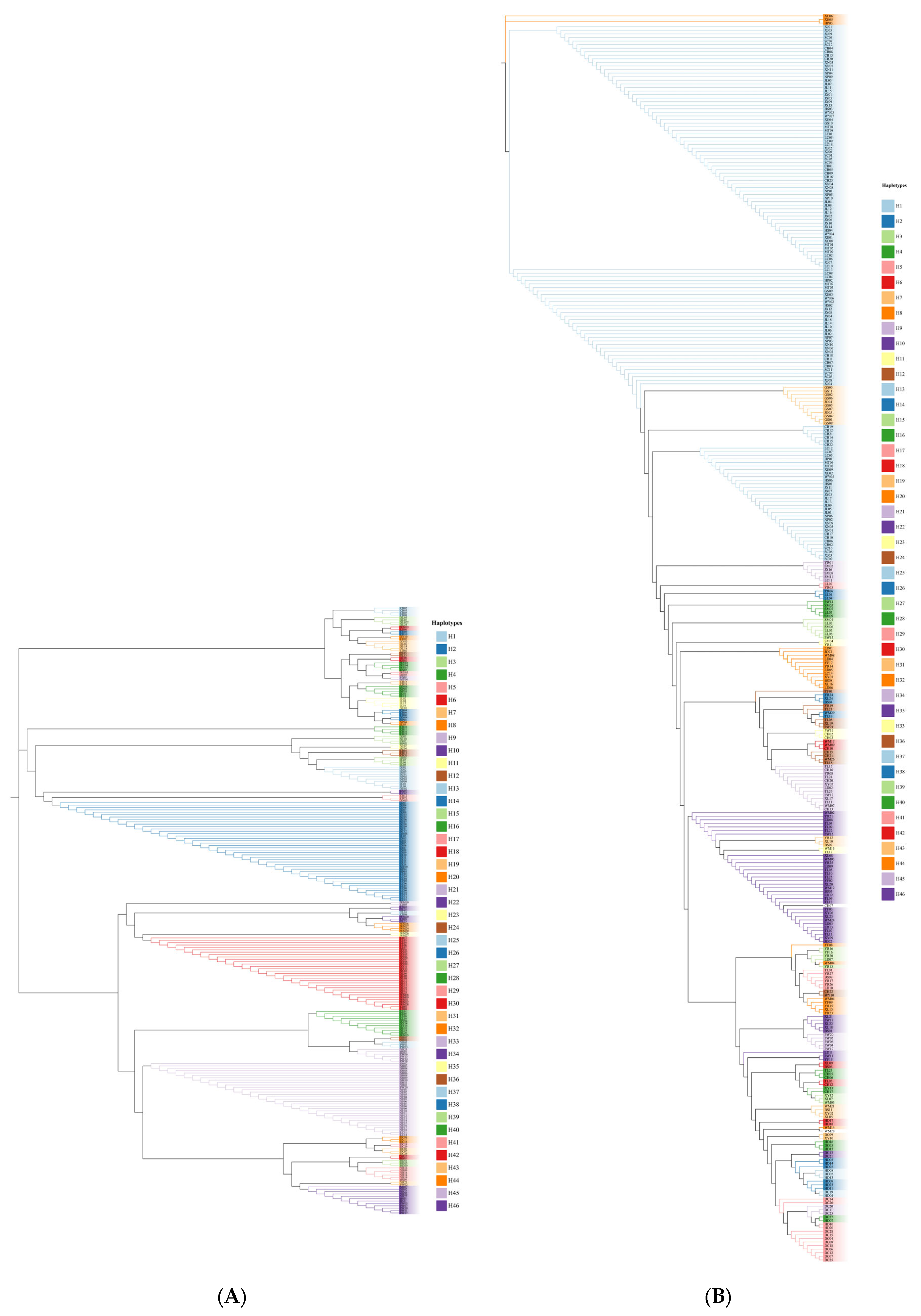
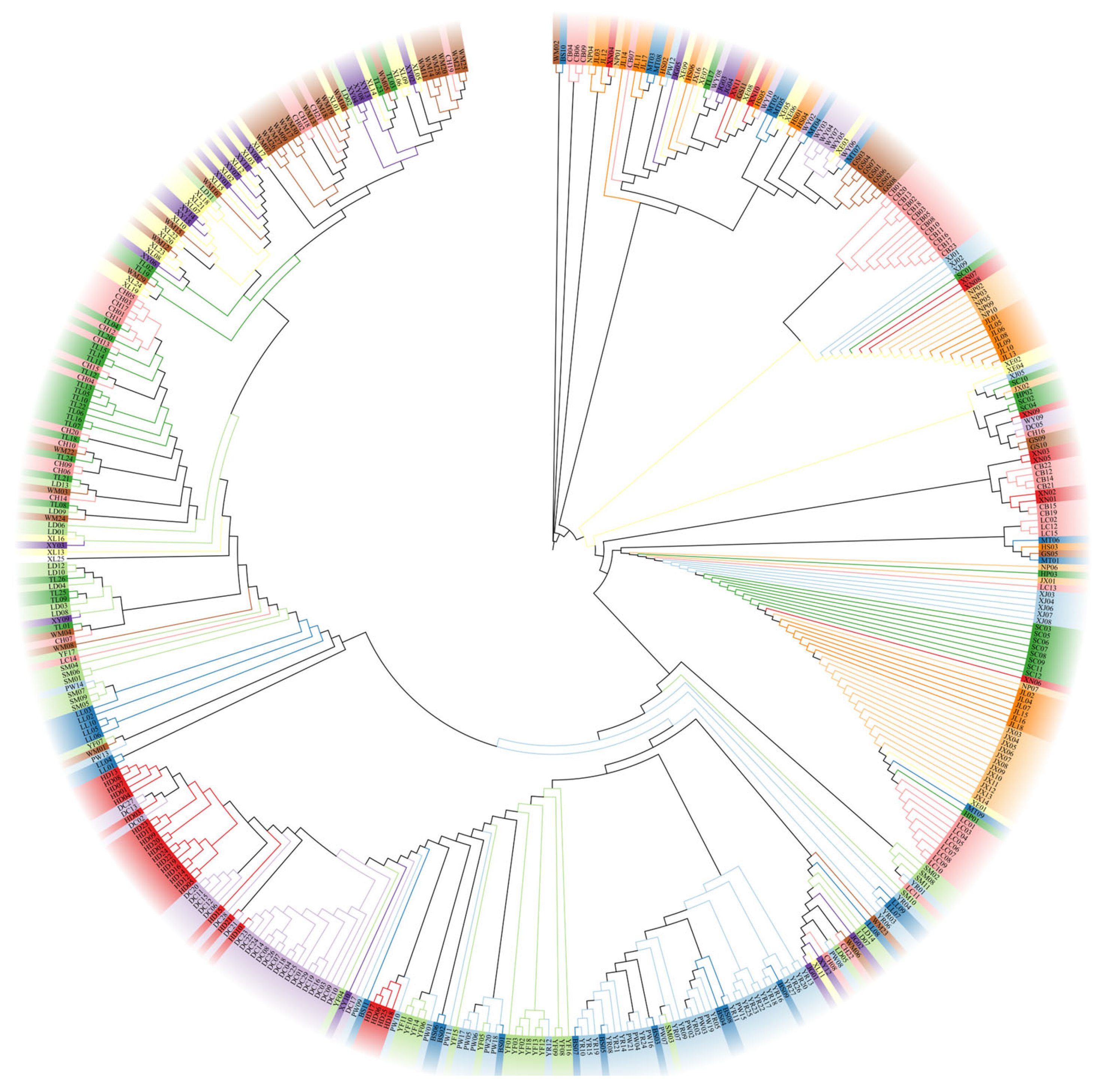
3.1. Haplotype Analysis
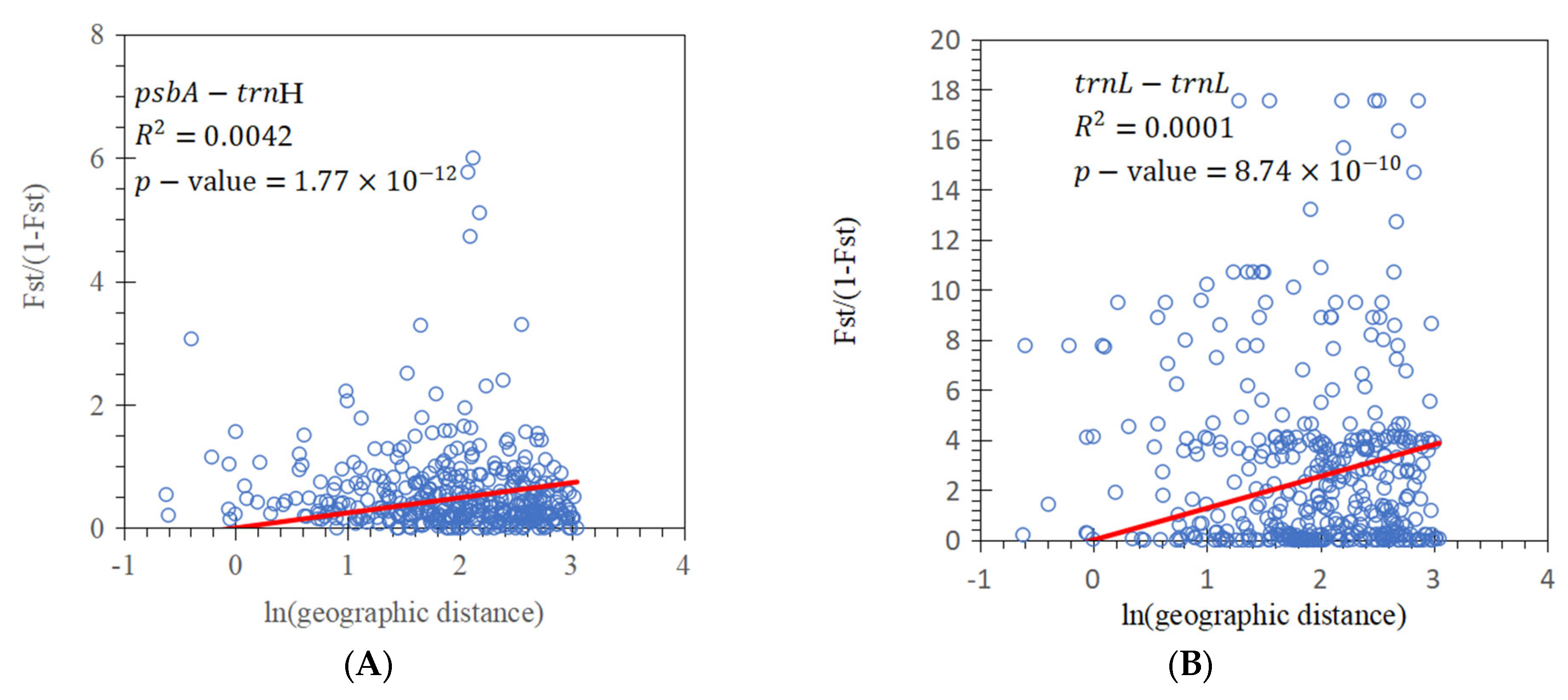
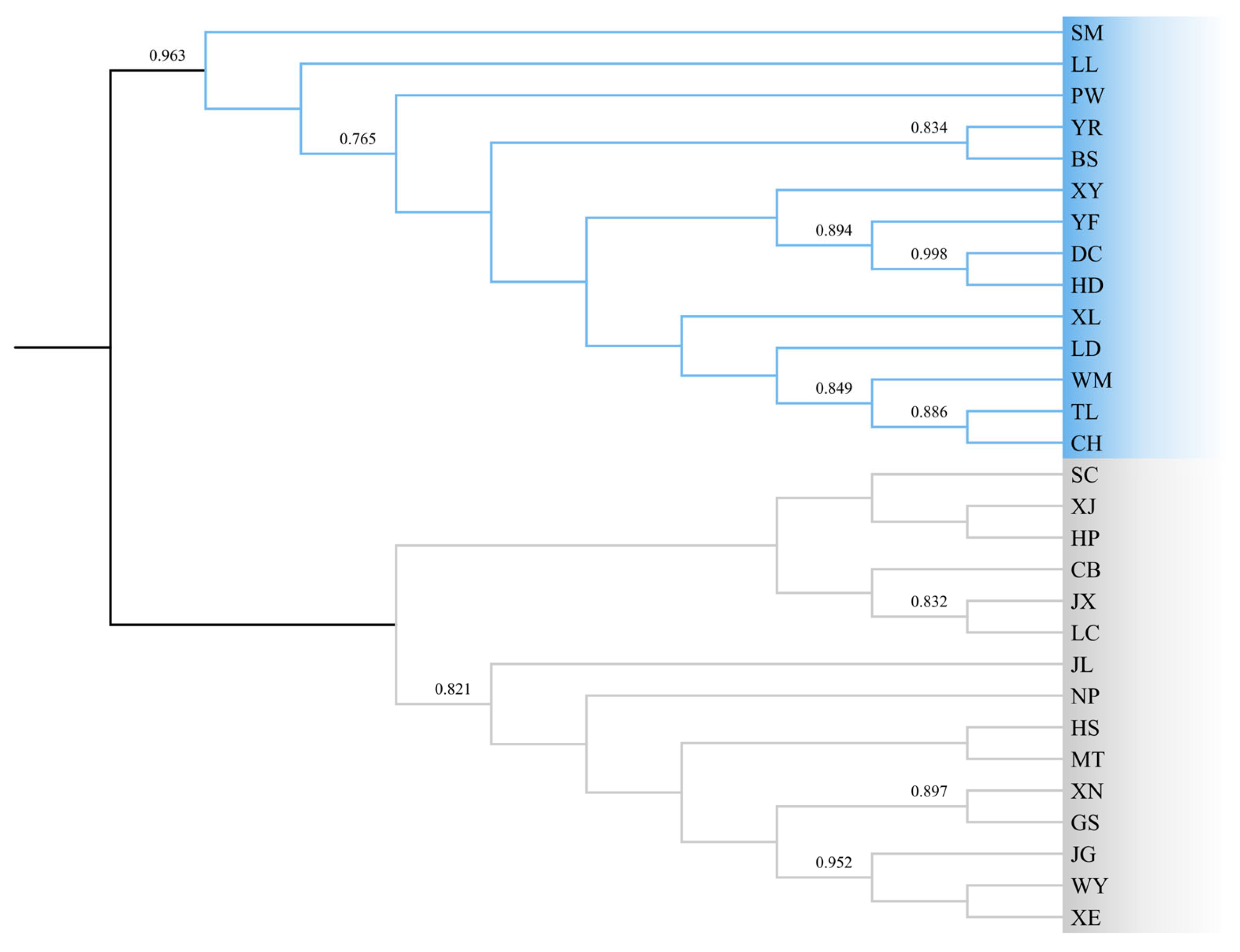
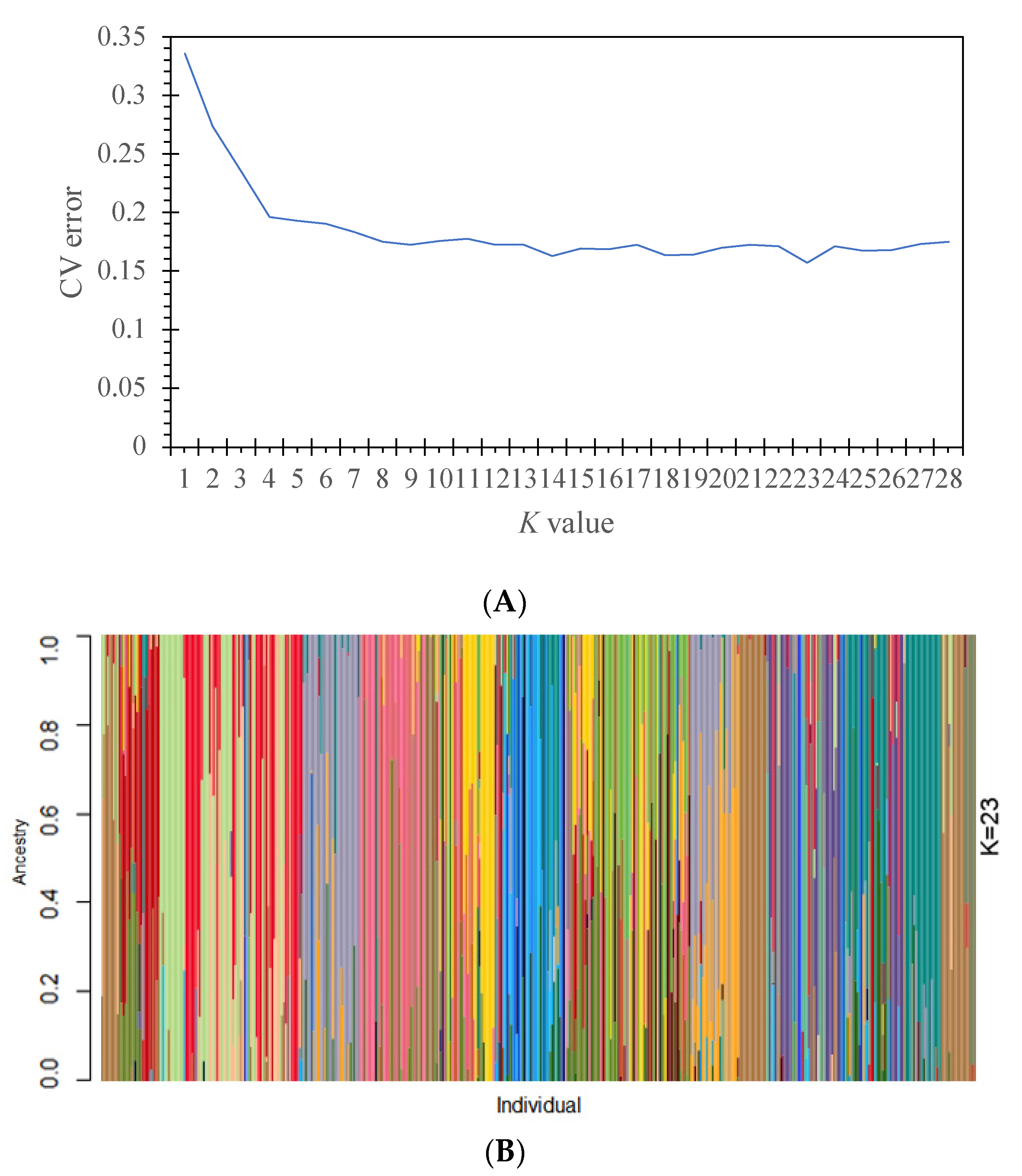
3.2. Population Genetic Structure
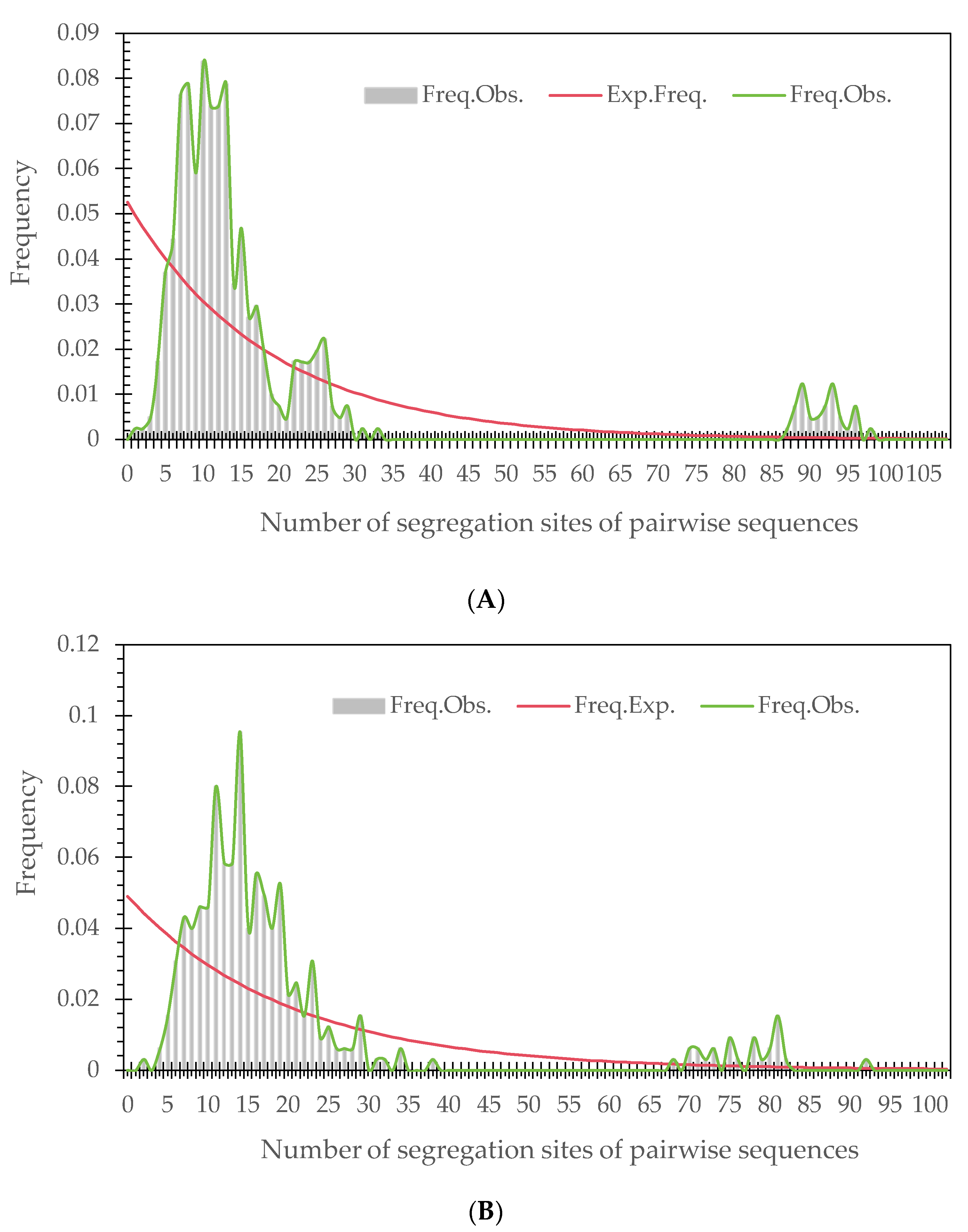
3.3. Analysis of Population Demography
4. Discussion
4.1. Genetic Diversity
4.2. Population Genetic Structure
4.3. Genetic Conservation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, S.K.; Li, H.; Chen, B.Y. Meliaceae. Flora Reipublicae Popularis Sinicae; Science Press: Beijing, China, 1997; Volume 43, pp. 40–43. [Google Scholar]
- Ferreira, D.M.; Palma-Silva, C.; Neri, J.; Medeiros, M.C.; Pinange, D.S.; Benko-Iseppon, A.M.; Louzada, R.B. Population genetic structure and species delimitation in the Cryptanthus zonatus complex (Bromeliaceae). Bot. J. Linn. Soc. 2021, 196, 123–140. [Google Scholar] [CrossRef]
- Freeland, J.R.; Petersen, S.D.; Kirk, H. Molecular Ecology; John Wiley & Sons, Ltd.: West Sussex, UK, 2011. [Google Scholar]
- Xiao, Y.; Zhang, X.X.; Hu, Y.; Wang, X.; Li, P.; He, Z.H.; Lv, Y.W.; Chen, X.Y.; Hu, X.S. Phylogeography of Toona ciliata (Meliaceae) complex in China inferred from cytonuclear markers. Genes 2023, 14, 116. [Google Scholar] [CrossRef]
- Edmonds, J.M. The potential value of Toona species (Meliaceae) as multipurpose and plantation trees in Southeast Asia. Commonw. For. Rev. 1993, 72, 181–186. [Google Scholar]
- Edmonds, J.M.; Staniforth, M. Toona sinensis (Meliaceae). Curtis’s Bot. Mag. 1998, 15, 186–193. [Google Scholar] [CrossRef]
- Styles, B.T. The flower biology of Meliaceae and its bearing on tree breeding. Silvae Genet. 1972, 21, 175–182. [Google Scholar]
- Liu, J.; Sun, Z.X.; Chen, Y.T.; Jiang, J.M. Isolation and characterization of microsatellite loci from an endangered tree species, Toona ciliata var. Pubescens. Genet. Mol. Res. 2012, 11, 4411–4417. [Google Scholar] [CrossRef] [PubMed]
- Cheng, D.S.; Cui, T.L. Utilization value and cultivation techniques of Toona sureni. For. By-Prod. Spec. China 2010, 4, 39–40. [Google Scholar]
- Liang, R.L.; Liao, R.Y.; Dai, J. Endangered causes and protection strategy of Toona ciliata. Guangxi For. Sci. 2011, 40, 201–203. [Google Scholar]
- Wu, J.Y.; Cheng, Y.; Wang, X.J.; Chen, R.; Liu, Q.; Liu, X.Y.; Yin, H.Y.; Wu, X.W. Softwood cutting propagation of Toona ciliata clones. Hunan For. Sci. Technol. 2012, 38, 5. [Google Scholar]
- Chen, K. Technology of Tonna Tissue Cultivation Research. Master’s Thesis, Central South University of Forestry and Technology, Changsha, China, 2016. [Google Scholar]
- Zhang, H.B.; Zhang, G.Y.; Huang, G.Y.; Wu, D.; Wang, X.Y. Research on Tonna ciliata seedling and silviculture technology. Sci. Technol. 2016, 26, 83. [Google Scholar]
