Development and Characterization of a Novel FVB-PrkdcR2140C Mouse Model for Adriamycin-Induced Nephropathy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Animals
2.3. Generation of Congenic FVB-PrkdcR2140C Mice
2.4. Administration of ADR
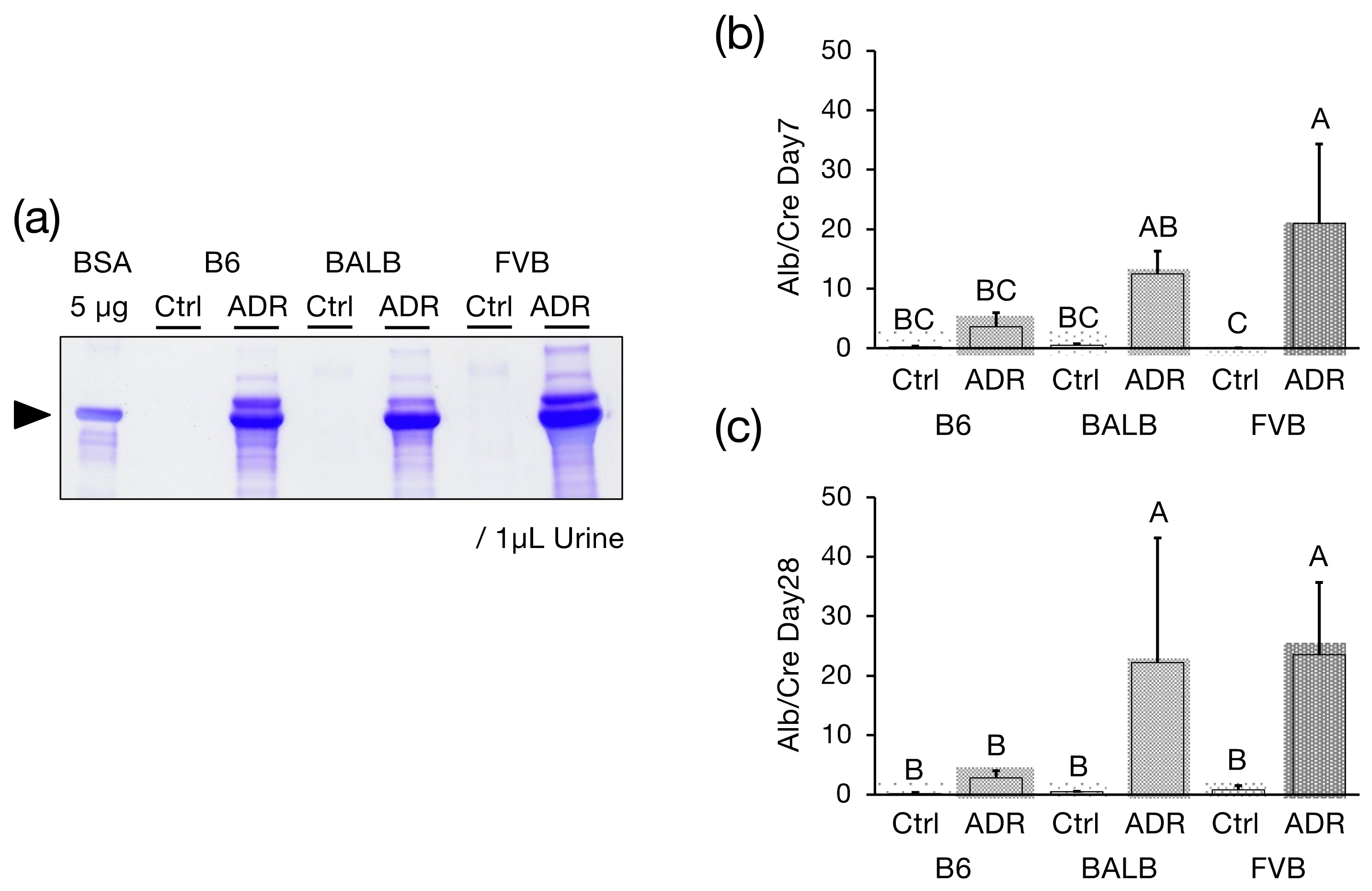
2.5. Detection of Urinary Albumin
2.6. Calculation of the Urinary Albumin-to-Creatinine Ratio
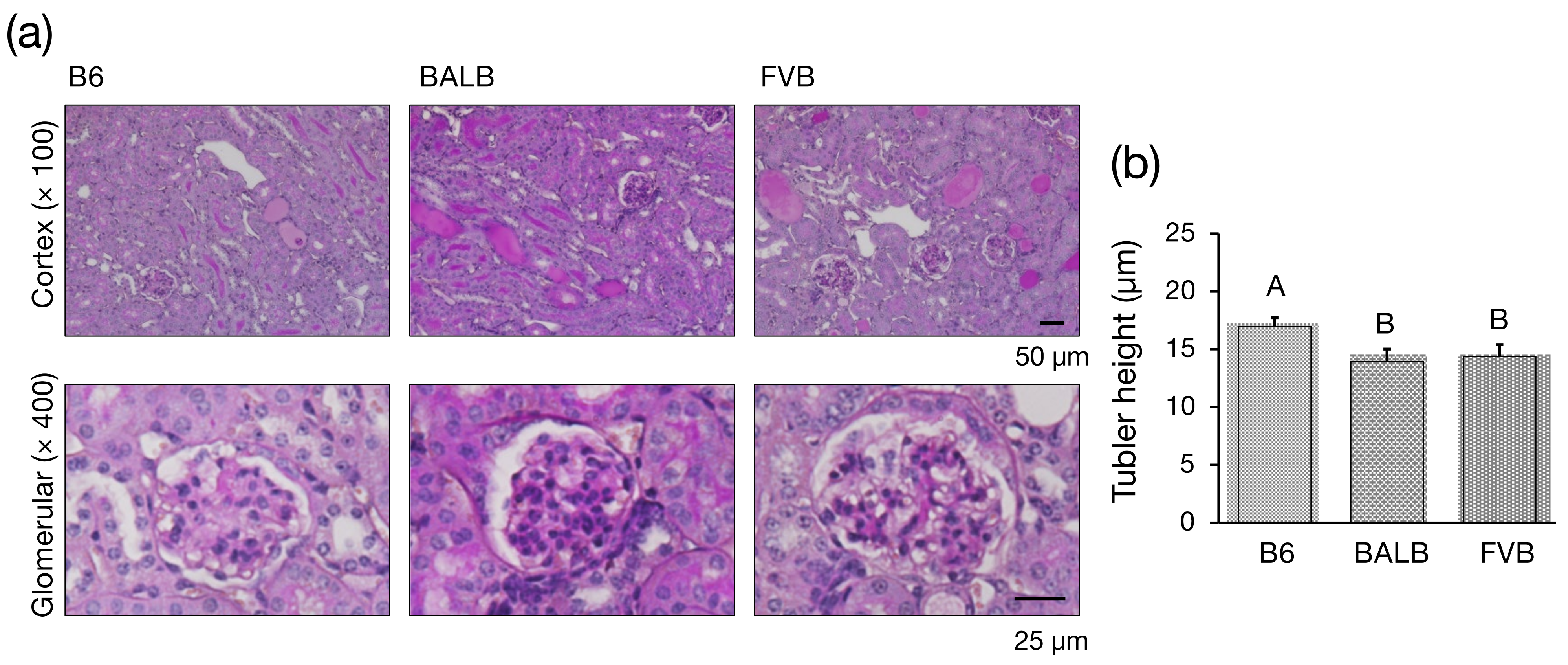
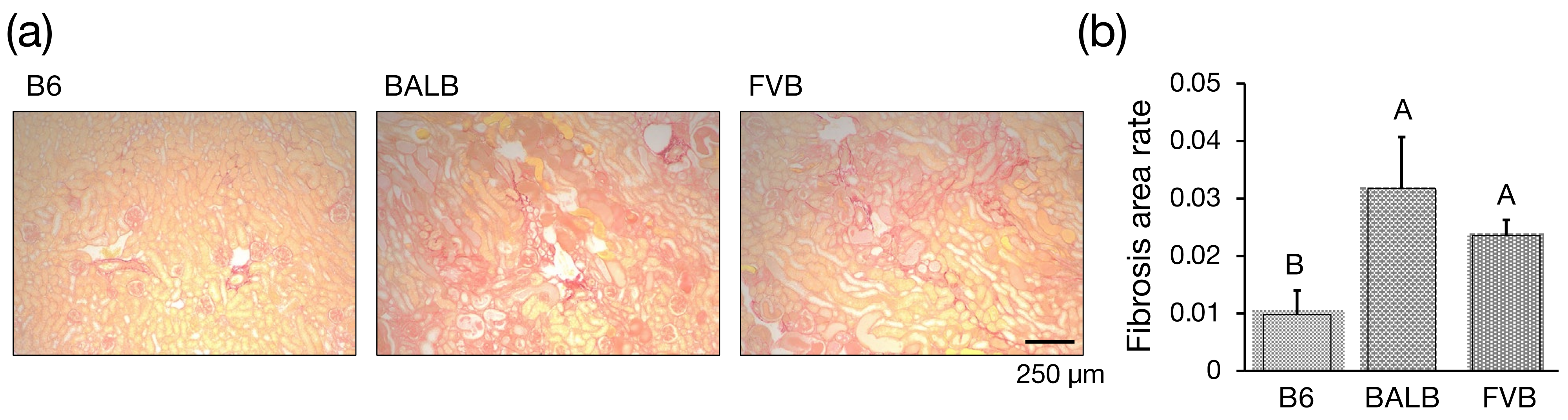
2.7. Histology
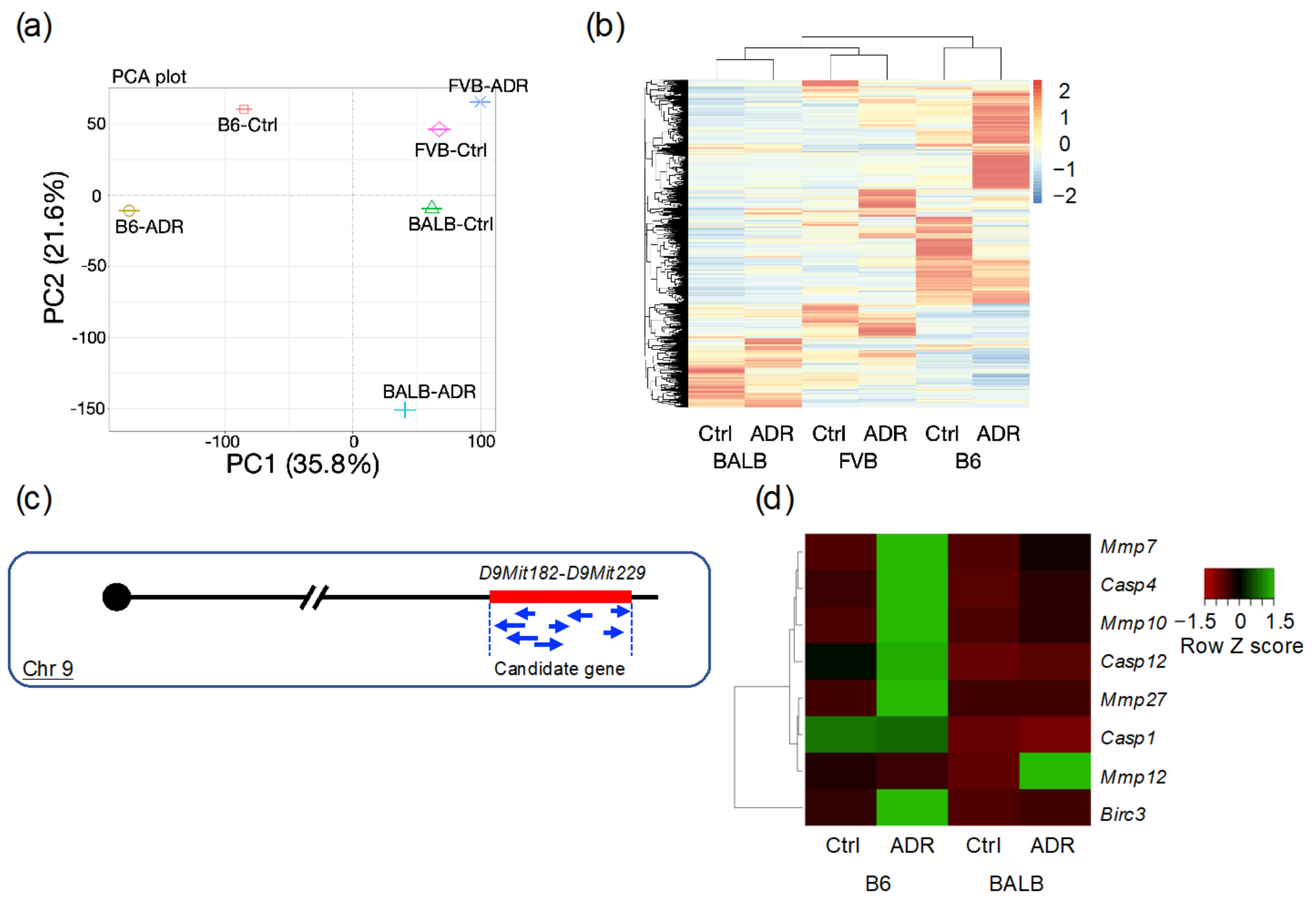
2.8. RNA Sequencing Analysis
2.9. Quantitative Reverse Transcription-PCR (qRT-PCR)
2.10. Statistics
3. Results
3.1. Urinary Alb Excretion
3.2. Histological Investigation
3.3. Assessment of Interstitial Fibrosis
3.4. Analysis of Differentially Expressed Genes through RNA-seq
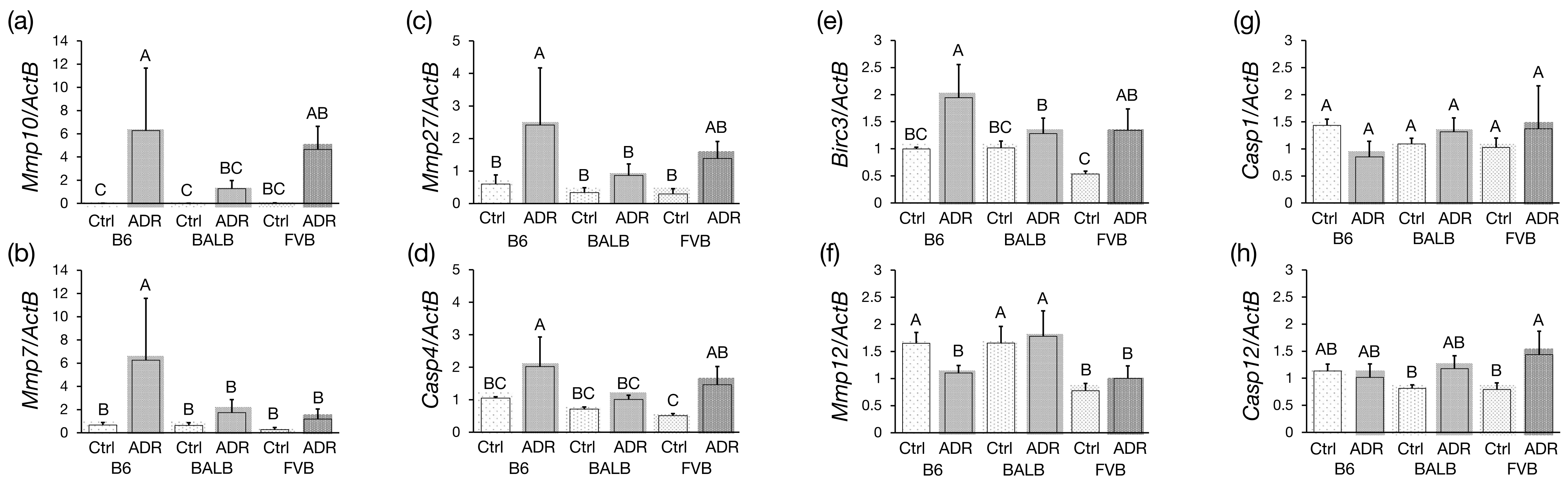
3.5. Analysis of Differentially Expressed Genes by qRT-PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cheo, S.W.; Low, Q.J.; Lim, T.H.; Mak, W.W.; Yip, C.A.K.; Wong, K.W. A practical approach to chronic kidney disease in primary care. Malays. Fam. Physician 2022, 17, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Liyanage, T.; Ninomiya, T.; Jha, V.; Neal, B.; Patrice, H.M.; Okpechi, I.; Zhao, M.; Lv, J.; Garg, A.X.; Knight, J.; et al. Worldwide access to treatment for end-atage kidney disease: A systematic review. Lancet 2015, 385, 1975–1982. [Google Scholar] [CrossRef] [PubMed]
- GBD Chronic Kidney Disease Collaboration. Global, regional, and national burden of chronic kidney disease, 1990–2017: A systematic analysis for the global burden of disease study 2017. Lancet 2020, 395, 709–733. [Google Scholar]
