A Summary of Two Decades of QTL and Candidate Genes That Control Seed Tocopherol Contents in Maize (Zea mays L.)
Abstract
:1. Introduction
2. Tocopherols and Tocotrienols Biosynthetic Pathway
3. QTLs That Control Seed Tocopherol Contents
- Thirteen QTL controlling seed α-Tocopherol (αT) on chromosomes 1, 2, 3, 4, 5, 6, 7, 8, and 9.
- Eighteen QTL regulating seed δ-Tocopherol (δT) on chromosomes 1, 2, 3, 4, 5, 6, 7, 9, and 10.
- Twenty-one QTL governing seed γ-Tocopherol (γT) on chromosomes 1, 2, 3, 4, 5, 6, 7, 8, 9, and 10.
- Eighteen QTL controlling seed Total tocopherols (ΣT) on chromosomes 1, 2, 3, 4, 5, 6, 8, 9, and 10.
- Seventeen QTL associated with seed α-Tocotrienol (αT3) on chromosomes 1, 2, 3, 4, 5, 6, 7, 8, 9, and 10.
- Twenty-one QTL regulating seed δ-Tocotrienol (δT3) on chromosomes 1, 3, 4, 5, 6, 7, 8, 9, and 10.
- Fourteen QTL governing seed γ-Tocotrienol (γT3) on chromosomes 1, 2, 3, 4, 5, 6, 7, 8, 9, and 10.
- Twelve QTL controlling seed total tocotrienols (ΣT3) on chromosomes 1, 2, 3, 4, 5, 6, 7, 8, 9, and 10.
- Twenty QTL governing seed total tocochromanols (ΣTT3) on chromosomes 1, 2, 3, 4, 5, 6, 7, 8, 9, and 10.
- Eight QTL regulating seed plastochromanol-8 (PC-8) on chromosomes 1, 2, 3, 5, 6, 7, and 9.
4. Seed Tocopherol Candidate Genes
5. Expression Analysis of the Candidate Gene Involved in the Tocopherol and Tocotrienol Biosynthetic Pathway
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Marwede, V.; Schierholt, A.; Möllers, C.; Becker, H.C. Genotype× environment interactions and heritability of tocopherol contents in canola. Crop Sci. 2004, 44, 728–731. [Google Scholar] [CrossRef]
- Kamal-Eldin, A.; Appelqvist, L.-Å. The chemistry and antioxidant properties of tocopherols and tocotrienols. Lipids 1996, 31, 671–701. [Google Scholar] [CrossRef] [PubMed]
- DellaPenna, D.; Pogson, B.J. Vitamin Synthesis in Plants: Tocopherols and Carotenoids. Annu. Rev. Plant Biol. 2006, 57, 711–738. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, K.M.; Weaver, L.M. The shikimate pathway. Annu. Rev. Plant Biol. 1999, 50, 473–503. [Google Scholar] [CrossRef] [PubMed]
- Kornsteiner, M.; Wagner, K.-H.; Elmadfa, I. Tocopherols and total phenolics in 10 different nut types. Food Chem. 2006, 98, 381–387. [Google Scholar] [CrossRef]
- Maeda, H.; DellaPenna, D. Tocopherol functions in photosynthetic organisms. Curr. Opin. Plant Biol. 2007, 10, 260–265. [Google Scholar] [CrossRef] [PubMed]
- Munné-Bosch, S. The role of α-tocopherol in plant stress tolerance. J. Plant Physiol. 2005, 162, 743–748. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Qiu, D.; Pang, Y.; Gao, H.; Wang, X.; Qin, Y. Diverse roles of tocopherols in response to abiotic and biotic stresses and strategies for genetic biofortification in plants. Mol. Breed. 2020, 40, 18. [Google Scholar] [CrossRef]
- Munné-Bosch, S.; Falk, J. New insights into the function of tocopherols in plants. Planta 2004, 218, 323–326. [Google Scholar] [CrossRef]