- Li, P.; Que, Q.M.; Wu, L.Y.; Zhu, Q.; Chen, X.Y. Growth rhythms of Toona ciliata seedlings from different provenances. J. South China Agri. Univ. 2017, 38, 96–102. [Google Scholar]
- Wen, W.H.; Wu, J.Y.; Chen, M.G.; Chen, J.F.; Cheng, Y.; Wang, X.J.; Liu, Q. Seedling growth performance of Toona ciliata elite trees progeny. China Agric. Sci. Bull. 2012, 28, 36–39. [Google Scholar]
- Liao, D.; Wu, J.; Shu, Y.; Cheng, Y.; Chen, M.; Chen, J.; Liu, Q.; Wu, Z. Tissue culture of Toona ciliata. Agric. Sci. Technol. 2017, 18, 2185–2187+2196. [Google Scholar]
- Lan, J.H.; Feng, L.X. Study on cutting propagation technology of rare timber species Toona ciliata Roem. J. Guangxi Agric. 2022, 37, 32–36+48. [Google Scholar]
- Li, P.; Zhan, X.; Que, Q.M.; Qu, W.T.; Liu, M.Q.; Ouyang, K.X.; Li, J.C.; Deng, X.M.; Zhang, J.J.; Liao, B.Y.; et al. Genetic diversity and population structure of Toona ciliata Roem. based on sequence-related amplified polymorphism (SRAP) markers. Forests 2015, 6, 1094–1106. [Google Scholar] [CrossRef]
- Xing, P.Y.; Liu, T.; Song, Z.Q.; Li, X.F. Genetic diversity of Toona sinensis Roem in China revealed by ISSR and SRAP markers. Genet. Mol. Res. 2016, 15, 15038387. [Google Scholar] [CrossRef]
- Zhan, X.; Li, P.; Hui, W.K.; Deng, Y.W.; Gan, S.M.; Sun, Y.; Zhao, X.H.; Chen, X.Y.; Deng, X.M. Genetic diversity and population structure of Toona ciliata revealed by simple sequence repeat markers. Biotechnol. Biotechnol. Equip. 2019, 33, 214–222. [Google Scholar] [CrossRef]
- Xiao, Y.; Wang, X.; He, Z.H.; Lv, Y.W.; Zhang, C.H.; Hu, X.S. Assessing phylogenetic relationships among varieties of Toona ciliata in sympatry with chloroplast genomes. Ecol. Evol. 2023, 13, e10828. [Google Scholar] [CrossRef]
- Liu, J.; Jiang, J.M.; Chen, Y.T. Genetic diversity of central and peripheral populations of Toona ciliata var. pubescens, an endangered tree species endemic to China. Genet. Mol. Res. 2014, 13, 4579–4590. [Google Scholar] [CrossRef]
- Liu, J.; Chen, Y.T.; Jiang, J.M.; He, G.P.; Yu, G.M. Study on population genetic structure in Toona ciliata var. pubescens with SSR. For. Res. 2009, 22, 37–41. [Google Scholar]
- Zhou, W.; Zhang, X.X.; Ren, Y.; Li, P.; Chen, X.Y.; Hu, X.S. Mating system and population structure in the natural distribution of Toona ciliata (Meliaceae) in South China. Sci. Rep. 2020, 10, 16998. [Google Scholar] [CrossRef]
- Wang, X.; Xiao, Y.; He, Z.H.; Li, L.L.; Song, H.Y.; Zhang, J.J.; Cheng, X.; Chen, X.Y.; Li, P.; Hu, X.S. A chromosome-level genome assembly of Toona ciliata (Meliaceae). Genome Biol. Evol. 2022, 14, evac121. [Google Scholar] [CrossRef]
