Insights for the Captive Management of South China Tigers Based on a Large-Scale Genetic Survey
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Mitochondrial DNA Analysis
2.3. Microsatellite Analysis
2.4. Genome Analysis
2.5. Statistical Analysis
3. Results
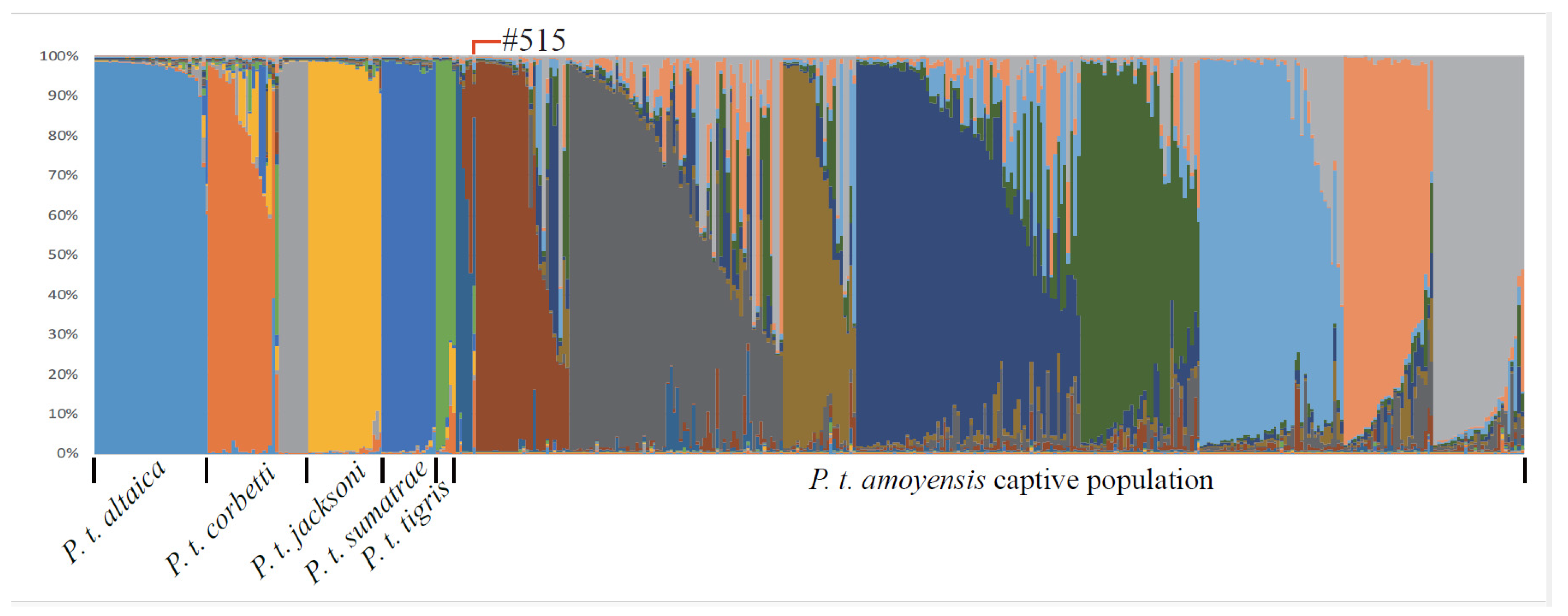
3.1. The Identification of Pureblood SCTs with Mitochondrial DNA and Microsatellite
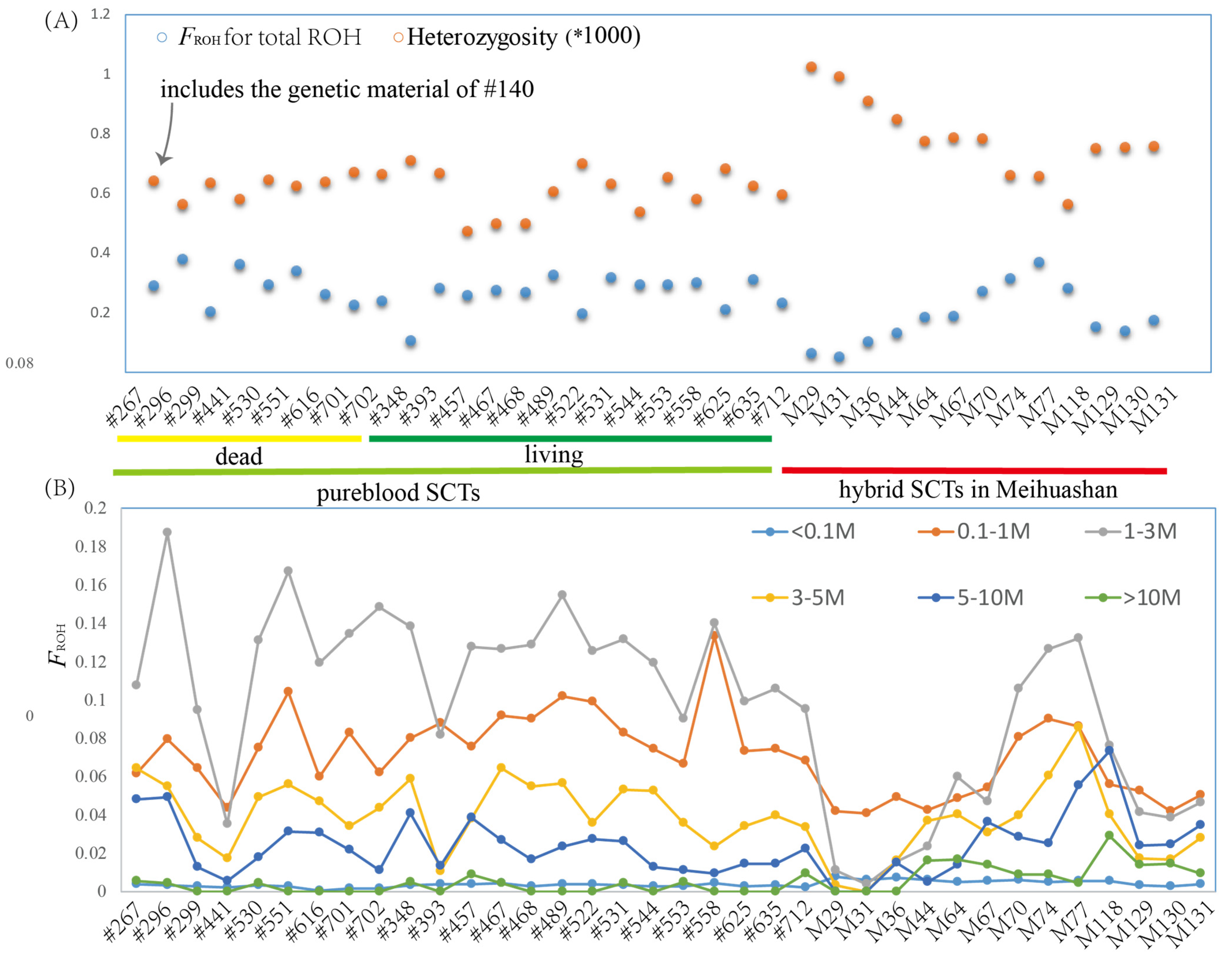
3.2. The Inbreeding of Pureblood SCTs
3.3. The Genomic Diversity of Pureblood SCTs
3.4. The Genomic Diversity of Hybrid SCTs
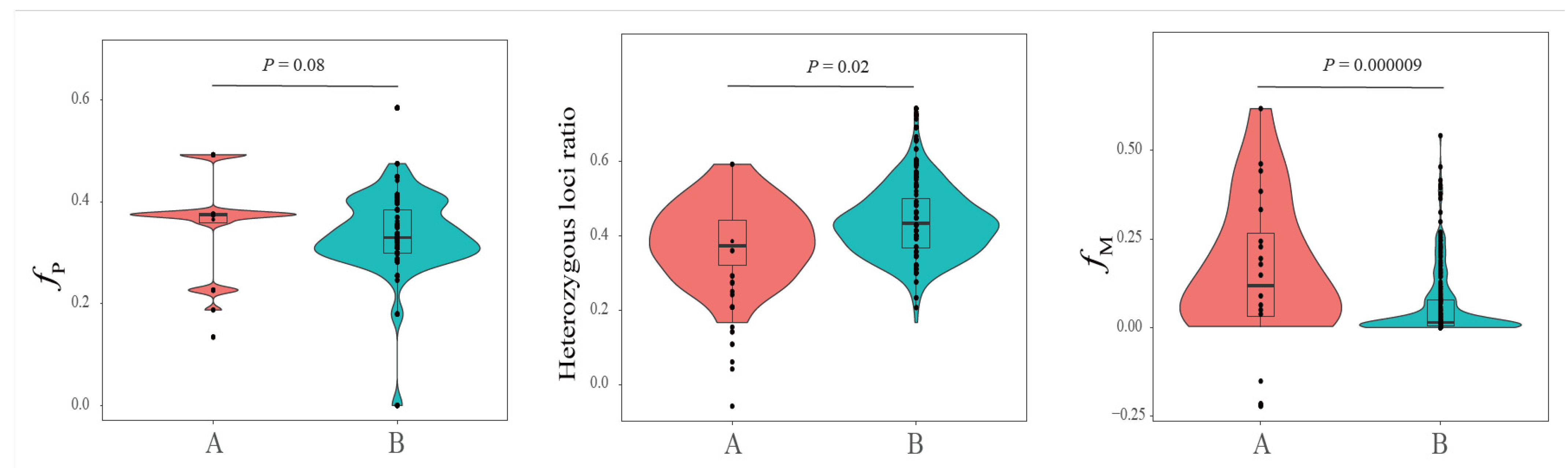
3.5. The Genomic Difference between Pureblood and Hybrid SCTs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kang, A.L.; Xie, Y.; Tang, J.R.; Sanderson, E.W.; Ginsberg, J.R.; Zhang, E.D. Historic distribution and recent loss of tigers in China. Integr. Zool. 2010, 5, 335–341. [Google Scholar] [CrossRef]
- Luo, S.J.; Kim, J.H.; Johnson, W.E.; van der Walt, J.; Martenson, J.; Yuhki, N.; Miquelle, D.G.; Uphyrkina, O.; Goodrich, J.M.; Quigley, H.B.; et al. Phylogeography and Genetic Ancestry of Tigers (Panthera tigris). PLoS Biol. 2004, 2, e442. [Google Scholar] [CrossRef]
- Liu, Y.C.; Sun, X.; Driscoll, C.; Miquelle, D.G.; Xu, X.; Martelli, P.; Uphyrkina, O.; Smith, J.L.D.; O’Brien, S.J.; Luo, S.J. Genome-Wide Evolutionary Analysis of Natural History and Adaptation in the World’s Tigers. Curr. Biol. 2018, 28, 3840–3849.e6. [Google Scholar] [CrossRef]
- Sun, X.; Liu, Y.C.; Tiunov, M.P.; Gimranov, D.O.; Zhuang, Y.; Han, Y.; Driscoll, C.A.; Pang, Y.; Li, C.; Pan, Y.; et al. Ancient DNA reveals genetic admixture in China during tiger evolution. Nat. Ecol. Evol. 2023, 7, 1914–1929. [Google Scholar] [CrossRef] [PubMed]
