Functional Roles and Genomic Impact of Miniature Inverted-Repeat Transposable Elements (MITEs) in Prokaryotes
Abstract
:1. Introduction
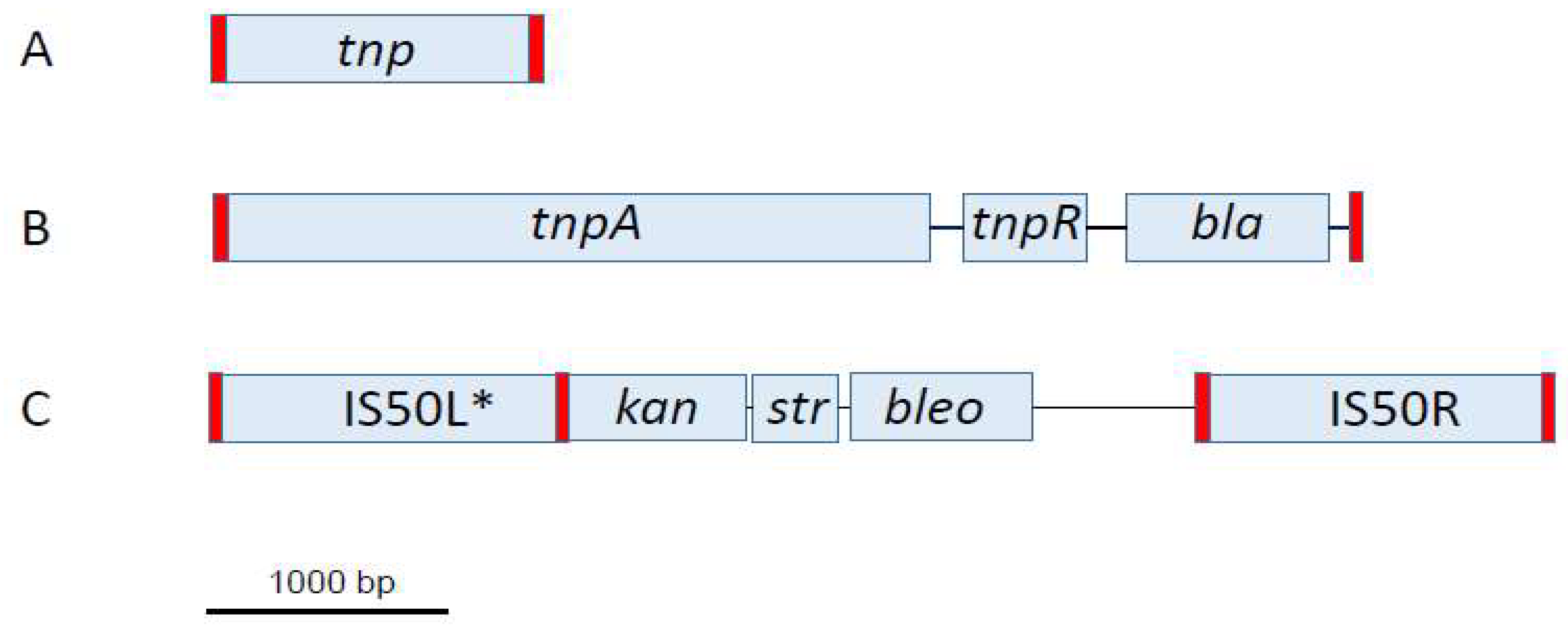
2. Prokaryotic MITEs
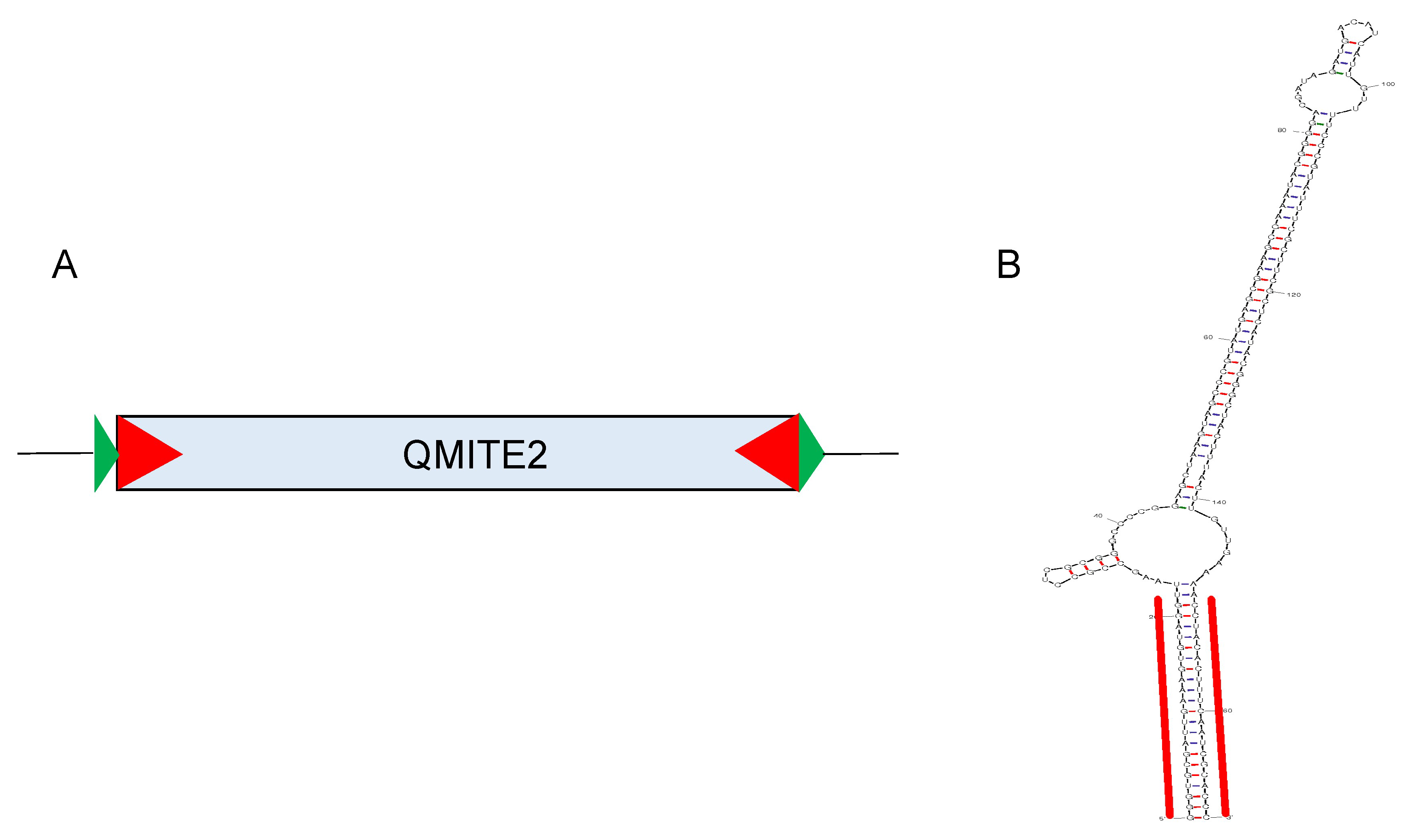
2.1. Conserved Structural Features
2.2. Size and Sequence Variation
3. Distribution across Prokaryotic Genomes
3.1. Taxonomic Distribution
3.2. Copy-Number Variation
3.3. Insertion Site Preferences
4. Functional Roles and Genomic Impact
4.1. Influence on Gene Expression
4.2. Role in Genome Plasticity and Evolution
5. Bioinformatic Tools for MITE Analysis
6. Conclusions and Future Directions
Funding
Conflicts of Interest
Abbreviations
References
- McClintock, B. The origin and behavior of mutable loci in maize. Proc. Natl. Acad. Sci. USA 1950, 36, 344–355. [Google Scholar] [CrossRef]
- McClintock, B. Induction of instability at selected loci in maize. Genetics 1953, 38, 579–599. [Google Scholar] [CrossRef]
- Peters, J.E.; Craig, N.L. Tn7: Smarter than we thought. Nat. Rev. Mol. Cell Biol. 2001, 2, 806–814. [Google Scholar] [CrossRef]
- Ahmed, A. Alternative mechanisms for Tn5 transposition. PLoS Genet. 2009, 5, e1000619. [Google Scholar] [CrossRef]
- Nicolas, E.; Lambin, M.; Dandoy, D.; Galloy, C.; Nguyen, N.; Oger, C.A.; Hallet, B. The Tn3-family of replicative transposons. Microbiol. Spectr. 2015, 3, 4. [Google Scholar] [CrossRef] [PubMed]
- Crescente, J.M.; Zavallo, D.; Del Vas, M.; Asurmendi, S.; Helguera, M.; Fernandez, E.; Vanzetti, L.S. Genome-wide identification of MITE-derived microRNAs and their targets in bread wheat. BMC Genom. 2022, 23, 154. [Google Scholar] [CrossRef]
- Guo, Z.; Kuang, Z.; Tao, Y.; Wang, H.; Wan, M.; Hao, C.; Shen, F.; Yang, X.; Li, L. Miniature inverted-repeat transposable elements drive rapid microRNA diversification in angiosperms. Mol. Biol. Evol. 2022, 39, msac224. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Chen, J.; Zhang, Y.; Hu, Q.; Su, W.; Kuang, H. Miniature inverted-repeat transposable elements (MITEs) have been accumulated through amplification bursts and play important roles in gene expression and species diversity in Oryza sativa. Mol. Biol. Evol. 2012, 29, 1005–1017. [Google Scholar] [CrossRef] [PubMed]
- Xin, Y.; Ma, B.; Xiang, Z.; He, N. Amplification of miniature inverted-repeat transposable elements and the associated impact on gene regulation and alternative splicing in mulberry (Morus notabilis). Mob. DNA 2019, 10, 27. [Google Scholar] [CrossRef]