- Wang, X.; Xiao, Y.; He, Z.H.; Li, L.L.; Lv, Y.W.; Hu, X.S. Evolutionary divergence between Toona ciliata and Toona sinensis assayed with their whole genome sequences. Genes 2022, 13, 1799. [Google Scholar] [CrossRef]
- Wolfe, K.H.; Li, W.H.; Sharp, P.M. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc. Natl. Acad. Sci. USA 1987, 84, 9054–9058. [Google Scholar] [CrossRef]
- Hu, Y.; Wang, X.; Zhang, X.X.; Zhou, W.; Chen, X.Y.; Hu, X.S. Advancing phylogeography with chloroplast DNA markers. Biodivers. Sci. 2019, 27, 219–234. [Google Scholar]
- Palmer, J.D. Comparative organization of chloroplast genomes. Ann. Rev. Genet. 1985, 19, 325–354. [Google Scholar] [CrossRef]
- Wright, S. Evolution and the Genetics of Populations; University Chicago Press: Chicago, IL, USA, 1969; pp. 1191–1192. [Google Scholar]
- Hu, X.S.; Ennos, R.A. Impacts of seed and pollen flow on population differentiation for plant genomes with three contrasting modes of inheritance. Genetics 1999, 152, 441–450. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf material. Phytochemistry 1987, 19, 11–15. [Google Scholar]
- Taberlet, P.; Fumagalli, L.; Wust-Saucy, A.-G.; Cosson, J.-F. Comparative phylogeography and postglacial colonization routes in Europe. Mol. Ecol. 1998, 7, 453–464. [Google Scholar] [CrossRef]
- Zhu, S.; Liu, Q.; Qiu, S.; Dai, J.; Gao, X. DNA barcoding: An efficient technology to authenticate plant species of traditional Chinese medicine and recent advances. Chin. Med. 2022, 17, 112. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Nei, M. Molecular Evolutionary Genetics; Columbia University Press: New York, NY, USA, 1987. [Google Scholar]
- Pons, O.; Petitt, R.J. Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics 1996, 144, 1237–1245. [Google Scholar] [CrossRef]
- Grivet, D.; Petit, R.J. Chloroplast DNA phylogeography of the hornbeam in Europe: Evidence for a bottleneck at the outset of postglacial colonization. Conserv. Genet. 2002, 4, 47–56. [Google Scholar] [CrossRef]
- Wright, S. The genetical structure of populations. Ann. Eugen. 1951, 15, 323–354. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Laval, G.; Schneider, S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol. Bioinform. 2005, 1, 47–50. [Google Scholar] [CrossRef]
- Meirmans, P.G. Using the AMOVA framework to estimate a standardized genetic differentiation measure. Evolution 2006, 60, 2399–2402. [Google Scholar] [CrossRef]
- Rousset, F. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 1997, 145, 1219–1228. [Google Scholar] [CrossRef] [PubMed]
- Mantel, N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967, 27, 109–220. [Google Scholar]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef] [PubMed]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Fu, Y.X. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 1997, 147, 915–925. [Google Scholar] [CrossRef]