- Tidière, M.; Müller, P.; Sliwa, A.; Siberchicot, A.; Douay, G. Sex-specific actuarial and reproductive senescence in zoo-housed tiger (Panthera tigris): The importance of sub-species for conservation. Zoo Biol. 2021, 40, 320–329. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, E.E.; Khan, A.; Taylor, R.W.; Gouy, A.; Greenbaum, G.; Thiéry, A.; Kang, J.T.; Redondo, S.A.; Prost, S.; Barsh, G.; et al. Recent Evolutionary History of Tigers Highlights Contrasting Roles of Genetic Drift and Selection. Mol. Biol. Evol. 2021, 38, 2366–2379. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Westbury, M.V.; Yuan, J.; Wang, C.; Xiao, B.; Chen, S.; Song, S.; Wang, L.; Lin, H.; Lai, X.; et al. An extinct and deeply divergent tiger lineage from northeastern China recognized through palaeogenomics. Proc. Biol. Sci. 2022, 289, 20220617. [Google Scholar] [CrossRef] [PubMed]
- Tilson, R.; Defu, H.; Muntifering, J.; Nyhus, P.J. Dramatic decline of wild South China tigers Panthera tigris amoyensis: Field survey of priority tiger reserves. Oryx 2004, 38, 40–47. [Google Scholar] [CrossRef]
- Qin, Y.; Nyhus, P.J.; Larson, C.L.; Carroll, C.J.W.; Muntifering, J.; Dahmer, T.D.; Jun, L.; Tilson, R.L. An assessment of South China tiger reintroduction potential in Hupingshan and Houhe National Nature Reserves, China. Biol. Conserv. 2015, 182, 72–86. [Google Scholar] [CrossRef]
- Zhang, W.; Xu, X.; Yue, B.; Hou, R.; Xie, J.; Zou, Z.T.; Han, Y.; Shen, F.; Zhang, L.; Xie, Z.; et al. Sorting Out the Genetic Background of the Last Surviving South China Tigers. J. Hered. 2019, 110, 641–650. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.Z. The International South China Tiger Studbook; Chongqing Zoological Garden: Chongqing, China, 2023. [Google Scholar]