- Boutanaev, A.M.; Osbourn, A.E. Multigenome analysis implicates miniature inverted-repeat transposable elements (MITEs) in metabolic diversification in eudicots. Proc. Natl. Acad. Sci. USA 2018, 115, E6650–E6658. [Google Scholar] [CrossRef]
- Chen, Y.; Zhou, F.; Li, G.; Xu, Y. A recently active miniature inverted-repeat transposable element, Chunjie, inserted into an operon without disturbing the operon structure in Geobacter uraniireducens Rf4. Genetics 2008, 179, 2291–2297. [Google Scholar] [CrossRef]
- Chen, Y.; Zhou, F.; Li, G.; Xu, Y. MUST: A system for identification of miniature inverted-repeat transposable elements and applications to Anabaena variabilis and Haloquadratum walsbyi. Gene 2009, 436, 1–7. [Google Scholar] [CrossRef]
- Tu, Z. Three novel families of miniature inverted-repeat transposable elements are associated with genes of the yellow fever mosquito, Aedes aegypti. Proc. Natl. Acad. Sci. USA 1997, 94, 7475–7480. [Google Scholar] [CrossRef]
- Wachter, S.; Raghavan, R.; Wachter, J.; Minnick, M.F. Identification of novel MITEs (miniature inverted-repeat transposable elements) in Coxiella burnetii: Implications for protein and small RNA evolution. BMC Genom. 2018, 19, 247. [Google Scholar] [CrossRef] [PubMed]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Siguier, P.; Filée, J.; Chandler, M. Insertion sequences in prokaryotic genomes. Curr. Opin. Microbiol. 2006, 9, 526–531. [Google Scholar] [CrossRef]
- Siguier, P.; Gourbeyre, E.; Chandler, M. Bacterial insertion sequences: Their genomic impact and diversity. FEMS Microbiol. Rev. 2014, 38, 865–891. [Google Scholar] [CrossRef]
- Delihas, N. Impact of small repeat sequences on bacterial genome evolution. Genome Biol. Evol. 2011, 3, 959–973. [Google Scholar] [CrossRef]
- Oosumi, T.; Garlick, B.; Belknap, W.R. Identification of putative nonautonomous transposable elements associated with several transposon families in Caenorhabditis elegans. J. Mol. Evol. 1996, 43, 11–18. [Google Scholar] [CrossRef]
- Correia, F.F.; Inouye, S.; Inouye, M. A family of small repeated elements with some transposon-like properties in the genome of Neisseria gonorrhoeae. J. Biol. Chem. 1988, 263, 12194–12198. [Google Scholar] [CrossRef] [PubMed]
- Snyder, L.A.; Cole, J.A.; Pallen, M.J. Comparative analysis of two Neisseria gonorrhoeae genome sequences reveals evidence of mobilization of Correia Repeat Enclosed Elements and their role in regulation. BMC Genom. 2009, 10, 70. [Google Scholar] [CrossRef] [PubMed]
- Hulton, C.S.; Higgins, C.F.; Sharp, P.M. ERIC sequences: A novel family of repetitive elements in the genomes of Escherichia coli, Salmonella typhimurium and other enterobacteria. Mol. Microbiol. 1991, 5, 825–834. [Google Scholar] [CrossRef] [PubMed]