- Havaux, M.; Lutz, C.; Grimm, B. Chloroplast membrane photostability in chlP transgenic tobacco plants deficient in tocopherols. Plant Physiol. 2003, 132, 300–310. [Google Scholar] [CrossRef]
- Li, Y.; Wang, Z.; Sun, X.; Tang, K. Current Opinions on the Functions of Tocopherol Based on the Genetic Manipulation of Tocopherol Biosynthesis in Plants. J. Integr. Plant Biol. 2008, 50, 1057–1069. [Google Scholar] [CrossRef] [PubMed]
- Sattler, S.E.; Cheng, Z.; DellaPenna, D. From Arabidopsis to agriculture: Engineering improved Vitamin E content in soybean. Trends Plant Sci. 2004, 9, 365–367. [Google Scholar] [CrossRef]
- Abbasi, A.-R.; Hajirezaei, M.; Hofius, D.; Sonnewald, U.; Voll, L.M. Specific roles of α-and γ-tocopherol in abiotic stress responses of transgenic tobacco. Plant Physiol. 2007, 143, 1720–1738. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Kang, I.; Fang, X.; Wang, W.; Lee, M.A.; Hollins, R.R.; Marshall, M.R.; Chung, S. γ-tocotrienol attenuates high-fat diet-induced obesity and insulin resistance by inhibiting adipose inflammation and M1 macrophage recruitment. Int. J. Obes. 2015, 39, 438–446. [Google Scholar] [CrossRef] [PubMed]
- Alcalá, M.; Sánchez-Vera, I.; Sevillano, J.; Herrero, L.; Serra, D.; Ramos, M.P.; Viana, M. Vitamin E reduces adipose tissue fibrosis, inflammation, and oxidative stress and improves metabolic profile in obesity. Obesity 2015, 23, 1598–1606. [Google Scholar] [CrossRef] [PubMed]
- Hasty, A.H.; Gruen, M.L.; Terry, E.S.; Surmi, B.K.; Atkinson, R.D.; Gao, L.; Morrow, J.D. Effects of vitamin E on oxidative stress and atherosclerosis in an obese hyperlipidemic mouse model. J. Nutr. Biochem. 2007, 18, 127–133. [Google Scholar] [CrossRef]
- Morrissey, P.A.; Buckley, D.J.; Sheehy, P.J.A.; Monahan, F.J. Vitamin E and meat quality. Proc. Nutr. Soc. 1994, 53, 289–295. [Google Scholar] [CrossRef] [PubMed]
- Upston, J.M.; Kritharides, L.; Stocker, R. The role of vitamin E in atherosclerosis. Prog. Lipid Res. 2003, 42, 405–422. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.K.; Chin, K.-Y.; Suhaimi, F.H.; Ahmad, F.; Ima-Nirwana, S. Vitamin E As a Potential Interventional Treatment for Metabolic Syndrome: Evidence from Animal and Human Studies. Front. Pharmacol. 2017, 8, 444. [Google Scholar] [CrossRef]
- Clarke, M.W.; Burnett, J.R.; Croft, K.D. Vitamin E in human health and disease. Crit. Rev. Clin. Lab. Sci. 2008, 45, 417–450. [Google Scholar] [CrossRef]
- Robinson, I.; de Serna, D.G.; Gutierrez, A.; Schade, D.S. Vitamin E in humans: An explanation of clinical trial failure. Endocr. Pract. 2006, 12, 576–582. [Google Scholar] [CrossRef] [PubMed]
- Heng, K.S.; Hejar, A.; Stanslas, J.J.; Ooi, C.; Loh, S.P. Potential of mixed tocotrienol supplementation to reduce cholesterol and cytokines level in adults with metabolic syndrome. Malays. J. Nutr. 2015, 21, 231–243. [Google Scholar]