- Yuan, Y.H.; Pei, E.L.; Liu, Q.X. Reproductive parameters of female South China Tigers in captivity. Eur. J. Wildl. Res. 2020, 66, 37. [Google Scholar] [CrossRef]
- Zhang, L.; Lan, T.; Lin, C.; Fu, W.; Yuan, Y.; Lin, K.; Li, H.; Sahu, S.K.; Liu, Z.; Chen, D.; et al. Chromosome-scale genomes reveal genomic consequences of inbreeding in the South China tiger: A comparative study with the Amur tiger. Mol. Ecol. Resour. 2023, 23, 330–347. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wu, D.D.; Yuan, Y.H.; Yao, M.C.; Han, J.L.; Wu, Y.J.; Shan, F.; Li, W.P.; Zhai, J.Q.; Huang, M.; et al. Population genomic analysis provides strong evidence of the past success and future strategies of South China tiger breeding. BMC Biol. 2023, 21, 64. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.J.; Johnson, W.E.; Martenson, J.; Antunes, A.; Martelli, P.; Uphyrkina, O.; Traylor-Holzer, K.; Smith, J.L.; O’Brien, S.J. Subspecies Genetic Assignments of Worldwide Captive Tigers Increase Conservation Value of Captive Populations. Curr. Biol. 2008, 18, 592–596. [Google Scholar] [CrossRef]
- Tilson, R.; Traylor-Holzer, K.; Jiang, Q.M. The decline and impending extinction of the South China tiger. Oryx 1997, 31, 243–252. [Google Scholar] [CrossRef]
- Xu, Y.C.; Fang, S.G.; Li, Z.K. Sustainability of the South China tiger: Implications of inbreeding depression and introgression. Conserv. Genet. 2007, 8, 1199–1207. [Google Scholar] [CrossRef]
- Yuan, Y.H.; Yin, Y.Z.; Liu, Q.X. Inbreeding depression and population viability analysis of the South China tigers (Panthera tigris amoyensis) in captivity. Mamm. Biol. 2021, 101, 803–809. [Google Scholar] [CrossRef]