- Georgianos, P.I.; Agarwal, R. Hypertension in chronic kidney disease: Treatment standard 2023. Nephrol. Dial. Transplant. 2023, 38, 2694–2703. [Google Scholar] [CrossRef]
- House, A.A.; Wanner, C.; Sarnak, M.J.; Piña, I.L.; McIntyre, C.W.; Komenda, P.; Kasiske, B.L.; Deswal, A.; deFilippi, C.R.; Cleland, J.G.F.; et al. Heart failure in chronic kidney disease: Conclusions from a Kidney Disease: Improving Global Outcomes (KDIGO) controversies conference. Kidney Int. 2019, 95, 1304–1317. [Google Scholar] [CrossRef] [PubMed]
- Thomas, M.C.; Brownlee, M.; Susztak, K.; Sharma, K.; Jandeleit-Dahm, K.A.M.; Zoungas, S.; Rossing, P.; Groop, P.; Cooper, M.E. Diabetic kidney disease. Nat. Rev. Dis. Primers 2015, 1, 15018. [Google Scholar] [CrossRef] [PubMed]
- Fogo, A.B. Animal models of FSGS: Lessons for pathogenesis and treatment. Semin. Nephrol. 2003, 23, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Rossini, M.; Fogo, A.B. Interpreting segmental glomerular sclerosis. Curr. Diagn. Pathol. 2004, 10, 1–10. [Google Scholar] [CrossRef]
- Fogo, A.B. Causes and pathogenesis of focal segmental glomerulosclerosis. Nat. Rev. Nephrol. 2015, 11, 76–87. [Google Scholar] [CrossRef] [PubMed]
- Stokes, M.B.; Valeri, A.M.; Markowitz, G.S.; D’Agati, V.D. Cellular focal segmental glomerulosclerosis: Clinical and pathologic features. Kidney Int. 2006, 70, 1783–1792. [Google Scholar] [CrossRef] [PubMed]