- Kieffer, N.; Nordmann, P.; Millemann, Y.; Poirel, L. Functional characterization of a miniature inverted transposable element at the origin of mcr-5 gene acquisition in Escherichia coli. Antimicrob. Agents Chemother. 2019, 63, e00559-19. [Google Scholar] [CrossRef]
- Zhou, F.; Tran, T.; Xu, Y. Nezha, a novel active miniature inverted-repeat transposable element in cyanobacteria. Biochem. Biophys. Res. Commun. 2008, 365, 790–794. [Google Scholar] [CrossRef]
- Kaneko, T.; Nakajima, N.; Okamoto, S.; Suzuki, I.; Tanabe, Y.; Tamaoki, M.; Nakamura, Y.; Kasai, F.; Watanabe, A.; Kawashima, K.; et al. Complete genomic structure of the bloom-forming toxic cyanobacterium Microcystis aeruginosa NIES-843. DNA Res. 2007, 14, 247–256. [Google Scholar] [CrossRef]
- Yui Eto, K.; Firth, N.; Davis, A.M.; Kwong, S.M.; Krysiak, M.; Lee, Y.T.; O’Brien, F.G.; Grubb, W.B.; Coombs, G.W.; Bond, C.S.; et al. Evolution of a 72-Kilobase cointegrant, conjugative multiresistance plasmid in community-associated methicillin-resistant Staphylococcus aureus isolates from the early 1990s. Antimicrob. Agents Chemother. 2019, 63, e01560-19. [Google Scholar] [CrossRef]
- Zong, Z. The complex genetic context of blaPER-1 flanked by miniature inverted-repeat transposable elements in Acinetobacter johnsonii. PLoS ONE 2014, 9, e90046. [Google Scholar] [CrossRef] [PubMed]
- Adams, F.G.; Brown, M.H. MITE Aba12, a novel mobile miniature inverted-repeat transposable element identified in Acinetobacter baumannii ATCC 17978 and its prevalence across the Moraxellaceae ramily. mSphere 2019, 4, e00028-19. [Google Scholar] [CrossRef]
- Robertson, A.E.; Wechter, W.P.; Denny, T.P.; Fortnum, B.A.; Kluepfel, D.A. Relationship between avirulence gene (avrA) diversity in Ralstonia solanacearum and bacterial wilt incidence. Mol Plant Microbe Interact. 2004, 17, 1376–1384. [Google Scholar] [CrossRef]
- Stavrinides, J.; Kirzinger, M.W.; Beasley, F.C.; Guttman, D.S. E622, a miniature, virulence-associated mobile element. J. Bacteriol. 2012, 194, 509–517. [Google Scholar] [CrossRef]
- Naito, M.; Hirakawa, H.; Yamashita, A.; Ohara, N.; Shoji, M.; Yukitake, H.; Nakayama, K.; Toh, H.; Yoshimura, F.; Kuhara, S.; et al. Determination of the genome sequence of Porphyromonas gingivalis strain ATCC 33277 and genomic comparison with strain W83 revealed extensive genome rearrangements in P. gingivalis. DNA Res. 2008, 15, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Klein, B.A.; Chen, T.; Scott, J.C.; Koenigsberg, A.L.; Duncan, M.J.; Hu, L.T. Identification and characterization of a minisatellite contained within a novel miniature inverted-repeat transposable element (MITE) of Porphyromonas gingivalis. Mob. DNA 2015, 6, 18. [Google Scholar] [CrossRef] [PubMed]
