Understanding the Variability of 22q11.2 Deletion Syndrome: The Role of Epigenetic Factors
Abstract
1. Introduction
2. Main Clinical Features
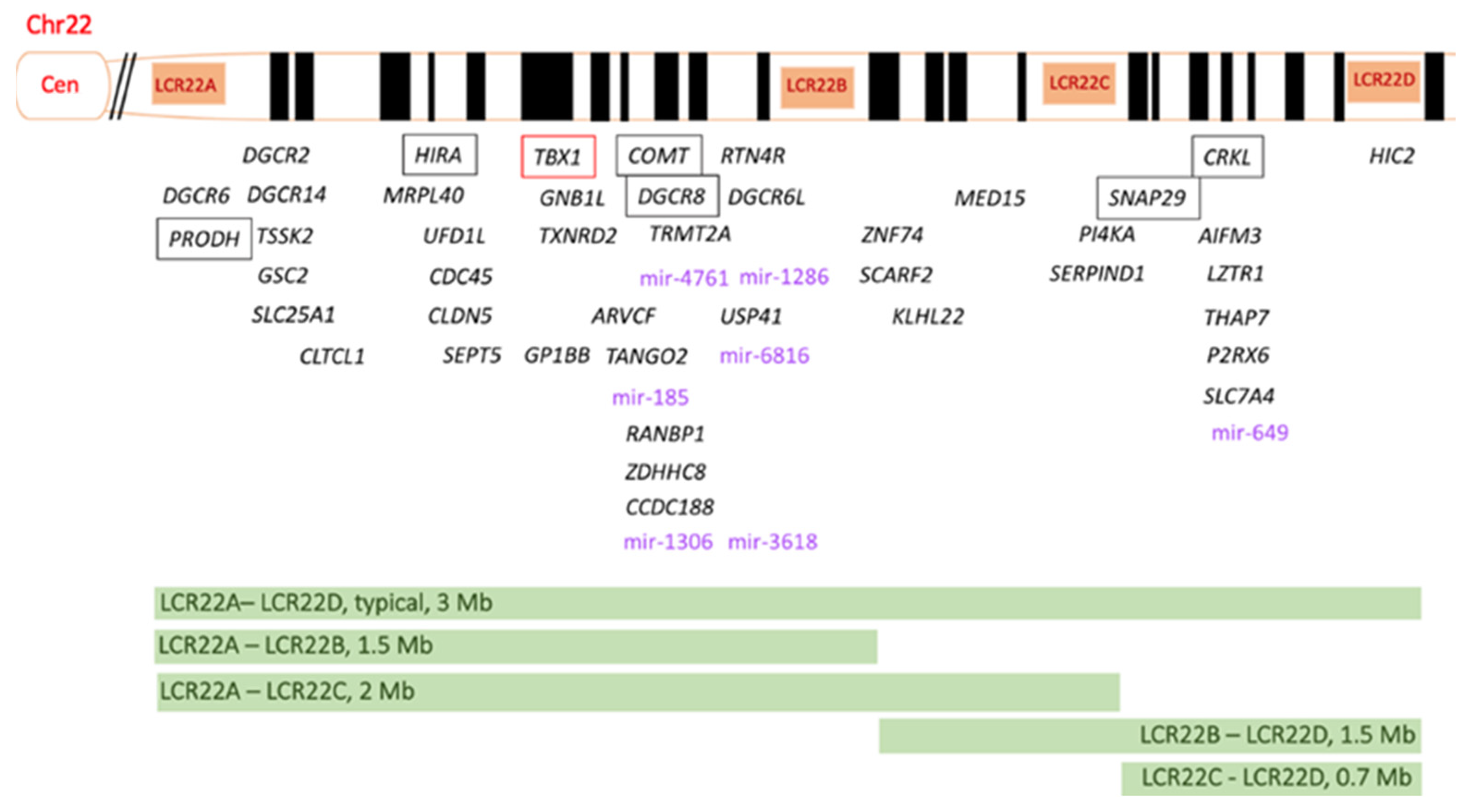
3. Genetic Features of 22q11.2DS
4. Epigenetic Mechanisms Implicated in the Syndrome
4.1. Micro-RNA Profile
4.2. Methylation Profile
5. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McDonald-McGinn, D.M.; Sullivan, K.E.; Marino, B.; Philip, N.; Swillen, A.; Vorstman, J.A.; Zackai, E.H.; Emanuel, B.S.; Vermeesch, J.R.; Morrow, B.E.; et al. 22q11.2 deletion syndrome. Nat. Rev. Dis. Prim. 2015, 1, 15071. [Google Scholar] [CrossRef]
- Grati, F.R.; Molina Gomes, D.; Ferreira, J.C.; Dupont, C.; Alesi, V.; Gouas, L.; Horelli-Kuitunen, N.; Choy, K.W.; García-Herrero, S.; de la Vega, A.G.; et al. Prevalence of recurrent pathogenic microdeletions and microduplications in over 9500 pregnancies. Prenat. Diagn. 2015, 35, 801–809. [Google Scholar] [CrossRef]
- Gross, S.J.; Stosic, M.; McDonald-McGinn, D.M.; Bassett, A.S.; Norvez, A.; Dhamankar, R.; Kobara, K.; Kirkizlar, E.; Zimmermann, B.; Wayham, N.; et al. Clinical experience with single-nucleotide polymorphism-based non-invasive prenatal screening for 22q11.2 deletion syndrome. Ultrasound Obstet. Gynecol. 2016, 47, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Blagojevic, C.; Heung, T.; Theriault, M.; Tomita-Mitchell, A.; Chakraborty, P.; Kernohan, K.; Bulman, D.E.; Bassett, A.S. Estimate of the contemporary live-birth prevalence of recurrent 22q11.2 deletions: A cross-sectional analysis from population-based newborn screening. CMAJ Open 2021, 9, E802–E809. [Google Scholar] [CrossRef]
- Bevilacqua, E.; Jani, J.C.; Chaoui, R.; Suk, E.A.; Palma-Dias, R.; Ko, T.M.; Warsof, S.; Stokowski, R.; Jones, K.J.; Grati, F.R.; et al. Performance of a targeted cell-free DNA prenatal test for 22q11.2 deletion in a large clinical cohort. Ultrasound Obstet. Gynecol. 2021, 58, 597–602. [Google Scholar] [CrossRef] [PubMed]
- Dar, P.; Norton, M.E. Performance of noninvasive prenatal screening for 22q11.2 deletion syndrome in the SMART study. Am. J. Obstet. Gynecol. 2022, 227, 124–125. [Google Scholar] [CrossRef]
- Kagan, K.O.; Hoopmann, M.; Pfaff, T.; Prodan, N.; Wagner, P.; Schmid, M.; Dufke, A.; Mau-Holzmann, U.; Brucker, S.; Marcato, L.; et al. First Trimester Screening for Common Trisomies and Microdeletion 22q11.2 Syndrome Using Cell-Free DNA: A Prospective Clinical Study. Fetal Diagn. Ther. 2020, 47, 841–852. [Google Scholar] [CrossRef]
- Blagowidow, N.; Nowakowska, B.; Schindewolf, E.; Grati, F.R.; Putotto, C.; Breckpot, J.; Swillen, A.; Crowley, T.B.; Loo, J.C.Y.; Lairson, L.A.; et al. Prenatal Screening and Diagnostic Considerations for 22q11.2 Microdeletions. Genes 2023, 14, 160. [Google Scholar] [CrossRef]
- Kwan, A.; Abraham, R.S.; Currier, R.; Brower, A.; Andruszewski, K.; Abbott, J.K.; Baker, M.; Ballow, M.; Bartoshesky, L.E.; Bonilla, F.A.; et al. Newborn screening for severe combined immunodeficiency in 11 screening programs in the United States. JAMA 2014, 312, 729–738. [Google Scholar] [CrossRef] [PubMed]
- Barry, J.C.; Crowley, T.B.; Jyonouchi, S.; Heimall, J.; Zackai, E.H.; Sullivan, K.E.; McDonald-McGinn, D.M. Identification of 22q11.2 Deletion Syndrome via Newborn Screening for Severe Combined Immunodeficiency. J. Clin. Immunol. 2017, 37, 476–485. [Google Scholar] [CrossRef]
- Palmer, L.D.; McManus, Z.; Heung, T.; McAlpine, G.; Blagojevic, C.; Corral, M.; Bassett, A.S. Reproductive Outcomes in Adults with 22q11.2 Deletion Syndrome. Genes 2022, 13, 2126. [Google Scholar] [CrossRef] [PubMed]
- Kapadia, R.K.; Bassett, A.S. Recognizing a common genetic syndrome: 22q11.2 deletion syndrome. CMAJ 2008, 178, 391–393. [Google Scholar] [CrossRef][Green Version]
- Delio, M.; Guo, T.; McDonald-McGinn, D.M.; Zackai, E.; Herman, S.; Kaminetzky, M.; Higgins, A.M.; Coleman, K.; Chow, C.; Jalbrzikowski, M.; et al. Enhanced maternal origin of the 22q11.2 deletion in velocardiofacial and DiGeorge syndromes. Am. J. Hum. Genet. 2013, 92, 439–447. [Google Scholar] [CrossRef]
- Costain, G.; Chow, E.W.; Silversides, C.K.; Bassett, A.S. Sex differences in reproductive fitness contribute to preferential maternal transmission of 22q11.2 deletions. J. Med. Genet. 2011, 48, 819–824. [Google Scholar] [CrossRef] [PubMed]
- Cancrini, C.; Puliafito, P.; Digilio, M.C.; Soresina, A.; Martino, S.; Rondelli, R.; Consolini, R.; Ruga, E.M.; Cardinale, F.; Finocchi, A.; et al. Clinical features and follow-up in patients with 22q11.2 deletion syndrome. J. Pediatr. 2014, 164, 1475–1480.e2. [Google Scholar] [CrossRef]
- Robin, N.H.; Shprintzen, R.J. Defining the clinical spectrum of deletion 22q11.2. J. Pediatr. 2005, 147, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Shprintzen, R.J.; Higgins, A.M.; Antshel, K.; Fremont, W.; Roizen, N.; Kates, W. Velo-cardio-facial syndrome. Curr. Opin. Pediatr. 2005, 17, 725–730. [Google Scholar] [CrossRef]
- McDonald-McGinn, D.M.; Tonnesen, M.K.; Laufer-Cahana, A.; Finucane, B.; Driscoll, D.A.; Emanuel, B.S.; Zackai, E.H. Phenotype of the 22q11.2 deletion in individuals identified through an affected relative: Cast a wide FISHing net! Genet. Med. 2001, 3, 23–29. [Google Scholar] [CrossRef]