- Rogers, A.R.; Harpending, H. Population growth makes waves in the distribution of pairwise genetic differences. Mol. Biol. Evol. 1992, 9, 552–569. [Google Scholar]
- Lu, C.X.; Zhang, D.C.; Wang, D.B. Origin and taxonomic position of Chinese Toona (Toona sinensis (A. Juss.) Roem.). Bull. Bot. Res. 2001, 21, 195–199. [Google Scholar]
- Li, S.; Liu, S.L.; Pei, S.Y.; Ning, M.M.; Tang, S.Q. Genetic diversity and population structure of Camellia huana (Theaceae), a limestone species with narrow geographic range, based on chloroplast DNA sequence and microsatellite markers. Plant Divers. 2020, 42, 343–350. [Google Scholar] [CrossRef]
- Deng, Y.; Liu, T.; Xie, Y.; Wei, Y.; Xie, Z.; Shi, Y.; Deng, X. High genetic diversity and low differentiation in Michelia shiluensis, an endangered Magnolia species in south China. Forests 2020, 11, 469. [Google Scholar] [CrossRef]
- Zhang, G.L.; Han, Y.; Wang, H.; Wang, Z.Y.; Xiao, H.X.; Sun, M.Z. Phylogeography of Iris loczyi (Iridaceae) in Qinghai–Tibet Plateau revealed by chloroplast DNA and microsatellite markers. AoB Plants 2021, 13, plab070. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, Y.H.; Wang, Y.H.; Shen, S.K. Genetic Diversity and Population Structure of Rhododendron rex Subsp. rex Inferred from Microsatellite Markers and Chloroplast DNA Sequences. Plants 2020, 9, 338. [Google Scholar] [CrossRef] [PubMed]
- Dutech, C.; Maggia, L.; Joly, H.I. Chloroplast diversity in Vouacapoua americana (Caesalpiniaceae), a neotropical forest tree. Mol. Ecol. 2000, 9, 1427–1432. [Google Scholar] [CrossRef]
- Fineschi, S.; Salvini, D.; Taurchini, D.; Carnevale, S.; Vendramin, G.G. Chloroplast DNA variation of Tilia cordata (Tiliaceae). Can. J. For. Res. 2003, 33, 2503–2508. [Google Scholar] [CrossRef]
- Hamrick, J.L.; Murawski, D.A. The breeding structure of tropical tree populations. Plant Species Biol. 1990, 5, 157–165. [Google Scholar] [CrossRef]
- Hu, X.S.; Yeh, F.C.; Hu, Y.; Deng, L.T.; Ennos, R.A.; Chen, X.Y. High mutation rates explain low population genetic divergence at copy-number-variable loci in Homo sapiens. Sci. Rep. 2017, 7, 43178. [Google Scholar] [CrossRef]
- Hamrick, J.L.; Godt, M.J.W.; Sherman-Broyles, S.L. Factors influencing levels of genetic diversity in woody plant species. New For. 1992, 6, 95–124. [Google Scholar] [CrossRef]