- Sternberg, S.S. Cross-striated fibrils and other ultrastructural alterations in glomeruli of rats with daunomycin nephrosis. Lab. Investig. 1970, 23, 39–51. [Google Scholar] [PubMed]
- Zheng, Z.; Pavlidis, P.; Chua, S.; D’Agati, V.D.; Gharavi, A.G. An ancestral haplotype defines susceptibility to doxorubicin nephropathy in the laboratory mouse. J. Am. Soc. Nephrol. 2006, 17, 1796–1800. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Schmidt-Ott, K.M.; Chua, S.; Foster, K.A.; Frankel, R.Z.; Pavlidis, P.; Barasch, J.; D’Agati, V.D.; Gharavi, A.G. A mendelian locus on chromosome 16 determines susceptibility to doxorubicin nephropathy in the mouse. Proc. Natl. Acad. Sci. USA 2005, 102, 2502–2507. [Google Scholar] [CrossRef] [PubMed]
- Papeta, N.; Zheng, Z.; Schon, E.A.; Brosel, S.; Altintas, M.M.; Nasr, S.H.; Reiser, J.; D’Agati, V.D.; Gharavi, A.G. Prkdc participates in mitochondrial genome maintenance and prevents adriamycin-induced nephropathy in mice. J. Clin. Investig. 2010, 120, 4055–4064. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Takahashi, Y.; Hiura, K.; Nakano, K.; Okamura, T.; Sasaki, H.; Sasaki, N. A Single Amino Acid Substitution in PRKDC is a Determinant of Sensitivity to Adriamycin-Induced Renal Injury in Mouse. Biochem. Biophys. Res. Commun. 2021, 556, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Laouari, D.; Burtin, M.; Phelep, A.; Martino, C.; Pillebout, E.; Montagutelli, X.; Friedlander, G.; Terzi, F. TGF-alpha mediates genetic susceptibility to chronic kidney disease. J. Am. Soc. Nephrol. 