- Coates, D.J.; Byrne, M.; Moritz, C. Genetic Diversity and Conservation Units: Dealing with the Species-Population Continuum in the Age of Genomics. Front. Ecol. Evol. 2018, 6, 165. [Google Scholar] [CrossRef]
- Frandsen, P.; Fontsere, C.; Nielsen, S.V.; Hanghøj, K.; Castejon-Fernandez, N.; Lizano, E.; Hughes, D.; Hernandez-Rodriguez, J.; Korneliussen, T.S.; Carlsen, F.; et al. Targeted conservation genetics of the endangered chimpanzee. Heredity 2020, 125, 15–27. [Google Scholar] [CrossRef]
- Blomqvist, D.; Pauliny, A.; Larsson, M.; Flodin, L.Å. Trapped in the extinction vortex? Strong genetic effects in a declining vertebrate population. BMC Evol. Biol. 2010, 10, 33. [Google Scholar] [CrossRef]
- Keller, L.; Waller, D.M. Inbreeding effects in wild populations. Trends Ecol. Evol. 2002, 17, 230–241. [Google Scholar] [CrossRef]
- Farquharson, K.A.; Hogg, C.J.; Grueber, C.E. Offspring survival changes over generations of captive breeding. Nat. Commun. 2021, 12, 3045. [Google Scholar] [CrossRef] [PubMed]
- Kalinowski, S.T.; Hedrick, P.W.; Miller, P.S. Inbreeding depression in the Speke’s gazelle captive breeding program. Conserv. Biol. 2000, 14, 1375–1384. [Google Scholar] [CrossRef]
- Doekes, H.P.; Curik, I.; Nagy, I.; Farkas, J.; Kövér, G.; Windig, J.J. Revised Calculation of Kalinowski’s Ancestral and New Inbreeding Coefficients. Diversity 2020, 12, 155. [Google Scholar] [CrossRef]
- Van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.M.; Shipley, P. Microchecker: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Minch, E.; Ruiz-Linares, A.; Goldstein, D.B. MICROSAT. 1995. Available online: http://hpgl.stanford.edu/projects/microsat/ (accessed on 14 October 2006).