- Mattes, T.E.; Alexander, A.K.; Richardson, P.M.; Munk, A.C.; Han, C.S.; Stothard, P.; Coleman, N.V. The genome of Polaromonas sp. strain JS666: Insights into the evolution of a hydrocarbon- and xenobiotic-degrading bacterium, and features of relevance to biotechnology. Appl. Environ. Microbiol. 2008, 74, 6405–6416. [Google Scholar] [CrossRef]
- Halter, T.; Hendrickx, F.; Horn, M.; Manzano-Marín, A. A novel widespread MITE element in the repeat-rich genome of the Cardinium endosymbiont of the spider Oedothorax gibbosus. Microbiol. Spectr. 2022, 10, e0262722. [Google Scholar] [CrossRef] [PubMed]
- Brügger, K.; Torarinsson, E.; Redder, P.; Chen, L.; Garrett, R.A. Shuffling of Sulfolobus genomes by autonomous and non-autonomous mobile elements. Biochem. Soc. Trans. 2004, 32 Pt 2, 179–183. [Google Scholar] [CrossRef]
- Guo, L.; Brügger, K.; Liu, C.; Shah, S.A.; Zheng, H.; Zhu, Y.; Wang, S.; Lillestøl, R.K.; Chen, L.; Frank, J.; et al. Genome analyses of Icelandic strains of Sulfolobus islandicus, model organisms for genetic and virus-host interaction studies. J. Bacteriol. 2011, 193, 1672–1680. [Google Scholar] [CrossRef]
- Jaubert, C.; Danioux, C.; Oberto, J.; Cortez, D.; Bize, A.; Krupovic, M.; She, Q.; Forterre, P.; Prangishvili, D.; Sezonov, G. Genomics and genetics of Sulfolobus islandicus LAL14/1, a model hyperthermophilic archaeon. Open Biol. 2013, 3, 130010. [Google Scholar] [CrossRef]
- Brügger, K.; Redder, P.; She, Q.; Confalonieri, F.; Zivanovic, Y.; Garrett, R.A. Mobile elements in archaeal genomes. FEMS Microbiol. Lett. 2002, 206, 131–141. [Google Scholar] [CrossRef]
- Ravin, N.V.; Mardanov, A.V.; Beletsky, A.V.; Kublanov, I.V.; Kolganova, T.V.; Lebedinsky, A.V.; Chernyh, N.A.; Bonch-Osmolovskaya, E.A.; Skryabin, K.G. Complete genome sequence of the anaerobic, protein-degrading hyperthermophilic crenarchaeon Desulfurococcus kamchatkensis. J. Bacteriol. 2009, 191, 2371–2379. [Google Scholar] [CrossRef]
- Kamoun, C.; Payen, T.; Hua-Van, A.; Filée, J. Improving prokaryotic transposable elements identification using a combination of de novo and profile HMM methods. BMC Genom. 2013, 14, 700. [Google Scholar] [CrossRef]