2011, 22, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Gharavi, A.G.; Ahmad, T.; Wong, R.D.; Hooshyar, R.; Vaughn, J.; Oller, S.; Frankel, R.Z.; Bruggeman, L.A.; D’Agati, V.D.; Klotman, P.E.; et al. Mapping a locus for susceptibility to HIV-1-associated nephropathy to mouse chromosome 3. Proc. Natl. Acad. Sci. USA 2004, 101, 2488–2493. [Google Scholar] [CrossRef] [PubMed]
- Baleato, R.M.; Guthrie, P.L.; Gubler, M.; Ashman, L.K.; Roselli, S. Deletion of CD151 results in a strain-dependent glomerular disease due to severe alterations of the glomerular basement membrane. Am. J. Pathol. 2008, 173, 927–937. [Google Scholar] [CrossRef] [PubMed]
- Kawai, S.; Takagi, Y.; Kaneko, S.; Kurosawa, T. Effect of three types of mixed anesthetic agents alternate to ketamine in mice. Exp. Anim. 2011, 60, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Hiura, K.; Sasaki, H.; Okamura, T.; Sasaki, N. Genetic background strongly influences the transition to chronic kidney disease of adriamycin nephropathy in mice. Exp. Anim. 2023, 72, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.A.; Kutlu, B.; Velmurugan, K.; Kizaka-Kondoh, S.; Lee, C.E.; Wong, R.; Valentine, A.; Davidson, H.W.; Hutton, J.C.; Pugazhenthi, S. Cytokine-mediated induction of anti-apoptotic genes that are linked to Nuclear Factor Kappa-B (NF-kappaB) signalling in human islets and in a mouse beta cell line. Diabetologia 2009, 52, 1092–1101. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Chen, J.; Geng, C.; Wang, X.; Wang, Y.; Sun, N.; Wang, P.; Han, L.; Li, Z.; Fan, H.; et al. Myoglobin promotes macrophage polarization to M1 type and pyroptosis Via the RIG-I/Caspase1/GSDMD signaling pathway in CS-AKI. Cell Death Discov. 2022, 8, 90. [Google Scholar] [CrossRef] [PubMed]