- Zhu, Q.; Liao, B.Y.; Li, P.; Li, J.C.; Deng, X.M.; Hu, X.S.; Chen, X.Y. Phylogeographic pattern suggests a major eastward dispersal in the distribution of Machilus pauhoi in South China. PLoS ONE 2017, 12, e0184456. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y. Phylogeographic Structure of Toona ciliata (Meliaceae) in China Inferred from cpDNA Markers and Ecological Niche Model Analyses. Master’s Thesis, South China Agricultural University, Guangzhou, China, 2019. [Google Scholar]
- Zhang, C.H.; He, J.; Sun, Y.Y.; Li, K. Prediction of distributional change of Toona ciliata var. ciliata and application in regionalization of introduction based on MaxEnt. J. Yunnan Univ. 2018, 40, 164–173. [Google Scholar]
- Zhang, C.H.; He, J.; Sun, Y.Y.; Li, K. Distributional change in suitable areas for T. ciliata var. pubescens based on MaxEnt. For. Res. 2018, 31, 120–126. [Google Scholar]

| Code | Location | Sample Size | Longitude (E) | Latitude (N) | Elevation (m) |
|---|---|---|---|---|---|
| XJ | Xianju, Zhejiang | 9 | 119.92 | 28.45 | 600 |
| LL | Longlin, Guangxi | 10 | 105.20 | 24.46 | 624 |
| SM | Simao, Yunnan | 11 | 100.58 | 22.46 | 1317 |
| SC | Suichang, Zhejiang | 12 | 119.25 | 28.59 | 510 |
| CB | Chengbu, Hunan | 23 | 111.28 | 27.14 | 737 |
| XN | Xinning, Hunan | 11 | 109.44 | 28.18 | 650 |
| NP | Nanping, Fujian | 9 | 118.10 | 26.38 | 800 |
| JL | Jiulianshan, Jiangxi | 18 | 114.47 | 24.54 | 610 |
| DC | Dechang, Sichuan | 29 | 102.32 | 26.40 | 1325 |
| XY | Xingyi, Guizhou | 15 | 104.54 | 25.06 | 1160 |
| XL | Xilin, Guangxi | 24 | 105.05 | 24.29 | 899 |
| WM | Wangmo, Guizhou | 30 | 106.05 | 25.10 | 500 |
| YR | Yongren, Yunnan | 27 | 101.32 | 25.01 | 1539 |
| BS | Baoshan, Yunnan | 11 | 99.06 | 25.04 | 1513 |
| LD | Luodian, Guizhou | 14 | 106.44 | 25.25 | 814 |
| TL | Tianlin, Guangxi | 26 | 106.13 | 24.17 | 792 |
| CH | Ceheng, Guizhou | 22 | 105.48 | 24.59 | 730 |
| HD | Huidong, Sichuan | 25 | 102.09 | 27.23 | 1862 |
| JX | Jingxian, Anhui | 15 | 118.24 | 30.41 | 366 |
| HS | Huangshan, Anhui | 6 | 118.08 | 30.16 | 499 |
| WY | Wuyishan, Fujian | 9 | 117.42 | 28.18 | 1200 |
| JG | Jinggangshan, Jiangxi | 5 | 114.17 | 26.44 | 400 |
| XE | Xuanen, Hubei | 9 | 109.28 | 30.17 | 632 |
| GS | Guanshan, Jiangxi | 11 | 115.26 | 27.19 | 337 |
| PW | Puwen, Yunnan | 21 | 101.04 | 22.23 | 890 |
| MT | Maotoushan, Jiangxi | 9 | 117.03 | 27.41 | 1000 |
| YF | Yunfu, Guangdong | 18 | 111.34 | 22.46 | 340 |
| HP | Hupingshan, Hunan | 3 | 111.41 | 29.01 | 436 |
| LC | Lechang, Guangdong | 15 | 113.20 | 25.07 | 359 |
| Primers | Sequence 5′-3′ |
|---|---|
| psbA-trnH | GTTATGCATGAACGTAATGCTC |
| CGCGCATGGTGGATTCACAATCC | |
| trnT-trnL | CATTACAAATGCGATGCTCT |
| TCTACCGATTTCGCCATATC | |
| trnL-trnL | CGAAATCGGTAGACGCTACG |
| GGGGATAGAGGGACTTGAAC | |
| trnL-trnF | GGTTCAAGTCCCTCTATCCC |
| ATTTGAACTGGTGACACGAG |
| Population | Observed Haplotype Diversity (h) | Nucleotide Diversity per Site (π) | Number of Haplotypes # | Types of Haplotypes |