- Cirillo, E.; Giardino, G.; Gallo, V.; Puliafito, P.; Azzari, C.; Bacchetta, R.; Cardinale, F.; Cicalese, M.P.; Consolini, R.; Martino, S.; et al. Intergenerational and intrafamilial phenotypic variability in 22q11.2 deletion syndrome subjects. BMC Med. Genet. 2014, 15, 1. [Google Scholar] [CrossRef]
- Vergaelen, E.; Swillen, A.; Van Esch, H.; Claes, S.; Van Goethem, G.; Devriendt, K. 3 generation pedigree with paternal transmission of the 22q11.2 deletion syndrome: Intrafamilial phenotypic variability. Eur. J. Med. Genet. 2015, 58, 244–248. [Google Scholar] [CrossRef]
- Morrow, B.E.; McDonald-McGinn, D.M.; Emanuel, B.S.; Vermeesch, J.R.; Scambler, P.J. Molecular genetics of 22q11.2 deletion syndrome. Am. J. Med. Genet. Part A 2018, 176, 2070–2081. [Google Scholar] [CrossRef]
- Szczawińska-Popłonyk, A.; Schwartzmann, E.; Chmara, Z.; Głukowska, A.; Krysa, T.; Majchrzycki, M.; Olejnicki, M.; Ostrowska, P.; Babik, J. Chromosome 22q11.2 Deletion Syndrome: A Comprehensive Review of Molecular Genetics in the Context of Multidisciplinary Clinical Approach. Int. J. Mol. Sci. 2023, 24, 8317. [Google Scholar] [CrossRef] [PubMed]
- McDonald-McGinn, D.M.; Sullivan, K.E. Chromosome 22q11.2 deletion syndrome (DiGeorge syndrome/velocardiofacial syndrome). Medicine 2011, 90, 1–18. [Google Scholar] [CrossRef]
- Repetto, G.M.; Guzmán, M.L.; Delgado, I.; Loyola, H.; Palomares, M.; Lay-Son, G.; Vial, C.; Benavides, F.; Espinoza, K.; Alvarez, P. Case fatality rate and associated factors in patients with 22q11 microdeletion syndrome: A retrospective cohort study. BMJ Open 2014, 4, e005041. [Google Scholar] [CrossRef]
- Sullivan, K.E. Chromosome 22q11.2 deletion syndrome and DiGeorge syndrome. Immunol. Rev. 2019, 287, 186–201. [Google Scholar] [CrossRef] [PubMed]
- Karbarz, M. Consequences of 22q11.2 Microdeletion on the Genome, Individual and Population Levels. Genes 2020, 11, 977. [Google Scholar] [CrossRef]
- Bertini, V.; Azzarà, A.; Legitimo, A.; Milone, R.; Battini, R.; Consolini, R.; Valetto, A. Deletion Extents Are Not the Cause of Clinical Variability in 22q11.2 Deletion Syndrome: Does the Interaction between DGCR8 and miRNA-CNVs Play a Major Role? Front. Genet. 2017, 8, 47. [Google Scholar] [CrossRef] [PubMed]
- Bartik, L.E.; Hughes, S.S.; Tracy, M.; Feldt, M.M.; Zhang, L.; Arganbright, J.; Kaye, A. 22q11.2 duplications: Expanding the clinical presentation. Am. J. Med. Genet. Part A 2022, 188, 779–787. [Google Scholar] [CrossRef]
- Yu, A.; Turbiville, D.; Xu, F.; Ray, J.W.; Britt, A.D.; Lupo, P.J.; Jain, S.K.; Shattuck, K.E.; Robinson, S.S.; Dong, J. Genotypic and phenotypic variability of 22q11.2 microduplications: An institutional experience. Am. J. Med. Genet. Part A 2019, 179, 2178–2189. [Google Scholar] [CrossRef]
- Meneses, Z.; Durant, J.; Ale, H. The Unique Experience of a New Multidisciplinary Program for 22q Deletion and Duplication Syndromes in a Community Hospital in Florida: A Reaffirmation That Multidisciplinary Care Is Essential for Best Outcomes in These Patients. Genes 2022, 13, 1949. [Google Scholar] [CrossRef]
- Goldmuntz, E. 22q11.2 deletion syndrome and congenital heart disease. Am. J. Med. Genet. Part C Semin. Med. Genet. 2020, 184, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Peyvandi, S.; Lupo, P.J.; Garbarini, J.; Woyciechowski, S.; Edman, S.; Emanuel, B.S.; Mitchell, L.E.; Goldmuntz, E. 22q11.2 deletions in patients with conotruncal defects: Data from 1610 consecutive cases. Pediatr. Cardiol. 2013, 34, 1687–1694. [Google Scholar] [CrossRef]
- Putotto, C.; Pugnaloni, F.; Unolt, M.; Maiolo, S.; Trezzi, M.; Digilio, M.C.; Cirillo, A.; Limongelli, G.; Marino, B.; Calcagni, G.; et al. 22q11.2 Deletion Syndrome: Impact of Genetics in the Treatment of Conotruncal Heart Defects. Children 2022, 9, 772. [Google Scholar] [CrossRef] [PubMed]
- Yeoh, T.Y.; Scavonetto, F.; Hamlin, R.J.; Burkhart, H.M.; Sprung, J.; Weingarten, T.N. Perioperative management of patients with DiGeorge syndrome undergoing cardiac surgery. J. Cardiothorac. Vasc. Anesth. 2014, 28, 983–989. [Google Scholar] [CrossRef] [PubMed]
- de Rinaldis, C.P.; Butensky, A.; Patel, S.; Edman, S.; Wasserman, M.; McGinn, D.E.; Bailey, A.; Zackai, E.H.; Crowley, T.B.; McDonald-McGinn, D.M.; et al. Aortic Root Dilation in Patients with 22q11.2 Deletion Syndrome without Intracardiac Anomalies. Pediatr. Cardiol. 2021, 42, 1594–1600. [Google Scholar] [CrossRef] [PubMed]
- Blagojevic, C.; Heung, T.; Malecki, S.; Ying, S.; Cancelliere, S.; Hegele, R.A.; Bassett, A.S. Hypertriglyceridemia in young adults with a 22q11.2 microdeletion. Eur. J. Endocrinol. 2022, 187, 91–99. [Google Scholar] [CrossRef] [PubMed]
- McDonald-McGinn, D.M.; Hain, H.S.; Emanuel, B.S.; Zackai, E.H. 22q11.2 Deletion Syndrome. In GeneReviews®; Adam, M.P., Feldman, J., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Gripp, K.W., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Campbell, I.M.; Sheppard, S.E.; Crowley, T.B.; McGinn, D.E.; Bailey, A.; McGinn, M.J.; Unolt, M.; Homans, J.F.; Chen, E.Y.; Salmons, H.I.; et al. What is new with 22q? An update from the 22q and You Center at the Children’s Hospital of Philadelphia. Am. J. Med. Genet. Part A 2018, 176, 2058–2069. [Google Scholar] [CrossRef]