- Sakurai, M.; Watanabe, T.; Suzuki, T.; Furihata, C. Time-course comparison of gene expression profiles induced by the genotoxic hepatocarcinogen, chrysene, in the mouse liver. Genes Environ. 2014, 5, 54–64. [Google Scholar] [CrossRef]
- Chen, C.; Wang, D.; Yu, Y.; Zhao, T.; Min, N.; Wu, Y.; Kang, L.; Zhao, Y.; Du, L.; Zhang, M.; et al. Legumain promotes tubular ferroptosis by facilitating chaperone-mediated autophagy of GPX4 in AKI. Cell Death Dis. 2021, 12, 65. [Google Scholar] [CrossRef] [PubMed]
- Kageyama, Y.; Nakamura, M.; Igari, Y.; Yamaguchi, S.; Oguchi, A.; Murakawa, Y.; Hattori, Y.; Sasano, Y. Expression of Matrix Metalloproteinase-3 and -10 is up-regulated in the periodontal tissues of aged mice. J. Periodontal. Res. 2022, 57, 733–741. [Google Scholar]
- Nuttall, R.K.; Sampieri, C.L.; Pennington, C.J.; Gill, S.E.; Schultz, G.A.; Edwards, D.R. Expression analysis of the entire MMP and TIMP gene families during mouse tissue development. FEBS Lett. 2004, 563, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Ranjan, M.; Lee, O.; Cottone, G.; Mirzaei Mehrabad, E.; Spike, B.T.; Zeng, Z.; Yadav, S.; Chatterton, R.; Kim, J.J.; Clare, S.E.; et al. Progesterone receptor antagonists reverse stem cell expansion and the paracrine effectors of progesterone action in the mouse mammary gland. Breast Cancer Res. 2021, 23, 78. [Google Scholar] [CrossRef]