|---|---|---|---|---|
| XJ | 0.5679 | 0.0015 | 3 + 0 | H1 H2 H3 |
| LL | 0.6600 | 0.0070 | 2 + 2 | H4 H6 H47 |
| SM | 0.3140 | 0.0011 | 2 + 1 | H9 H10 H47 |
| SC | 0.5139 | 0.0017 | 4 + 0 | H1 H2 H3 H11 |
| CB | 0.8809 | 0.0084 | 6 + 5 | H12 H13 H14 H16 H17 H19 H47 |
| XN | 0.9091 | 0.0712 | 5 + 6 | H2 H17 H26 H27 H28 H47 |
| NP | 0.8025 | 0.0070 | 4 + 2 | H1 H26 H28 H32 H47 |
| JL | 0.8765 | 0.0118 | 6 + 5 | H1 H2 H27 H28 H32 H35 H47 |
| DC | 0.9489 | 0.0186 | 3 + 21 | H5 H43 H44 H47 |
| XY | 0.5867 | 0.0046 | 4 + 1 | H4 H6 H7 H8 H47 |
| XL | 0.7396 | 0.0088 | 4 + 4 | H4 H6 H7 H15 H47 |
| WM | 0.9289 | 0.0321 | 7 + 15 | H4 H6 H18 H20 H21 H22 H23 H47 |
| YR | 0.9163 | 0.0087 | 8 + 11 | H9 H10 H24 H25 H29 H30 H31 H33 H47 |
| BS | 0.8926 | 0.0526 | 4 + 6 | H10 H29 H30 H33 H47 |
| LD | 0.8980 | 0.0154 | 6 + 6 | H6 H7 H18 H21 H22 H34 H47 |
| TL | 0.9586 | 0.0257 | 4 + 21 | H22 H34 H36 H37 H47 |
| CH | 0.9504 | 0.0263 | 3 + 18 | H8 H37 H38 H47 |
| HD | 0.9504 | 0.0214 | 1 + 22 | H39 H47 |
| JX | 0.3467 | 0.0161 | 2 + 2 | H2 H3 H47 |
| HS | 0.8333 | 0.1398 | 0 + 6 | H47 |
| WY | 0.8148 | 0.0922 | 2 + 5 | H40 H41 H47 |
| JG | 0.8000 | 0.0768 | 0 + 5 | H47 |
| XE | 0.8889 | 0.0817 | 4 + 5 | H1 H35 H41 H42 H47 |
| GS | 0.9091 | 0.1133 | 4 + 7 | H40 H42 H45 H46 H47 |
| PW | 0.8753 | 0.0144 | 5 + 8 | H9 H10 H25 H31 H33 H47 |
| MT | 0.8889 | 0.0164 | 2 + 7 | H2 H45 H47 |
| YF | 0.0000 | 0.0000 | 1 + 0 | H9 |
| HP | 0.4444 | 0.0027 | 2 + 0 | H2 H3 |
| LC | 0.3378 | 0.0013 | 3 + 0 | H2 H9 H46 |
| Total | 0.9767 | 0.0303 | 46 + 193 | H1–H47 |
| Population | Observed Haplotype Diversity (h) | Nucleotide Diversity per Site (π) | Number of Haplotypes # | Types of Haplotypes |
|---|---|---|---|---|
| XJ | 0.0000 | 0.0000 | 1 + 0 | H1 |
| LL | 0.8200 | 0.0031 | 4 + 3 | H2 H3 H4 H5 H47 |
| SM | 0.7934 | 0.0076 | 4 + 2 | H3 H4 H9 H11 H47 |
| SC | 0.0000 | 0.0000 | 1 + 0 | H1 |
| CB | 0.3856 | 0.0008 | 2 + 0 | H1 H13 |
| XN | 0.0000 | 0.0000 | 1 + 0 | H1 |
| NP | 0.0000 | 0.0000 | 1 + 0 | H1 |
| JL | 0.0000 | 0.0000 | 1 + 0 | H1 |
| DC | 0.8252 | 0.0049 | 7 + 9 | H16 H17 H19 H21 H22 H25 H28 H47 |
| XY | 0.9244 | 0.0084 | 7 + 7 | H19 H31 H32 H34 H35 H39 H40 H47 |
| XL | 0.9375 | 0.0073 | 11 + 9 | H6 H7 H8 H10 H12 H14 H31 H32 H34 H35 H39 H47 |
| WM | 0.9489 | 0.0085 | 12 + 13 | H8 H15 H18 H20 H23 H24 H26 H31 H32 H34 H35 H39 H47 |
| YR | 0.9465 | 0.0076 | 14 + 8 | H2 H5 H7 H8 H9 H11 H12 H14 H15 H27 H29 H32 H34 H35 H47 |
| BS | 0.9091 | 0.0066 | 8 + 3 | H6 H7 H10 H14 H29 H31 H32 H35 H47 |
| LD | 0.7653 | 0.0028 | 6 + 1 | H10 H15 H29 H32 H34 H35 H47 |
| TL | 0.7781 | 0.0032 | 9 + 3 | H12 H23 H24 H26 H29 H30 H34 H35 H40 H47 |