- Funato, N. Craniofacial Phenotypes and Genetics of DiGeorge Syndrome. J. Dev. Biol. 2022, 10, 18. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.H.; Chien, Y.H.; Lu, M.Y.; Hu, Y.C.; Lee, J.H.; Wang, L.C.; Lin, Y.T.; Yang, Y.H.; Chiang, B.L. Clinical and Immunological Defects and Outcomes in Patients with Chromosome 22q11.2 Deletion Syndrome. J. Clin. Immunol. 2022, 42, 1721–1729. [Google Scholar] [CrossRef]
- Levy-Shraga, Y.; Gothelf, D.; Goichberg, Z.; Katz, U.; Somech, R.; Pinhas-Hamiel, O.; Modan-Moses, D. Growth characteristics and endocrine abnormalities in 22q11.2 deletion syndrome. Am. J. Med. Genet. Part A 2017, 173, 1301–1308. [Google Scholar] [CrossRef]
- Óskarsdóttir, S.; Boot, E.; Crowley, T.B.; Loo, J.C.Y.; Arganbright, J.M.; Armando, M.; Baylis, A.L.; Breetvelt, E.J.; Castelein, R.M.; Chadehumbe, M.; et al. Updated clinical practice recommendations for managing children with 22q11.2 deletion syndrome. Genet. Med. 2023, 25, 100338. [Google Scholar] [CrossRef]
- Boot, E.; Óskarsdóttir, S.; Loo, J.C.Y.; Crowley, T.B.; Orchanian-Cheff, A.; Andrade, D.M.; Arganbright, J.M.; Castelein, R.M.; Cserti-Gazdewich, C.; de Reuver, S.; et al. Updated clinical practice recommendations for managing adults with 22q11.2 deletion syndrome. Genet. Med. 2023, 25, 100344. [Google Scholar] [CrossRef]
- Shugar, A.L.; Shapiro, J.M.; Cytrynbaum, C.; Hedges, S.; Weksberg, R.; Fishman, L. An increased prevalence of thyroid disease in children with 22q11.2 deletion syndrome. Am. J. Med. Genet. Part A 2015, 167, 1560–1564. [Google Scholar] [CrossRef]
- Weinzimer, S.A. Endocrine aspects of the 22q11.2 deletion syndrome. Genet. Med. 2001, 3, 19–22. [Google Scholar] [CrossRef]
- Pignata, L.; Sparago, A.; Palumbo, O.; Andreucci, E.; Lapi, E.; Tenconi, R.; Carella, M.; Riccio, A.; Cerrato, F. Mosaic Segmental and Whole-Chromosome Upd(11)mat in Silver-Russell Syndrome. Genes 2021, 12, 581. [Google Scholar] [CrossRef]
- Mustillo, P.J.; Sullivan, K.E.; Chinn, I.K.; Notarangelo, L.D.; Haddad, E.; Davies, E.G.; de la Morena, M.T.; Hartog, N.; Yu, J.E.; Hernandez-Trujillo, V.P.; et al. Clinical Practice Guidelines for the Immunological Management of Chromosome 22q11.2 Deletion Syndrome and Other Defects in Thymic Development. J. Clin. Immunol. 2023, 43, 247–270. [Google Scholar] [CrossRef] [PubMed]
- Giardino, G.; Radwan, N.; Koletsi, P.; Morrogh, D.M.; Adams, S.; Ip, W.; Worth, A.; Jones, A.; Meyer-Parsonson, I.; Gaspar, H.B.; et al. Clinical and immunological features in a cohort of patients with partial DiGeorge syndrome followed at a single center. Blood 2019, 133, 2586–2596. [Google Scholar] [CrossRef] [PubMed]
- McLean-Tooke, A.; Spickett, G.P.; Gennery, A.R. Immunodeficiency and autoimmunity in 22q11.2 deletion syndrome. Scand. J. Immunol. 2007, 66, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, D.R.; Demirdag, Y.Y.; Marsh, R.A.; Sullivan, K.E.; Orange, J.S.; The USIDNET Consortium. Relationship Between Severity of T Cell Lymphopenia and Immune Dysregulation in Patients with DiGeorge Syndrome (22q11.2 Deletions and/or Related TBX1 Mutations): A USIDNET Study. J. Clin. Immunol. 2021, 41, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Markert, M.L.; Gupton, S.E.; McCarthy, E.A. Experience with cultured thymus tissue in 105 children. J. Allergy Clin. Immunol. 2022, 149, 747–757. [Google Scholar] [CrossRef] [PubMed]
- Markert, M.L.; Alexieff, M.J.; Li, J.; Sarzotti, M.; Ozaki, D.A.; Devlin, B.H.; Sempowski, G.D.; Rhein, M.E.; Szabolcs, P.; Hale, L.P.; et al. Complete DiGeorge syndrome: Development of rash, lymphadenopathy, and oligoclonal T cells in 5 cases. J. Allergy Clin. Immunol. 2004, 113, 734–741. [Google Scholar] [CrossRef] [PubMed]
- Hankey, P.B.; Ghulmiyyah, J.; Yeh, H.W.; Tracy, M.; Arganbright, J. Airway anomalies in patients with 22q11.2 deletion syndrome: A scoping review. Int. J. Pediatr. Otorhinolaryngol. 2022, 163, 111373. [Google Scholar] [CrossRef] [PubMed]
- Framme, J.L.; Lundqvist, C.; Lundell, A.C.; van Schouwenburg, P.A.; Lemarquis, A.L.; Thörn, K.; Lindgren, S.; Gudmundsdottir, J.; Lundberg, V.; Degerman, S.; et al. Long-Term Follow-Up of Newborns with 22q11 Deletion Syndrome and Low TRECs. J. Clin. Immunol. 2022, 42, 618–633. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, K.E.; McDonald-McGinn, D.M.; Driscoll, D.A.; Zmijewski, C.M.; Ellabban, A.S.; Reed, L.; Emanuel, B.S.; Zackai, E.H.; Athreya, B.H.; Keenan, G. Juvenile rheumatoid arthritis-like polyarthritis in chromosome 22q11.2 deletion syndrome (DiGeorge anomalad/velocardiofacial syndrome/conotruncal anomaly face syndrome). Arthritis Rheum. 1997, 40, 430–436. [Google Scholar] [CrossRef]
- Kratz, C.P.; Niehues, T.; Lyding, S.; Heusch, A.; Janssen, G.; Göbel, U. Evans syndrome in a patient with chromosome 22q11.2 deletion syndrome: A case report. Pediatr. Hematol. Oncol. 2003, 20, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, S.; McDonald-McGinn, D.M.; Zackai, E.; Sullivan, K.E. Thrombocytopenia in patients with chromosome 22q11.2 deletion syndrome. J. Pediatr. 2003, 143, 277–278. [Google Scholar] [CrossRef]
- Kawame, H.; Adachi, M.; Tachibana, K.; Kurosawa, K.; Ito, F.; Gleason, M.M.; Weinzimer, S.; Levitt-Katz, L.; Sullivan, K.; McDonald-McGinn, D.M. Graves’ disease in patients with 22q11.2 deletion. J. Pediatr. 2001, 139, 892–895. [Google Scholar] [CrossRef]