- Asanuma, K.; Akiba-Takagi, M.; Kodama, F.; Asao, R.; Nagai, Y.; Lydia, A.; Fukuda, H.; Tanaka, E.; Shibata, T.; Takahara, H.; et al. Dendrin location in podocytes is associated with disease progression in animal and human glomerulopathy. Am. J. Nephrol. 2011, 33, 537–549. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Watanabe, M.; Hiura, K.; Isobe, A.; Sasaki, H.; Sasaki, N. Positive correlation between renal tubular flattening and renal tubular injury/interstitial fibrosis in murine kidney disease models. J. Vet. Med. Sci. 2021, 83, 397–402. [Google Scholar] [CrossRef] [PubMed]
- Bohle, A.; Müller, G.A.; Wehrmann, M.; Mackensen-Haen, S.; Xiao, J.C. Pathogenesis of chronic renal failure in the primary glomerulopathies, renal vasculopathies, and chronic interstitial nephritides. Kidney Int. Suppl. 1996, 54, S2. [Google Scholar] [PubMed]
- Sasaki, H.; Marusugi, K.; Kimura, J.; Kitamura, H.; Nagasaki, K.; Torigoe, D.; Agui, T.; Sasaki, N. Genetic background-dependent diversity in renal failure caused by the Tensin2 gene deficiency in the mouse. Biomed. Res. 2015, 36, 323–330. [Google Scholar] [CrossRef] [PubMed]
- Nishino, T.; Sasaki, N.; Nagasaki, K.; Ahmad, Z.; Agui, T. Genetic background strongly influences the severity of glomerulosclerosis in mice. J. Vet. Med. Sci. 2010, 72, 1313–1318. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Sasaki, H.; Okawara, S.; Sasaki, N. Genetic loci for resistance to podocyte injury caused by the Tensin2 gene deficiency in mice. BMC Genet. 2018, 19, 24. [Google Scholar] [CrossRef] [PubMed]