- Siddiqui, I.A.; Jaleel, A.; Al’kadri, H.M.; Akram, S.; Tamimi, W. Biomarkers of oxidative stress in women with pre-eclampsia. Biomark. Med. 2013, 7, 229–234. [Google Scholar] [CrossRef] [PubMed]
- Chander, S.; Guo, Y.Q.; Yang, X.H.; Yan, J.B.; Zhang, Y.R.; Song, T.M.; Li, J.S. Genetic dissection of tocopherol content and composition in maize grain using quantitative trait loci analysis and the candidate gene approach. Mol. Breed. 2008, 22, 353–365. [Google Scholar] [CrossRef]
- Carrão-Panizzi, M.C.; Erhan, S.Z. Environmental and genetic variation of soybean tocopherol content under Brazilian growing conditions. J. Am. Oil Chem. Soc. 2007, 84, 921–928. [Google Scholar] [CrossRef]
- Carrera, C.S.; Seguin, P. Factors Affecting Tocopherol Concentrations in Soybean Seeds. J. Agric. Food Chem. 2016, 64, 9465–9474. [Google Scholar] [CrossRef] [PubMed]
- Forgey, W. Inheritence of the Isomers of Vitamin E in Zea mays; University of Illinois at Urbana-Champaign: Champaign, IL, USA, 1974. [Google Scholar]
- Rocheford, T.R.; Wong, J.C.; Egesel, C.O.; Lambert, R.J. Enhancement of Vitamin E Levels in Corn. J. Am. Coll. Nutr. 2002, 21 (Suppl. S3), 191S–198S. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.C.; Lambert, R.J.; Tadmor, Y.; Rocheford, T.R. QTL Associated with Accumulation of Tocopherols in Maize. Crop Sci. 2003, 43, 2257–2266. [Google Scholar] [CrossRef]
- Fritsche, S.; Wang, X.; Li, J.; Stich, B.; Kopisch-Obuch, F.J.; Endrigkeit, J.; Leckband, G.; Dreyer, F.; Friedt, W.; Meng, J.; et al. A candidate gene-based association study of tocopherol content and composition in rapeseed (Brassica napus). Front. Plant Sci. 2012, 3, 129. [Google Scholar] [CrossRef]
- Wang, S.; Basten, C.J.; Zeng, Z.-B. Windows QTL Cartographer 2.5; Department of Statistics, North Carolina State University: Raleigh, NC, USA, 2012. [Google Scholar]
- Gajardo, H.A.; Wittkop, B.; Soto-Cerda, B.; Higgins, E.E.; Parkin, I.A.P.; Snowdon, R.J.; Federico, M.L.; Iniguez-Luy, F.L. Association mapping of seed quality traits in Brassica napus L. using GWAS and candidate QTL approaches. Mol. Breed. 2015, 35, 143. [Google Scholar]
- Del Moral, L.; Fernández-Martínez, J.M.; Velasco, L.; Pérez-Vich, B. Quantitative Trait Loci for Seed Tocopherol Content in Sunflower. Crop Sci. 2012, 52, 786–794. [Google Scholar] [CrossRef]
- Haddadi, P.; Ebrahimi, A.; Langlade, N.B.; Yazdi-Samadi, B.; Berger, M.; Calmon, A.; Naghavi, M.R.; Vincourt, P.; Sarrafi, A. Genetic dissection of tocopherol and phytosterol in recombinant inbred lines of sunflower through quantitative trait locus analysis and the candidate gene approach. Mol. Breed. 2012, 29, 717–729. [Google Scholar] [CrossRef]
- Sui, M.; Jing, Y.; Li, H.; Zhan, Y.; Luo, J.; Teng, W.; Qiu, L.; Zheng, H.; Li, W.; Zhao, X.; et al. Identification of Loci and Candidate Genes Analyses for Tocopherol Concentration of Soybean Seed. Front. Plant Sci. 2020, 11, 539460. [Google Scholar] [CrossRef] [PubMed]