- Ricci, S.; Sarli, W.M.; Lodi, L.; Canessa, C.; Lippi, F.; Azzari, C.; Stagi, S. Characterization of Autoimmune Thyroid Disease in a Cohort of 73 Paediatric Patients Affected by 22q11.2 Deletion Syndrome: Longitudinal Single-Centre Study. Genes 2022, 13, 1552. [Google Scholar] [CrossRef]
- Murphy, K.C.; Jones, L.A.; Owen, M.J. High rates of schizophrenia in adults with velo-cardio-facial syndrome. Arch. Gen. Psychiatry 1999, 56, 940–945. [Google Scholar] [CrossRef]
- Green, T.; Gothelf, D.; Glaser, B.; Debbane, M.; Frisch, A.; Kotler, M.; Weizman, A.; Eliez, S. Psychiatric disorders and intellectual functioning throughout development in velocardiofacial (22q11.2 deletion) syndrome. J. Am. Acad. Child. Adolesc. Psychiatry 2009, 48, 1060–1068. [Google Scholar] [CrossRef]
- Bayat, M.; Bayat, A. Neurological manifestation of 22q11.2 deletion syndrome. Neurol. Sci. 2022, 43, 1695–1700. [Google Scholar] [CrossRef]
- Homans, J.F.; Tromp, I.N.; Colo, D.; Schlösser, T.P.C.; Kruyt, M.C.; Deeney, V.F.X.; Crowley, T.B.; McDonald-McGinn, D.M.; Castelein, R.M. Orthopaedic manifestations within the 22q11.2 Deletion syndrome: A systematic review. Am. J. Med. Genet. Part A 2018, 176, 2104–2120. [Google Scholar] [CrossRef] [PubMed]
- Giardino, G.; Cirillo, E.; Maio, F.; Gallo, V.; Esposito, T.; Naddei, R.; Grasso, F.; Pignata, C. Gastrointestinal involvement in patients affected with 22q11.2 deletion syndrome. Scand. J. Gastroenterol. 2014, 49, 274–279. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Rivera, E.; Liu, Y.P.; Verbitsky, M.; Anderson, B.R.; Capone, V.P.; Otto, E.A.; Yan, Z.; Mitrotti, A.; Martino, J.; Steers, N.J.; et al. Genetic Drivers of Kidney Defects in the DiGeorge Syndrome. N. Engl. J. Med. 2017, 376, 742–754. [Google Scholar] [CrossRef]
- Cortés-Martín, J.; Peñuela, N.L.; Sánchez-García, J.C.; Montiel-Troya, M.; Díaz-Rodríguez, L.; Rodríguez-Blanque, R. Deletion Syndrome 22q11.2: A Systematic Review. Children 2022, 9, 1168. [Google Scholar] [CrossRef]
- Van, L.; Heung, T.; Graffi, J.; Ng, E.; Malecki, S.; Van Mil, S.; Boot, E.; Corral, M.; Chow, E.W.C.; Hodgkinson, K.A.; et al. All-cause mortality and survival in adults with 22q11.2 deletion syndrome. Genet. Med. 2019, 21, 2328–2335. [Google Scholar] [CrossRef] [PubMed]
- Wahrmann, S.; Kainulainen, L.; Kytö, V.; Lempainen, J. Childhood manifestations of 22q11.2 deletion syndrome: A Finnish nationwide register-based cohort study. Acta Paediatr. 2023, 112, 1312–1318. [Google Scholar] [CrossRef]
- Cheung, E.N.; George, S.R.; Costain, G.A.; Andrade, D.M.; Chow, E.W.; Silversides, C.K.; Bassett, A.S. Prevalence of hypocalcaemia and its associated features in 22q11·2 deletion syndrome. Clin. Endocrinol. 2014, 81, 190–196. [Google Scholar] [CrossRef]
- Piliero, L.M.; Sanford, A.N.; McDonald-McGinn, D.M.; Zackai, E.H.; Sullivan, K.E. T-cell homeostasis in humans with thymic hypoplasia due to chromosome 22q11.2 deletion syndrome. Blood 2004, 103, 1020–1025. [Google Scholar] [CrossRef]
- Jyonouchi, S.; McDonald-McGinn, D.M.; Bale, S.; Zackai, E.H.; Sullivan, K.E. CHARGE (coloboma, heart defect, atresia choanae, retarded growth and development, genital hypoplasia, ear anomalies/deafness) syndrome and chromosome 22q11.2 deletion syndrome: A comparison of immunologic and nonimmunologic phenotypic features. Pediatrics 2009, 123, e871–e877. [Google Scholar] [CrossRef]
- Lewyllie, A.; Roosenboom, J.; Indencleef, K.; Claes, P.; Swillen, A.; Devriendt, K.; Carels, C.; Cadenas De Llano-Pérula, M.; Willems, G.; Hens, G.; et al. A Comprehensive Craniofacial Study of 22q11.2 Deletion Syndrome. J. Dent. Res. 2017, 96, 1386–1391. [Google Scholar] [CrossRef]
- Vervoort, L.; Dierckxsens, N.; Pereboom, Z.; Capozzi, O.; Rocchi, M.; Shaikh, T.H.; Vermeesch, J.R. 22q11.2 Low Copy Repeats Expanded in the Human Lineage. Front. Genet. 2021, 12, 706641. [Google Scholar] [CrossRef]
- Demaerel, W.; Mostovoy, Y.; Yilmaz, F.; Vervoort, L.; Pastor, S.; Hestand, M.S.; Swillen, A.; Vergaelen, E.; Geiger, E.A.; Coughlin, C.R.; et al. The 22q11 low copy repeats are characterized by unprecedented size and structural variability. Genome Res. 2019, 29, 1389–1401. [Google Scholar] [CrossRef]
- Adeyinka, A.; Stockero, K.J.; Flynn, H.C.; Lorentz, C.P.; Ketterling, R.P.; Jalal, S.M. Familial 22q11.2 deletions in DiGeorge/velocardiofacial syndrome are predominantly smaller than the commonly observed 3Mb. Genet. Med. 2004, 6, 517–520. [Google Scholar] [CrossRef] [PubMed]
- Guna, A.; Butcher, N.J.; Bassett, A.S. Comparative mapping of the 22q11.2 deletion region and the potential of simple model organisms. J. Neurodev. Disord. 2015, 7, 18. [Google Scholar] [CrossRef] [PubMed]
- Jerome, L.A.; Papaioannou, V.E. DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat. Genet. 2001, 27, 286–291. [Google Scholar] [CrossRef] [PubMed]
- Fulcoli, F.G.; Franzese, M.; Liu, X.; Zhang, Z.; Angelini, C.; Baldini, A. Rebalancing gene haploinsufficiency in vivo by targeting chromatin. Nat. Commun. 2016, 7, 11688. [Google Scholar] [CrossRef] [PubMed]