- Chen, J.; Hu, Q.; Zhang, Y.; Lu, C.; Kuang, H. P-MITE: A database for plant miniature inverted-repeat transposable elements. Nucleic Acids Res. 2014, 42, D1176–D1181. [Google Scholar] [CrossRef]
- Redder, P.; She, Q.; Garrett, R.A. Non-autonomous mobile elements in the crenarchaeon Sulfolobus solfataricus. J. Mol. Biol. 2001, 306, 1–6. [Google Scholar] [CrossRef]
- Zo, Y.G. Reductive divergence of enterobacterial repetitive intergenic consensus sequences among Gammaproteobacteria genomes. J. Microbiol. 2011, 49, 35–45. [Google Scholar] [CrossRef]
- Wilson, L.A.; Sharp, P.M. Enterobacterial repetitive intergenic consensus (ERIC) sequences in Escherichia coli: Evolution and implications for ERIC-PCR. Mol. Biol. Evol. 2006, 23, 1156–1168. [Google Scholar] [CrossRef] [PubMed]
- Duchaud, E.; Rusniok, C.; Frangeul, L.; Buchrieser, C.; Givaudan, A.; Taourit, S.; Bocs, S.; Boursaux-Eude, C.; Chandler, M.; Charles, J.F.; et al. The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat. Biotechnol. 2003, 21, 1307–1313. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.L.; Shapiro, L. Identification of long intergenic repeat sequences associated with DNA methylation sites in Caulobacter crescentus and other α-proteobacteria. J. Bacteriol. 2003, 185, 4997–5002. [Google Scholar] [CrossRef] [PubMed]
- Nelson, W.C.; Bhaya, D.; Heidelberg, J.F. Novel miniature transposable elements in thermophilic Synechococcus strains and their impact on an environmental population. J. Bacteriol. 2012, 194, 3636–3642. [Google Scholar] [CrossRef] [PubMed]
- Delihas, N. Enterobacterial small mobile sequences carry open reading frames and are found intragenically--evolutionary implications for formation of new peptides. Gene Regul. Syst. Bio. 2007, 1, 191–205. [Google Scholar] [CrossRef]
- Croucher, N.J.; Vernikos, G.S.; Parkhill, J.; Bentley, S.D. Identification, variation and transcription of pneumococcal repeat sequences. BMC Genom. 2011, 12, 120. [Google Scholar] [CrossRef] [PubMed]
- Black, C.G.; Fyfe, J.A.; Davies, J.K. A promoter associated with the neisserial repeat can be used to transcribe the uvrB gene from Neisseria gonorrhoeae. J. Bacteriol. 1995, 177, 1952–1958. [Google Scholar] [CrossRef] [PubMed]
- Siddique, A.; Buisine, N.; Chalmers, R. The transposon-like Correia elements encode numerous strong promoters and provide a potential new mechanism for phase variation in the meningococcus. PLoS Genet. 2011, 7, e1001277. [Google Scholar] [CrossRef]