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Wang, J.L. Triadic IBD coefficients and applications to estimating pairwise relatedness. Genet Res. 2007, 89, 135–153. [Google Scholar] [CrossRef]
- Wang, J.L. Coancestry: A program for simulating, estimating and analysing relatedness and inbreeding coefficients. Mol. Ecol. Resour. 2015, 11, 141–145. [Google Scholar] [CrossRef]
- Marshall, T.C.; Coltman, D.W.; Pemberton, J.M.; Slate, J.; Spalton, J.A.; Guinness, F.E.; Smith, J.A.; Pilkington, J.G.; Clutton-Brock, T.H. Estimating the prevalence of inbreeding from incomplete pedigrees. Proc. Biol. Sci. 2002, 269, 1533–1539. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- DePristo, M.A.; Banks, E.; Poplin, R.; Garimella, K.V.; Maguire, J.R.; Hartl, C.; Philippakis, A.A.; del Angel, G.; Rivas, M.A.; Hanna, M.; et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011, 43, 491–498. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Alexander, D.H.; Lange, K. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinform. 2011, 12, 246. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Li, L.-H.; Ho, S.-F.; Chen, C.-H.; Wei, C.-Y.; Wong, W.-C.; Li, L.-Y.; Hung, S.-I.; Chung, W.-H.; Pan, W.-H.; Lee, M.-T.; et al. Long contiguous stretches of homozygosity in the human genome. Hum. Mutat. 2006, 27, 1115–1121. [Google Scholar] [CrossRef]
- Kardos, M.; Åkesson, M.; Fountain, T.; Flagstad, Ø.; Liberg, O.; Olason, P.; Sand, H.; Wabakken, P.; Wikenros, C.; Ellegren, H. Genomic consequences of intensive inbreeding in an isolated wolf population. Nat. Ecol. Evol. 2018, 2, 124–131. [Google Scholar] [CrossRef]
- Keller, M.C.; Visscher, P.M.; Goddard, M.E. Quantification of Inbreeding Due to Distant Ancestors and Its Detection Using Dense Single Nucleotide Polymorphism Data. Genetics 2011, 189, 237–249. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w 1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Shan, L.; Hu, Y.; Zhu, L.; Yan, L.; Wang, C.; Li, D.; Jin, X.; Zhang, C.; Wei, F. Large-Scale Genetic Survey Provides Insights into the Captive Management and Reintroduction of Giant Pandas. Mol. Biol. Evol. 2014, 31, 2663–2671. [Google Scholar] [CrossRef] [PubMed]
- Howrigan, D.P.; Simonson, M.A.; Keller, M.C. Detecting autozygosity through runs of homozygosity: A comparison of three autozygosity detection algorithms. BMC Genom. 2011, 12, 460. [Google Scholar] [CrossRef]
- Teixeira, J.C.; Huber, C.D. The inflated significance of neutral genetic diversity in conservation genetics. Proc. Natl. Acad. Sci. USA 2021, 118, e2015096118. [Google Scholar] [CrossRef]
- Kyriazis, C.C.; Wayne, R.K.; Lohmueller, K.E. Strongly deleterious mutations are a primary determinant of extinction risk due to inbreeding depression. Evol. Lett. 2021, 5, 33–47. [Google Scholar] [CrossRef]
- Kuderna, L.F.K.; Gao, H.; Janiak, M.C.; Kuhlwilm, M.; Orkin, J.D.; Bataillon, T.; Manu, S.; Valenzuela, A.; Bergman, J.; Rousselle, M.; et al. A global catalog of whole-genome diversity from 233 primate species. Science 2023, 380, 906–913. [Google Scholar] [CrossRef] [PubMed]