- Yagi, H.; Furutani, Y.; Hamada, H.; Sasaki, T.; Asakawa, S.; Minoshima, S.; Ichida, F.; Joo, K.; Kimura, M.; Imamura, S.; et al. Role of TBX1 in human del22q11.2 syndrome. Lancet 2003, 362, 1366–1373. [Google Scholar] [CrossRef] [PubMed]
- Baldini, A.; Fulcoli, F.G.; Illingworth, E. Tbx1: Transcriptional and Developmental Functions. Curr. Top. Dev. Biol. 2017, 122, 223–243. [Google Scholar] [CrossRef] [PubMed]
- Roberts, C.; Ivins, S.M.; James, C.T.; Scambler, P.J. Retinoic acid down-regulates Tbx1 expression in vivo and in vitro. Dev. Dyn. 2005, 232, 928–938. [Google Scholar] [CrossRef]
- Ryckebüsch, L.; Bertrand, N.; Mesbah, K.; Bajolle, F.; Niederreither, K.; Kelly, R.G.; Zaffran, S. Decreased levels of embryonic retinoic acid synthesis accelerate recovery from arterial growth delay in a mouse model of DiGeorge syndrome. Circ. Res. 2010, 106, 686–694. [Google Scholar] [CrossRef]
- Mark, M.; Ghyselinck, N.B.; Chambon, P. Retinoic acid signalling in the development of branchial arches. Curr. Opin. Genet. Dev. 2004, 14, 591–598. [Google Scholar] [CrossRef]
- Lania, G.; Bresciani, A.; Bisbocci, M.; Francone, A.; Colonna, V.; Altamura, S.; Baldini, A. Vitamin B12 ameliorates the phenotype of a mouse model of DiGeorge syndrome. Hum. Mol. Genet. 2016, 25, 4369–4375. [Google Scholar] [CrossRef] [PubMed]
- Cioffi, S.; Martucciello, S.; Fulcoli, F.G.; Bilio, M.; Ferrentino, R.; Nusco, E.; Illingworth, E. Tbx1 regulates brain vascularization. Hum. Mol. Genet. 2014, 23, 78–89. [Google Scholar] [CrossRef] [PubMed]
- Giacomelli, M.; Kumar, R.; Soresina, A.; Tamassia, N.; Lorenzini, T.; Moratto, D.; Gasperini, S.; Cassatella, M.; Plebani, A.; Lougaris, V.; et al. Reduction of CRKL expression in patients with partial DiGeorge syndrome is associated with impairment of T-cell functions. J. Allergy Clin. Immunol. 2016, 138, 229–240.e3. [Google Scholar] [CrossRef] [PubMed]
- Zheng, P.; Noroski, L.M.; Hanson, I.C.; Chen, Y.; Lee, M.E.; Huang, Y.; Zhu, M.X.; Banerjee, P.P.; Makedonas, G.; Orange, J.S.; et al. Molecular mechanisms of functional natural killer deficiency in patients with partial DiGeorge syndrome. J. Allergy Clin. Immunol. 2015, 135, 1293–1302. [Google Scholar] [CrossRef] [PubMed]
- Ray-Gallet, D.; Quivy, J.P.; Scamps, C.; Martini, E.M.; Lipinski, M.; Almouzni, G. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol. Cell 2002, 9, 1091–1100. [Google Scholar] [CrossRef] [PubMed]
- Romano, R.; Cillo, F.; Moracas, C.; Pignata, L.; Nannola, C.; Toriello, E.; De Rosa, A.; Cirillo, E.; Coppola, E.; Giardino, G.; et al. Epigenetic Alterations in Inborn Errors of Immunity. J. Clin. Med. 2022, 11, 1261. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.L.; Crowley, T.B.; McGinn, D.E.; McDougall, C.; Unolt, M.; Lambert, M.P.; Emanuel, B.S.; Zackai, E.H.; McDonald-McGinn, D.M. 22q and two: 22q11.2 deletion syndrome and coexisting conditions. Am. J. Med. Genet. Part A 2018, 176, 2203–2214. [Google Scholar] [CrossRef]
- Cecere, F.; Pignata, L.; Hay Mele, B.; Saadat, A.; D’Angelo, E.; Palumbo, O.; Palumbo, P.; Carella, M.; Scarano, G.; Rossi, G.B.; et al. Co-Occurrence of Beckwith-Wiedemann Syndrome and Early-Onset Colorectal Cancer. Cancers 2023, 15, 1944. [Google Scholar] [CrossRef]
- Carli, D.; Operti, M.; Russo, S.; Cocchi, G.; Milani, D.; Leoni, C.; Prada, E.; Melis, D.; Falco, M.; Spina, J.; et al. Clinical and molecular characterization of patients affected by Beckwith-Wiedemann spectrum conceived through assisted reproduction techniques. Clin. Genet. 2022, 102, 314–323. [Google Scholar] [CrossRef]
- Guo, T.; Repetto, G.M.; McDonald-McGinn, D.M.; Chung, J.H.; Nomaru, H.; Campbell, C.L.; Blonska, A.; Bassett, A.S.; Chow, E.W.C.; Mlynarski, E.E.; et al. Genome-Wide Association Study to Find Modifiers for Tetralogy of Fallot in the 22q11.2 Deletion Syndrome Identifies Variants in the GPR98 Locus on 5q14.3. Circ. Cardiovasc. Genet. 2017, 10, e001690. [Google Scholar] [CrossRef] [PubMed]
- León, L.E.; Benavides, F.; Espinoza, K.; Vial, C.; Alvarez, P.; Palomares, M.; Lay-Son, G.; Miranda, M.; Repetto, G.M. Partial microduplication in the histone acetyltransferase complex member KANSL1 is associated with congenital heart defects in 22q11.2 microdeletion syndrome patients. Sci. Rep. 2017, 7, 1795. [Google Scholar] [CrossRef] [PubMed]
- Mlynarski, E.E.; Sheridan, M.B.; Xie, M.; Guo, T.; Racedo, S.E.; McDonald-McGinn, D.M.; Gai, X.; Chow, E.W.; Vorstman, J.; Swillen, A.; et al. Copy-Number Variation of the Glucose Transporter Gene SLC2A3 and Congenital Heart Defects in the 22q11.2 Deletion Syndrome. Am. J. Hum. Genet. 2015, 96, 753–764. [Google Scholar] [CrossRef] [PubMed]
- Chisaka, O.; Capecchi, M.R. Regionally restricted developmental defects resulting from targeted disruption of the mouse homeobox gene HOX-1.5. Nature 1991, 350, 473–479. [Google Scholar] [CrossRef]
- Stalmans, I.; Lambrechts, D.; De Smet, F.; Jansen, S.; Wang, J.; Maity, S.; Kneer, P.; von der Ohe, M.; Swillen, A.; Maes, C.; et al. VEGF: A modifier of the del22q11 (DiGeorge) syndrome? Nat. Med. 2003, 9, 173–182. [Google Scholar] [CrossRef]
- Cirillo, E.; Prencipe, M.R.; Giardino, G.; Romano, R.; Scalia, G.; Genesio, R.; Nitsch, L.; Pignata, C. Clinical Phenotype, Immunological Abnormalities, and Genomic Findings in Patients with DiGeorge Spectrum Phenotype without 22q11.2 Deletion. J. Allergy Clin. Immunol. Pract. 2020, 8, 3112–3120. [Google Scholar] [CrossRef]