| CH | 0.9215 | 0.0043 | 8 + 8 | H18 H24 H30 H33 H34 H35 H36 H40 H47 |
| HD | 0.8928 | 0.0079 | 7 + 8 | H16 H25 H28 H37 H38 H41 H42 H47 |
| JX | 0.1244 | 0.0005 | 2 + 0 | H1 H9 |
| HS | 0.2778 | 0.0019 | 1 + 1 | H1 H47 |
| WY | 0.4938 | 0.0086 | 2 + 2 | H1 H36 H47 |
| JG | 0.7200 | 0.0154 | 3 + 1 | H32 H35 H43 H47 |
| XE | 0.4938 | 0.0041 | 2 + 1 | H1 H44 H47 |
| GS | 0.2975 | 0.0006 | 2 + 0 | H1 H43 |
| PW | 0.9070 | 0.0077 | 9 + 8 | H3 H4 H10 H12 H33 H34 H35 H45 H46 H47 |
| MT | 0.0000 | 0.0000 | 1 + 0 | H1 |
| YF | 0.9321 | 0.0084 | 6 + 10 | H8 H12 H27 H32 H35 H46 H47 |
| HP | 0.7778 | 0.0013 | 2 + 0 | H1 H44 |
| LC | 0.2400 | 0.0027 | 3 + 0 | H1 H9 H32 |
| Total | 0.8999 | 0.0189 | 46 + 97 | H1–H47 |
| Marker | Source of Variation | d.f. | Sum of Squares | Variance Component | Percentage of Variance (%) | Φst | p-Value |
|---|---|---|---|---|---|---|---|
| psbA-trnH+ trnL-trnL | Among population | 28 | 2110.429 | 4.5357 | 42.84 | 0.4284 | 0.0000 |
| Within population | 418 | 2529.542 | 6.0515 | 57.16 | |||
| Total | 446 | 4639.971 | 10.5873 | ||||
| psbA-trnH | Among population | 28 | 910.769 | 1.8021 | 26.55 | 0.2655 | 0.0000 |
| Within population | 418 | 2083.932 | 4.9855 | 73.45 | |||
| Total | 446 | 2994.7 | 6.7876 | ||||
| trnL-trnL | Among population | 28 | 1202.878 | 2.7344 | 70.04 | 0.7004 | 0.0000 |
| Within population | 418 | 488.911 | 1.1696 | 29.96 | |||
| Total | 446 | 1691.79 | 3.9040 |
| Population | Tajima’s D (p-Value) | Fu’s F (p-Value) | Mismatch Distribution | ||||
|---|---|---|---|---|---|---|---|
| SSD (p-Value) | Rag (p-Value) | θ0 | θ1 | τ | |||
| XJ | −0.0638 (0.4) | −0.2393 (0.346) | 0.0443 (0.15) | 0.2677 (0.15) | 0 | 3407.185 | 0.936 |
| LL | −1.6802 (0.038) | 1.6590 (0.811) | 0.0394 (0.4) | 0.0523 (0.8) | 3.029 | 6914.97 | 1.172 |
| SM | −1.5999 (0.044) | −0.5370 (0.129) | 0.0219 (0.55) | 0.0575 (0.45) | 0 | 14.16 | 5.23 |
| SC | 0.6879 (0.796) | −1.0476 (0.106) | 0.0079 (0.7) | 0.0870 (0.85) | 0 | 1.988 | 1.479 |
| CB | 0.4359 (0.726) | −2.046 (0.161) | 0.0043 (0.85) | 0.0168 (0.95) | 0 | 6.392 | 3.711 |
| XN | −0.6741 (0.244) | −1.441 (0.145) | 0.2121 (0) | 0.0519 (1) | 3.025 | 3641.256 | 1.045 |
| NP | 0.7724 (0.794) | −0.8396 (0.24) | 0.0149 (0.45) | 0.0709 (0.95) | 0.002 | 4.962 | 2.135 |
| JL | −1.1649 (0.108) | −1.967 (0.179) | 0.0541 (0.55) | 0.1139 (0.4) | 0 | 2.317 | 15.604 |
| DC | −1.8304 (0.015) | −12.3959 (0) | 0.0031 (0.6) | 0.0098 (0.65) | 3.7 | 6934.956 | 3.33 |
| XY | −0.9887 (0.143) | 0.2473 (0.572) | 0.0055 (0.6) | 0.2176 (0.6) | 0.083 | 55.029 | 6.111 |
| XL | −0.8636 (0.202) | 0.4259 (0.616) | 0.0038 (0.6) | 0.0146 (0.85) | 1.9 | 3414.978 | 3.029 |
| WM | −2.4485 (0) | −3.9285 (0.092) | 0.0423 (0) | 0.0072 (1) | 3.938 | 8040.089 | 6.094 |