- Elbeyioglu, F.; Roberts, S.B.; Spencer-Smith, R.; Pulijala, M.; Zelewska, M.A.; Nebel, J.C.; Snyder, L.A.S. Inversion of Correia repeat enclosed elements in Neisseria gonorrhoeae. Microbiology (Reading) 2017, 163, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Romine, M.F.; Carlson, T.S.; Norbeck, A.D.; McCue, L.A.; Lipton, M.S. Identification of mobile elements and pseudogenes in the Shewanella oneidensis MR-1 genome. Appl. Environ. Microbiol. 2008, 74, 3257–3265. [Google Scholar] [CrossRef]
- Goosen, N.; van de Putte, P. The regulation of transcription initiation by integration host factor. Mol. Microbiol. 1995, 16, 1–7. [Google Scholar] [CrossRef]
- Rouquette-Loughlin, C.E.; Balthazar, J.T.; Hill, S.A.; Shafer, W.M. Modulation of the mtrCDE-encoded efflux pump gene complex of Neisseria meningitidis due to a Correia element insertion sequence. Mol. Microbiol. 2004, 54, 731–741. [Google Scholar] [CrossRef] [PubMed]
- De Gregorio, E.; Abrescia, C.; Carlomagno, M.S.; Di Nocera, P.P. The abundant class of nemis repeats provides RNA substrates for ribonuclease III in Neisseriae. Biochim. Biophys. Acta 2002, 1576, 39–44. [Google Scholar] [CrossRef]
- Mazzone, M.; De Gregorio, E.; Lavitola, A.; Pagliarulo, C.; Alifano, P.; Di Nocera, P.P. Whole-genome organization and functional properties of miniature DNA insertion sequences conserved in pathogenic Neisseriae. Gene 2001, 278, 211–222. [Google Scholar] [CrossRef] [PubMed]
- De Gregorio, E.; Silvestro, G.; Petrillo, M.; Carlomagno, M.S.; Di Nocera, P.P. Enterobacterial repetitive intergenic consensus sequence repeats in yersiniae: Genomic organization and functional properties. J. Bacteriol. 2005, 187, 7945–7954. [Google Scholar] [CrossRef]
- Wright, M.S.; Jacobs, M.R.; Bonomo, R.A.; Adams, M.D. Transcriptome remodeling of Acinetobacter baumannii during infection and treatment. mBio 2017, 8, e02193-16. [Google Scholar] [CrossRef] [PubMed]
- Deveson Lucas, D.; Crane, B.; Wright, A.; Han, M.L.; Moffatt, J.; Bulach, D.; Gladman, S.L.; Powell, D.; Aranda, J.; Seemann, T.; et al. Emergence of high-level colistin resistance in an Acinetobacter baumannii clinical isolate mediated by inactivation of the global regulator H-NS. Antimicrob. Agents Chemother. 2018, 62, e02442-17. [Google Scholar] [CrossRef]
- Pegler, J.L.; Oultram, J.M.J.; Mann, C.W.G.; Carroll, B.J.; Grof, C.P.L.; Eamens, A.L. Miniature inverted-repeat transposable elements: Small DNA transposons that have contributed to plant MICRORNA gene evolution. Plants 2023, 12, 1101. [Google Scholar] [CrossRef]
- Wolk, C.P.; Lechno-Yossef, S.; Jäger, K.M. The insertion sequences of Anabaena sp. strain PCC 7120 and their effects on its open reading frames. J. Bacteriol. 2010, 192, 5289–5303. [Google Scholar] [CrossRef]