- Webster, A.C.; Nagler, E.V.; Morton, R.L.; Masson, P. Chronic kidney disease. Lancet 2017, 389, 1238–1252. [Google Scholar] [CrossRef] [PubMed]
- Ra, H.; Parks, W.C. Control of matrix metalloproteinase catalytic activity. Matrix Biol. 2007, 26, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Liu, Y. Matrix Metalloproteinase-10 in kidney injury repair and disease. Int. J. Mol. Sci. 2022, 23, 2131. [Google Scholar] [CrossRef]
- Hu, C.; Zuo, Y.; Ren, Q.; Sun, X.; Zhou, S.; Liao, J.; Hong, X.; Miao, J.; Zhou, L.; Liu, Y. Matrix Metalloproteinase-10 protects against acute kidney injury by augmenting epidermal growth factor receptor signaling. Cell Death Dis. 2021, 12, 70. [Google Scholar] [CrossRef] [PubMed]
- Frazzi, R. BIRC3 and BIRC5: Multi-faceted inhibitors in cancer. Cell Biosci. 2021, 11, 8. [Google Scholar] [CrossRef] [PubMed]
- Hitomi, J.; Katayama, T.; Eguchi, Y.; Kudo, T.; Taniguchi, M.; Koyama, Y.; Manabe, T.; Yamagishi, S.; Bando, Y.; Imaizumi, K.; et al. Involvement of Caspase-4 in endoplasmic reticulum stress-induced apoptosis and Aβ-Induced cell death. J. Cell Biol. 2004, 165, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Niu, H.; Li, Y.; Li, H.; Chi, Y.; Zhuang, M.; Zhang, T.; Liu, M.; Nie, L. Matrix Metalloproteinase 12 modulates high-fat-diet induced glomerular fibrogenesis and inflammation in a mouse model of obesity. Sci. Rep. 2016, 6, 20171. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Zhou, D.; Zhu, H.; Liao, J.; Lin, L.; Hong, X.; Hou, F.F.; Liu, Y. Matrix Metalloproteinase-7 protects against acute kidney injury by priming renal tubules for survival and regeneration. Kidney Int. 2019, 95, 1167–1180. [Google Scholar] [CrossRef]