- Ralls, K.; Sunnucks, P.; Lacy, R.C.; Frankham, R. Genetic rescue: A critique of the evidence supports maximizing genetic diversity rather than minimizing the introduction of putatively harmful genetic variation. Biol. Conserv. 2020, 251, 108784. [Google Scholar] [CrossRef]
- Charlesworth, D.; Willis, J.H. The genetics of inbreeding depression. Nat. Rev. Genet. 2009, 10, 783–796. [Google Scholar] [CrossRef]
- Miquelle, D.G.; Smirnov, E.N.; Zaumyslova, O.Y.; Soutyrina, S.V.; Johnson, D.H. Population dynamics of Amur tigers (Panthera tigris altaica) in Sikhote-Alin Biosphere Zapovednik: 1966–2012. Integr. Zool. 2015, 10, 315–328. [Google Scholar] [CrossRef]

| Location | Living SCTs Numbers | Living SCTs Numbers for Microsatellite | Living SCTs Numbers for Genome |
|---|---|---|---|
| Canton Zoological Garden | 14 | 14 (100%) | 4 |
| Changsha Zoological Garden | 12 | 10 (83%) | 0 |
| Chengchou Zoological Garden | 2 | 2 (100%) | 0 |
| Chengdu Zoological Garden | 2 | 2 (100%) | 0 |
| Chongqing Zoological Garden | 5 | 4 (80%) | 0 |
| Fuzhou Zoological Garden | 1 | 1 (100%) | 0 |
| Hangzhou Wild Animal Park | 17 | 17 (100%) | 0 |
| Kueiyang Qianling Park | 1 | 1 (100%) | 0 |
| Laohu Valley Reserve Africa | 18 | 1 (6%) | 0 |
| Linyi Botanical Garden | 1 | 1 (100%) | 0 |
| Luoyang Wangcheng Park Zoo | 57 | 56 (98%) | 1 |
| Meihuashan Natural Reserve | 9 | 9 (100%) | 0 |
| Nanchang Zoological Garden | 44 | 40 (91%) | 6 |
| Panyu Xiangjiang Safari Park | 5 | 5 (100%) | 0 |
| Shanghai Zoological Garden | 30 | 23 (77%) | 3 |
| Shaoguan Zoological Garden | 11 | 10 (91%) | 0 |
| Suchou Zoological Garden | 17 | 15 (88%) | 0 |
| Total | 246 | 211 (86%) | 14 (6%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, W.; Lin, K.; Fu, W.; Xie, J.; Fan, X.; Zhang, M.; Luo, H.; Yin, Y.; Guo, Q.; Huang, H.; et al. Insights for the Captive Management of South China Tigers Based on a Large-Scale Genetic Survey. Genes 2024, 15, 398. https://doi.org/10.3390/genes15040398
Zhang W, Lin K, Fu W, Xie J, Fan X, Zhang M, Luo H, Yin Y, Guo Q, Huang H, et al. Insights for the Captive Management of South China Tigers Based on a Large-Scale Genetic Survey. Genes. 2024; 15(4):398. https://doi.org/10.3390/genes15040398
Chicago/Turabian StyleZhang, Wenping, Kaixiong Lin, Wenyuan Fu, Junjin Xie, Xueyang Fan, Mingchun Zhang, Hongxing Luo, Yuzhong Yin, Qiang Guo, He Huang, and et al. 2024. "Insights for the Captive Management of South China Tigers Based on a Large-Scale Genetic Survey" Genes 15, no. 4: 398. https://doi.org/10.3390/genes15040398
APA StyleZhang, W., Lin, K., Fu, W., Xie, J., Fan, X., Zhang, M., Luo, H., Yin, Y., Guo, Q., Huang, H., Chen, T., Lin, X., Yuan, Y., Huang, C., & Du, S. (2024). Insights for the Captive Management of South China Tigers Based on a Large-Scale Genetic Survey. Genes, 15(4), 398. https://doi.org/10.3390/genes15040398