- Ogata, H.; Audic, S.; Barbe, V.; Artiguenave, F.; Fournier, P.E.; Raoult, D.; Claverie, J.M. Selfish DNA in protein-coding genes of Rickettsia. Science 2000, 290, 347–350. [Google Scholar] [CrossRef]
- Ogata, H.; Audic, S.; Abergel, C.; Fournier, P.E.; Claverie, J.M. Protein coding palindromes are a unique but recurrent feature in Rickettsia. Genome Res. 2002, 12, 808–816. [Google Scholar] [CrossRef]
- Fedoroff, N.; Wessler, S.; Shure, M. Isolation of the transposable maize controlling elements Ac and Ds. Cell 1983, 35, 235–242. [Google Scholar] [CrossRef]
- Fewer, D.P.; Halinen, K.; Sipari, H.; Bernardová, K.; Mänttäri, M.; Eronen, E.; Sivonen, K. Non-autonomous transposable elements associated with inactivation of microcystin gene clusters in strains of the genus Anabaena isolated from the Baltic Sea. Environ. Microbiol. Rep. 2011, 3, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Bardaji, L.; Añorga, M.; Jackson, R.W.; Martínez-Bilbao, A.; Yanguas-Casás, N.; Murillo, J. Miniature transposable sequences are frequently mobilized in the bacterial plant pathogen Pseudomonas syringae pv. phaseolicola. PLoS ONE 2011, 6, e25773. [Google Scholar] [CrossRef] [PubMed]
- Blount, Z.D.; Grogan, D.W. New insertion sequences of Sulfolobus: Functional properties and implications for genome evolution in hyperthermophilic archaea. Mol. Microbiol. 2005, 55, 312–325. [Google Scholar] [CrossRef] [PubMed]
- Gillings, M.R. Integrons: Past, present, and future. Microbiol. Mol. Biol. Rev. 2014, 78, 257–277. [Google Scholar] [CrossRef] [PubMed]
- Domingues, S.; Nielsen, K.M.; da Silva, G.J. The blaIMP-5-carrying integron in a clinical Acinetobacter baumannii strain is flanked by miniature inverted-repeat transposable elements (MITEs). J. Antimicrob. Chemother. 2011, 66, 2667–2668. [Google Scholar] [CrossRef] [PubMed]
- Gillings, M.R.; Labbate, M.; Sajjad, A.; Giguère, N.J.; Holley, M.P.; Stokes, H.W. Mobilization of a Tn402-like class 1 integron with a novel cassette array via flanking miniature inverted-repeat transposable element-like structures. Appl. Environ. Microbiol. 2009, 75, 6002–6004. [Google Scholar] [CrossRef]
- Mendes, R.E.; Castanheira, M.; Toleman, M.A.; Sader, H.S.; Jones, R.N.; Walsh, T.R. Characterization of an integron carrying blaIMP-1 and a new aminoglycoside resistance gene, aac(6′)-31, and its dissemination among genetically unrelated clinical isolates in a Brazilian hospital. Antimicrob. Agents Chemother. 2007, 51, 2611–2614. [Google Scholar] [CrossRef] [PubMed]