| Gene Names | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| ActB | cagaaggagattactgctctggct | tactcctgcttgctgatccacatc |
| Birc3 | cagaggtcattgctggcgtt | tggtcggttttactgctaggc |
| Casp1 | acaaggcacgggacctatg | tcccagtcagtcctggaaatg |
| Casp4 | gaagacttaggctacgatgtggtg | tgtctgatgtctggtgttctgag |
| Casp12 | gcccatgtggagacagattt | atagtgggcatctgggtcag |
| Mmp7 | cttacctcggatcgtagtgga | ccccaactaaccctcttgaagt |
| Mmp10 | tccaggaattgagccacaag | gggtcaaactcgaactgtgat |
| Mmp12 | gcagtcctctatttcaaaagacac | ccaaaccagcttgtacttttcaatg |
| Mmp27 | aggataataaagtgcttcccagga | aagaaatagaggaatccattatgttgg |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Watanabe, M.; Ishii, Y.; Hashimoto, K.; Takimoto, H.R.; Sasaki, N. Development and Characterization of a Novel FVB-PrkdcR2140C Mouse Model for Adriamycin-Induced Nephropathy. Genes 2024, 15, 456. https://doi.org/10.3390/genes15040456
Watanabe M, Ishii Y, Hashimoto K, Takimoto HR, Sasaki N. Development and Characterization of a Novel FVB-PrkdcR2140C Mouse Model for Adriamycin-Induced Nephropathy. Genes. 2024; 15(4):456. https://doi.org/10.3390/genes15040456
Chicago/Turabian StyleWatanabe, Masaki, Yuki Ishii, Kazuki Hashimoto, Hayato R. Takimoto, and Nobuya Sasaki. 2024. "Development and Characterization of a Novel FVB-PrkdcR2140C Mouse Model for Adriamycin-Induced Nephropathy" Genes 15, no. 4: 456. https://doi.org/10.3390/genes15040456