- Du, Q.; de la Morena, M.T.; van Oers, N.S.C. The Genetics and Epigenetics of 22q11.2 Deletion Syndrome. Front. Genet. 2020, 10, 1365. [Google Scholar] [CrossRef]
- Phelan, K.; Rogers, R.C.; Boccuto, L. Phelan-McDermid Syndrome. In GeneReviews®; Adam, M.P., Feldman, J., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Gripp, K.W., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Butensky, A.; de Rinaldis, C.P.; Patel, S.; Edman, S.; Bailey, A.; McGinn, D.E.; Zackai, E.; Crowley, T.B.; McDonald-McGinn, D.M.; Min, J.; et al. Cardiac evaluation of patients with 22q11.2 duplication syndrome. Am. J. Med. Genet. Part A 2021, 185, 753–758. [Google Scholar] [CrossRef]
- Liotti, A.; Ferrara, A.L.; Loffredo, S.; Galdiero, M.R.; Varricchi, G.; Di Rella, F.; Maniscalco, G.T.; Belardo, M.; Vastano, R.; Prencipe, R.; et al. Epigenetics: An opportunity to shape innate and adaptive immune responses. Immunology 2022, 167, 451–470. [Google Scholar] [CrossRef]
- Saliminejad, K.; Khorram Khorshid, H.R.; Soleymani Fard, S.; Ghaffari, S.H. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J. Cell Physiol. 2019, 234, 5451–5465. [Google Scholar] [CrossRef]
- García-López, J.; Brieño-Enríquez, M.A.; Del Mazo, J. MicroRNA biogenesis and variability. Biomol. Concepts 2013, 4, 367–380. [Google Scholar] [CrossRef]
- Kim, K.; Nguyen, T.D.; Li, S.; Nguyen, T.A. SRSF3 recruits DROSHA to the basal junction of primary microRNAs. RNA 2018, 24, 892–898. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef] [PubMed]
- Chevalier, D.; Borchert, G.M. Genome-Wide Analysis of MicroRNA-Regulated Transcripts. In Bioinformatics in microRNA Research; Methods in Molecular Biology; Humana: New York, NY, USA, 2017; Volume 1617, pp. 93–107. [Google Scholar] [CrossRef]
- Sellier, C.; Hwang, V.J.; Dandekar, R.; Durbin-Johnson, B.; Charlet-Berguerand, N.; Ander, B.P.; Sharp, F.R.; Angkustsiri, K.; Simon, T.J.; Tassone, F. Decreased DGCR8 expression and miRNA dysregulation in individuals with 22q11.2 deletion syndrome. PLoS ONE 2014, 9, e103884. [Google Scholar] [CrossRef] [PubMed]
- de la Morena, M.T.; Eitson, J.L.; Dozmorov, I.M.; Belkaya, S.; Hoover, A.R.; Anguiano, E.; Pascual, M.V.; van Oers, N.S.C. Signature MicroRNA expression patterns identified in humans with 22q11.2 deletion/DiGeorge syndrome. Clin. Immunol. 2013, 147, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Stark, K.L.; Xu, B.; Bagchi, A.; Lai, W.S.; Liu, H.; Hsu, R.; Wan, X.; Pavlidis, P.; Mills, A.A.; Karayiorgou, M.; et al. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat. Genet. 2008, 40, 751–760. [Google Scholar] [CrossRef] [PubMed]
- Merico, D.; Costain, G.; Butcher, N.J.; Warnica, W.; Ogura, L.; Alfred, S.E.; Brzustowicz, L.M.; Bassett, A.S. MicroRNA Dysregulation, Gene Networks, and Risk for Schizophrenia in 22q11.2 Deletion Syndrome. Front. Neurol. 2014, 5, 238. [Google Scholar] [CrossRef]
- Ouchi, Y.; Banno, Y.; Shimizu, Y.; Ando, S.; Hasegawa, H.; Adachi, K.; Iwamoto, T. Reduced adult hippocampal neurogenesis and working memory deficits in the Dgcr8-deficient mouse model of 22q11.2 deletion-associated schizophrenia can be rescued by IGF2. J. Neurosci. 2013, 33, 9408–9419. [Google Scholar] [CrossRef]
- Barr, I.; Weitz, S.H.; Atkin, T.; Hsu, P.; Karayiorgou, M.; Gogos, J.A.; Weiss, S.; Guo, F. Cobalt(III) Protoporphyrin Activates the DGCR8 Protein and Can Compensate microRNA Processing Deficiency. Chem. Biol. 2015, 22, 793–802. [Google Scholar] [CrossRef]
- Mendell, J.T.; Olson, E.N. MicroRNAs in Stress Signaling and Human Disease. Cell 2012, 148, 1172–1187. [Google Scholar] [CrossRef] [PubMed]
- Fénelon, K.; Mukai, J.; Xu, B.; Hsu, P.K.; Drew, L.J.; Karayiorgou, M.; Fischbach, G.D.; Macdermott, A.B.; Gogos, J.A. Deficiency of Dgcr8, a gene disrupted by the 22q11.2 microdeletion, results in altered short-term plasticity in the prefrontal cortex. Proc. Natl. Acad. Sci. USA 2011, 108, 4447–4452. [Google Scholar] [CrossRef] [PubMed]
- Ying, S.; Heung, T.; Zhang, Z.; Yuen, R.K.C.; Bassett, A.S. Schizophrenia Risk Mediated by microRNA Target Genes Overlapped by Genome-Wide Rare Copy Number Variation in 22q11.2 Deletion Syndrome. Front. Genet. 2022, 13, 812183. [Google Scholar] [CrossRef] [PubMed]
- Pong, S.K.; Gullerova, M. Noncanonical functions of microRNA pathway enzymes–Drosha, DGCR8, Dicer and Ago proteins. FEBS Lett. 2018, 592, 2973–2986. [Google Scholar] [CrossRef] [PubMed]
- Cirera-Salinas, D.; Yu, J.; Bodak, M.; Ngondo, R.P.; Herbert, K.M.; Ciaudo, C. Noncanonical function of DGCR8 controls mESC exit from pluripotency. J. Cell Biol. 2017, 216, 355–366. [Google Scholar] [CrossRef] [PubMed]
- Britzolaki, A.; Saurine, J.; Flaherty, E.; Thelen, C.; Pitychoutis, P.M. The SERCA2: A Gatekeeper of Neuronal Calcium Homeostasis in the Brain. Cell. Mol. Neurobiol. 2018, 38, 981–994. [Google Scholar] [CrossRef] [PubMed]
- Forstner, A.J.; Basmanav, F.B.; Mattheisen, M.; Böhmer, A.C.; Hollegaard, M.V.; Janson, E.; Strengman, E.; Priebe, L.; Degenhardt, F.; Hoffmann, P.; et al. Investigation of the involvement of MIR185 and its target genes in the development of schizophrenia. J. Psychiatry Neurosci. 2014, 39, 386–396. [Google Scholar] [CrossRef] [PubMed]