- Domingues, S.; Toleman, M.A.; Nielsen, K.M.; da Silva, G.J. Identical miniature inverted repeat transposable elements flank class 1 integrons in clinical isolates of Acinetobacter spp. J. Clin. Microbiol. 2013, 51, 2382–2384. [Google Scholar] [CrossRef] [PubMed]
- Minnick, M.F.; Raghavan, R. Genetics of Coxiella burnetii: On the path of specialization. Future Microbiol. 2011, 6, 1297–1314. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Zheng, Y.; Shang, X. MiteFinderII: A novel tool to identify miniature inverted-repeat transposable elements hidden in eukaryotic genomes. BMC Med. Genom. 2018, 11 (Suppl. S5), 101. [Google Scholar] [CrossRef] [PubMed]
- Crescente, J.M.; Zavallo, D.; Helguera, M.; Vanzetti, L.S. MITE Tracker: An accurate approach to identify miniature inverted-repeat transposable elements in large genomes. BMC Bioinform. 2018, 19, 348. [Google Scholar] [CrossRef] [PubMed]
- Ge, R.; Mai, G.; Zhang, R.; Wu, X.; Wu, Q.; Zhou, F. MUSTv2: An improved de novo detection program for recently active miniature inverted repeat transposable elements (MITEs). J. Integr. Bioinform. 2017, 14, 20170029. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.; Ji, G.; Liang, C. detectMITE: A novel approach to detect miniature inverted repeat transposable elements in genomes. Sci. Rep. 2016, 6, 19688. [Google Scholar] [CrossRef] [PubMed]
- Yang, G. MITE Digger, an efficient and accurate algorithm for genome wide discovery of miniature inverted repeat transposable elements. BMC Bioinform. 2013, 14, 186. [Google Scholar] [CrossRef]
- Han, Y.; Wessler, S.R. MITE-Hunter: A program for discovering miniature inverted-repeat transposable elements from genomic sequences. Nucleic Acids Res. 2010, 38, e199. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Minnick, M.F. Functional Roles and Genomic Impact of Miniature Inverted-Repeat Transposable Elements (MITEs) in Prokaryotes. Genes 2024, 15, 328. https://doi.org/10.3390/genes15030328
Minnick MF. Functional Roles and Genomic Impact of Miniature Inverted-Repeat Transposable Elements (MITEs) in Prokaryotes. Genes. 2024; 15(3):328. https://doi.org/10.3390/genes15030328
Chicago/Turabian StyleMinnick, Michael F. 2024. "Functional Roles and Genomic Impact of Miniature Inverted-Repeat Transposable Elements (MITEs) in Prokaryotes" Genes 15, no. 3: 328. https://doi.org/10.3390/genes15030328
APA StyleMinnick, M. F. (2024). Functional Roles and Genomic Impact of Miniature Inverted-Repeat Transposable Elements (MITEs) in Prokaryotes. Genes, 15(3), 328. https://doi.org/10.3390/genes15030328