| YR | −0.3648 (0.424) | −14.8439 (0) | 0.0009 (0.85) | 0.0072 (0.95) | 1.22 | 3045 | 6.717 |
| BS | −2.0701 (0.001) | −0.3533 (0.356) | 0.0367 (0.2) | 0.0893 (0.05) | 5.5 | 3414.978 | 7.451 |
| LD | −0.8212 (0.217) | −4.4225 (0.018) | 0.0085 (0.8) | 0.0200 (0.7) | 7.3 | 6854.957 | 2.77 |
| TL | −1.6168 (0.028) | −15.1810 (0) | 0.0421 (0) | 0.0079 (1) | 3.825 | 7174.983 | 5.74 |
| CH | −1.0965 (0.129) | −10.8408 (0.001) | 0.0086 (0.5) | 0.0141 (0.55) | 0 | 64.688 | 11.434 |
| HD | −0.4642 (0.378) | −13.3933 (0) | 0.0137 (0.05) | 0.0089 (0.7) | 5.4 | 3935.014 | 10.283 |
| JX | −2.0409 (0) | 6.0964 (0.989) | 0.0195 (0.4) | 0.2302 (0.55) | 0 | 0.523 | 3 |
| HS | −0.1419 (0.456) | 1.2436 (0.423) | 0.0583 (0.6) | 0.0889 (0.9) | 67.4 | 219.363 | 12.262 |
| WY | −0.3811 (0.385) | 3.2794 (0.923) | 0.0613 (0.7) | 0.0439 (0.85) | 0.002 | 9.545 | 80.68 |
| JG | 0.8909 (0.805) | 1.1153 (0.466) | 0.0949 (0.35) | 0.2200 (0.2) | 72 | 157.478 | 43.17 |
| XE | −0.2181 (0.446) | −0.4294 (0.26) | 0.0371 (0.8) | 0.0525 (0.75) | 28.8 | 3515.191 | 0.895 |
| GS | −1.1527 (0.114) | −0.6481 (0.222) | 0.0562 (0.25) | 0.0856 (0.25) | 5.05 | 3474.992 | 1.227 |
| PW | −1.7853 (0.018) | −2.7789 (0.092) | 0.0029 (0.8) | 0.0063 (0.95) | 2.602 | 61.328 | 6.117 |
| MT | −0.1726 (0.459) | −4.0639 (0.008) | 0.0152 (0.3) | 0.0478 (0.35) | 1.9 | 80.986 | 5.871 |
| YF | 0 (1) | 0 (1) | 0.0686 (0) | 0.0212 (0.9) | 2.406 | 7354.997 | 2.4 |
| HP | 0 (1) | 0.2007 (0.377) | 0.1643 (0.3) | 0.6667 (0.6) | 0 | 6829.957 | 1.629 |
| LC | −0.9481 (0.22) | −0.0064 (0.36) | 0.3878 (0) | 0.0863 (1) | 1.143 | 3414.622 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.-Y.; Hu, Y.; Lv, Y.-W.; Xiao, Y.; He, Z.-H.; Wu, C.; Hu, X.-S. Population Structure and Genetic Diversity of the Toona ciliata (Meliaceae) Complex Assayed with Chloroplast DNA Markers. Genes 2024, 15, 320. https://doi.org/10.3390/genes15030320
Wang Z-Y, Hu Y, Lv Y-W, Xiao Y, He Z-H, Wu C, Hu X-S. Population Structure and Genetic Diversity of the Toona ciliata (Meliaceae) Complex Assayed with Chloroplast DNA Markers. Genes. 2024; 15(3):320. https://doi.org/10.3390/genes15030320
Chicago/Turabian StyleWang, Zi-Yun, Ying Hu, Yan-Wen Lv, Yu Xiao, Zi-Han He, Chao Wu, and Xin-Sheng Hu. 2024. "Population Structure and Genetic Diversity of the Toona ciliata (Meliaceae) Complex Assayed with Chloroplast DNA Markers" Genes 15, no. 3: 320. https://doi.org/10.3390/genes15030320
APA StyleWang, Z.-Y., Hu, Y., Lv, Y.-W., Xiao, Y., He, Z.-H., Wu, C., & Hu, X.-S. (2024). Population Structure and Genetic Diversity of the Toona ciliata (Meliaceae) Complex Assayed with Chloroplast DNA Markers. Genes, 15(3), 320. https://doi.org/10.3390/genes15030320