- Earls, L.R.; Fricke, R.G.; Yu, J.; Berry, R.B.; Baldwin, L.T.; Zakharenko, S.S. Age-dependent microRNA control of synaptic plasticity in 22q11 deletion syndrome and schizophrenia. J. Neurosci. 2012, 32, 14132–14144. [Google Scholar] [CrossRef]
- Belver, L.; de Yébenes, V.G.; Ramiro, A.R. MicroRNAs prevent the generation of autoreactive antibodies. Immunity 2010, 33, 713–722. [Google Scholar] [CrossRef]
- Flach, H.; Rosenbaum, M.; Duchniewicz, M.; Kim, S.; Zhang, S.L.; Cahalan, M.D.; Mittler, G.; Grosschedl, R. Mzb1 protein regulates calcium homeostasis, antibody secretion, and integrin activation in innate-like B cells. Immunity 2010, 33, 723–735. [Google Scholar] [CrossRef]
- Belkaya, S.; Murray, S.E.; Eitson, J.L.; de la Morena, M.T.; Forman, J.A.; van Oers, N.S.C. Transgenic expression of microRNA-185 causes a developmental arrest of T cells by targeting multiple genes including Mzb1. J. Biol. Chem. 2013, 288, 30752–30762. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, B.J.; De Windt, L.J.; Bueno, O.F.; Braz, J.C.; Glascock, B.J.; Kimball, T.F.; Molkentin, J.D. Targeted disruption of NFATc3, but not NFATc4, reveals an intrinsic defect in calcineurin-mediated cardiac hypertrophic growth. Mol. Cell. Biol. 2002, 22, 7603–7613. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.O.; Song, D.W.; Kwon, E.J.; Hong, S.E.; Song, H.K.; Min, C.K.; Kim, D.H. miR-185 plays an anti-hypertrophic role in the heart via multiple targets in the calcium-signaling pathways. PLoS ONE 2015, 10, e0122509. [Google Scholar] [CrossRef]
- Meechan, D.W.; Maynard, T.M.; Gopalakrishna, D.; Wu, Y.; LaMantia, A.S. When half is not enough: Gene expression and dosage in the 22q11 deletion syndrome. Gene Expr. 2007, 13, 299–310. [Google Scholar] [CrossRef]
- Jeanne, M.; Vuillaume, M.L.; Ung, D.C.; Vancollie, V.E.; Wagner, C.; Collins, S.C.; Vonwill, S.; Haye, D.; Chelloug, N.; Pfundt, R.; et al. Haploinsufficiency of the HIRA gene located in the 22q11 deletion syndrome region is associated with abnormal neurodevelopment and impaired dendritic outgrowth. Hum. Genet. 2021, 140, 885–896. [Google Scholar] [CrossRef]
- Aref-Eshghi, E.; Kerkhof, J.; Pedro, V.P.; Di France, G.; Barat-Houari, M.; Ruiz-Pallares, N.; Andrau, J.C.; Lacombe, D.; Van-Gils, J.; Fergelot, P.; et al. Evaluation of DNA Methylation Episignatures for Diagnosis and Phenotype Correlations in 42 Mendelian Neurodevelopmental Disorders. Am. J. Hum. Genet. 2021, 108, 1161–1163. [Google Scholar] [CrossRef]
- Aref-Eshghi, E.; Schenkel, L.C.; Lin, H.; Skinner, C.; Ainsworth, P.; Paré, G.; Rodenhiser, D.; Schwartz, C.; Sadikovic, B. The defining DNA methylation signature of Kabuki syndrome enables functional assessment of genetic variants of unknown clinical significance. Epigenetics 2017, 12, 923–933. [Google Scholar] [CrossRef]
- Pignata, L.; Palumbo, O.; Cerrato, F.; Acurzio, B.; de Álava, E.; Roma, J.; Gallego, S.; Mora, J.; Carella, M.; Riccio, A.; et al. Both Epimutations and Chromosome Aberrations Affect Multiple Imprinted Loci in Aggressive Wilms Tumors. Cancers 2020, 12, 3411. [Google Scholar] [CrossRef]
- Carmel, M.; Michaelovsky, E.; Weinberger, R.; Frisch, A.; Mekori-Domachevsky, E.; Gothelf, D.; Weizman, A. Differential methylation of imprinting genes and MHC locus in 22q11.2 deletion syndrome-related schizophrenia spectrum disorders. World J. Biol. Psychiatry 2021, 22, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Shi, L.; Song, L.; Maurer, K.; Zhao, X.; Zackai, E.H.; McGinn, D.E.; Crowley, T.B.; McGinn, D.M.M.; Sullivan, K.E. Chromatin Modifications in 22q11.2 Deletion Syndrome. J. Clin. Immunol. 2021, 41, 1853–1864. [Google Scholar] [CrossRef]
- Rooney, K.; Levy, M.A.; Haghshenas, S.; Kerkhof, J.; Rogaia, D.; Tedesco, M.G.; Imperatore, V.; Mencarelli, A.; Squeo, G.M.; Di Venere, E.; et al. Identification of a DNA Methylation Episignature in the 22q11.2 Deletion Syndrome. Int. J. Mol. Sci. 2021, 22, 8611. [Google Scholar] [CrossRef] [PubMed]
- Mokhtari, R.; Lachman, H.M. The Major Histocompatibility Complex (MHC) in Schizophrenia: A Review. J. Clin. Cell. Immunol. 2016, 7, 479. [Google Scholar] [CrossRef] [PubMed]

| Apparatus Involved | Clinical Features | Percentage | References |
|---|---|---|---|
| Congenital Heart Disease (CHD) | Interrupted aortic arch type B Truncus arteriosus Tetralogy of Fallot Conoventricular septal defects Isolated aortic arch anomaly Double outlet right ventricle Transposition of the great arteries Hypoplastic left ventricle Pulmonary arteries hypoplasia | 75% | [1,31,32,33] |
| Hypocalcemia (hypoparathyroidism) | 35% | [69] | |
| Immune Deficiency | Athymia Thymic hypoplasia/ectopy Humoral immunity impairment | 50–70% | [70,71] |
| Craniofacial dysmorphisms | Elongated face Hooded eyelids Epicanthus Wide nasal bridge Short philtrum Micrognathia and retrognathia Low-set small ears | 50% | [22,72] |
| Palatal anomalies | Velopharyngeal insufficiency Overt cleft palate Submucosal cleft palate Bifid uvula | 69–100% | [37,38,39] |
| Renal anomalies | Renal agenesis Multicystic kidney Hydronephrosis Duplicated collecting system | 14% | [65] |
| Skeletal defect | Spine and vertebral anomalies Fingers anomalies | 60% | [63] |
| Learning problems Developmental delay | 70% | [42] | |
| Psychiatric disorders | Anxiety Autism spectrum disorders Schizophrenia Behavior disorders Parkinson’s disease | 30% | [60,61] |
| Gastrointestinal abnormalities | Esophageal atresia Esophageal reflux Hirschsprung disease Imperforated anus | 30% | [64] |
| Associated Phenotype | Genomic Coordinates | Inheritance | |
|---|---|---|---|
| DGCR6 | - | 22:18,906,319-18,912,087 | - |
| PRODH | Hyperprolinemia type 1 | 22:18,912,780-18,936,552 | AR |
| DGCR2 | - | 22:19,036,285-19,122,453 | - |
| DGCR14 | - | 22:19,130,278-19,144,725 | - |
| TSSK2 | - | 22:19,131,307-19,132,621 | - |
| GSC2 | - | 22:19,146,992-19,150,291 | - |
| SLC25A1 | Combined D-2, L-2 hydroxyglutaric aciduria; Presynaptic Congenital Myasthenic Syndrome 23 | 22:19,175,580-19,178,735 | AR AR |
| CLTCL1 | - | 22:19,179,472-19,291,718 | - |
| HIRA | - | 22:19,330,697-19,431,732 | - |
| MRPL40 | - | 22:19,432,544-19,436,074 | - |
| UFD1L | - | 22:19,449,910-19,479,192 | - |
| CDC45 | Meier-Gorlin Syndrome | 22:19,479,293-19,520,611 | AR |
| CLDN5 | - | 22:19,523,023-19,525,336 | - |
| SEPT5 | - | 22:19,714,502-19,723,318 | - |
| TBX1 | - | 22:19,756,702-19,783,592 | - |
| GNB1L | - | 22:19,783,222-19,854,873 | - |
| TXNRD2 | Glucocorticoid deficiency? | 22:19,875,521-19,941,817 | - |
| GP1BB | Bernard-Soulier Syndrome, type B; Giant platelet disorder | 22:19,723,538-19,724,770 | AR AR |
| COMT | schizophrenia, susceptibility | 22:19,941,771-19,969,97 | AD |
| ARVCF | - | 22:19,966,726-20,016,822 | - |
| TANGO2 | Metabolic encephalomyopathic crises, recurrent, with rhabdomyolisis, cardiac arrhythmias and neurodegeneration | 22:20,016,999-20,067,163 | AR |
| DGCR8 | - | 22:20,080,240-20,111,871 | - |
| TRMT2A | - | 22:20,111,871-20,117,253 | - |
| RANBP1 | - | 22:20,116,103-20,127,354 | - |
| ZDHHC8 | - | 22:20,131,803-20,148,006 | - |
| CCDC188 | - | 22:20,148,113-20,151,828 | - |
| RTN4R | schizophrenia, susceptibility | 22:20,241,414-20,268,317 | AD |
| DGCR6L | - | 22:20,314,237-20,320,059 | - |
| USP41 | - | 22:20,350,578-20,390,758 | - |
| ZNF74 | - | 22:20,394,150-20,408,454 | - |
| SCARF2 | Van den Ende-Gupta Syndrome | 22:20,424,583-20,437,824 | AR |
| KLHL22 | - | 22:20,441,518-20,497,304 | - |
| MED15 | - | 22:20,507,581-20,587,620 | - |
| PI4KA | Gastrointestinal defects and immunodeficiency syndrome 2; perisylvian polymicrogyria with cerebellar hypoplasia and arthrogryposis; spastic paraplegia 84 | 22:20,707,690-20,858,811 | AR |
| SERPIND1 | Thrombophilia 10 due to heparin cofactor II deficiency | 22:20,774,112-20,787,719 | AD |
| SNAP29 | CEDNIK Syndrome | 22:20,859,006-20,891,213 | AR |
| CRKL | - | 22:20,917,406-20,953,746 | - |
| AIFM3 | - | 22:20,965,171-20,981,357 | - |
| LZTR1 | Noonan Syndrome 10; Noonan Syndrome 2; Schwannomatosis 2, susceptibility | 22:20,982,296-20,999,031 | AD AR AD |
| THAP7 | - | 22:20,999,103-21,002,117 | - |
| P2RX6 | - | 22:21,009,699-21,028,013 | - |
| SLC7A4 | - | 22:21,028,717-21,032,560 | - |
| HIC2 | - | 22:21,417,370-21,451,462 | - |
| Clinical Manifestations | 10p13-14 DGS2 Locus | 3p10.3 | 4q34.1-35.2 | Del2p11.2 | Microdup 22q11.2 | Del22q13.33 Phelan-McDermid Syndrome |
|---|---|---|---|---|---|---|
| Congenital Heart Disease (CHD) | 82% | Yes | 15% | No | Yes | 3–25% |
| Hypocalcemia (hypoparathyroidism) | 22% | Yes | Na | Yes | Yes | Na |
| Immune Deficiency | 17% | Yes | Na | Yes | Yes | Na |
| Craniofacial dysmorphisms | 50% | Yes | 95–99% | Yes | Yes | >75% |
| Renal anomalies | 5% | Yes | Na | No | Yes | 38% |
| Skeletal defects | 30–80% | Na | 88% | Yes | Yes | >75% |
| Learning problems/ Developmental delay | 80–99% | Yes | 65% | Yes | Yes | >75% |
| Psychiatric disorders | Na | Na | Na | Na | Yes | >75% |
| Gastrointestinal abnormalities | Na | Na | Na | No | Yes | >25% |
| Genes mapping in the region | Ni | FOXI3 | See Table 3 | SHANK3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cillo, F.; Coppola, E.; Habetswallner, F.; Cecere, F.; Pignata, L.; Toriello, E.; De Rosa, A.; Grilli, L.; Ammendola, A.; Salerno, P.; et al. Understanding the Variability of 22q11.2 Deletion Syndrome: The Role of Epigenetic Factors. Genes 2024, 15, 321. https://doi.org/10.3390/genes15030321
Cillo F, Coppola E, Habetswallner F, Cecere F, Pignata L, Toriello E, De Rosa A, Grilli L, Ammendola A, Salerno P, et al. Understanding the Variability of 22q11.2 Deletion Syndrome: The Role of Epigenetic Factors. Genes. 2024; 15(3):321. https://doi.org/10.3390/genes15030321
Chicago/Turabian StyleCillo, Francesca, Emma Coppola, Federico Habetswallner, Francesco Cecere, Laura Pignata, Elisabetta Toriello, Antonio De Rosa, Laura Grilli, Antonio Ammendola, Paolo Salerno, and et al. 2024. "Understanding the Variability of 22q11.2 Deletion Syndrome: The Role of Epigenetic Factors" Genes 15, no. 3: 321. https://doi.org/10.3390/genes15030321
APA StyleCillo, F., Coppola, E., Habetswallner, F., Cecere, F., Pignata, L., Toriello, E., De Rosa, A., Grilli, L., Ammendola, A., Salerno, P., Romano, R., Cirillo, E., Merla, G., Riccio, A., Pignata, C., & Giardino, G. (2024). Understanding the Variability of 22q11.2 Deletion Syndrome: The Role of Epigenetic Factors. Genes, 15(3), 321. https://doi.org/10.3390/genes15030321