- Baseggio, M.; Murray, M.; Magallanes-Lundback, M.; Kaczmar, N.; Chamness, J.; Buckler, E.S.; Smith, M.E.; DellaPenna, D.; Tracy, W.F.; Gore, M.A. Genome-Wide Association and Genomic Prediction Models of Tocochromanols in Fresh Sweet Corn Kernels. Plant Genome 2019, 12, 180038. [Google Scholar] [CrossRef] [PubMed]
- Fenton, M.E.; Owens, B.F.; Lipka, A.E.; Ortiz, D.; Tiede, T.; Mateos-Hernandez, M.; Ferruzzi, M.G.; Rocheford, T. High-density linkage mapping of vitamin E content in maize grain. Mol. Breed. 2018, 38, 31. [Google Scholar] [CrossRef]
- Li, Q.; Yang, X.; Xu, S.; Cai, Y.; Zhang, D.; Han, Y.; Li, L.; Zhang, Z.; Gao, S.; Li, J.; et al. Genome-wide association studies identified three independent polymorphisms associated with α-tocopherol content in maize kernels. PLoS ONE 2012, 7, e36807. [Google Scholar] [CrossRef]
- Xu, S.; Zhang, D.; Cai, Y.; Zhou, Y.; Trushar, S.; Farhan, A.; Li, Q.; Li, Z.; Wang, W.; Li, J.; et al. Dissecting tocopherols content in maize (Zea mays L.), using two segregating populations and high-density single nucleotide polymorphism markers. BMC Plant Biol. 2012, 12, 201. [Google Scholar] [CrossRef]
- Feng, F.; Deng, F.; Zhou, P.; Yan, J.; Wang, Q.; Yang, R.; Li, X. QTL mapping for the tocopherols at milk stage of kernel development in sweet corn. Euphytica 2013, 193, 409–417. [Google Scholar] [CrossRef]
- Lipka, A.E.; Gore, M.A.; Magallanes-Lundback, M.; Mesberg, A.; Lin, H.; Tiede, T.; Chen, C.; Buell, C.R.; Buckler, E.S.; Rocheford, T.; et al. Genome-wide association study and pathway-level analysis of tocochromanol levels in maize grain. G3 Genes Genomes Genet. 2013, 3, 1287–1299. [Google Scholar] [CrossRef]
- Feng, F.; Wang, Q.; Liang, C.; Yang, R.; Li, X. Enhancement of tocopherols in sweet corn by marker-assisted backcrossing of ZmVTE4. Euphytica 2015, 206, 513–521. [Google Scholar] [CrossRef]
- Diepenbrock, C.H.; Kandianis, C.B.; Lipka, A.E.; Magallanes-Lundback, M.; Vaillancourt, B.; Góngora-Castillo, E.; Wallace, J.G.; Cepela, J.; Mesberg, A.; Bradbury, P.J.; et al. Novel loci underlie natural variation in vitamin E levels in maize grain. Plant Cell 2017, 29, 2374–2392. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Xu, S.; Fan, Y.; Liu, N.; Zhan, W.; Liu, H.; Xiao, Y.; Li, K.; Pan, Q.; Li, W.; et al. Beyond pathways: Genetic dissection of tocopherol content in maize kernels by combining linkage and association analyses. Plant Biotechnol. J. 2018, 16, 1464–1475. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Yu, Y.; Li, G.; Xie, L.; Guo, X.; Li, J.; Li, Y.; Hu, J. Genome-wide association study of vitamin E in sweet corn kernels. Crop J. 2020, 8, 341–350. [Google Scholar] [CrossRef]
- Alves, M.L.; Bento-Silva, A.; Carbas, B.; Gaspar, D.; Paulo, M.; Brites, C.; Mendes-Moreira, P.; Brites, C.M.; Bronze, M.D.R.; Malosetti, M.; et al. Alleles to Enhance Antioxidant Content in Maize—A Genome-Wide Association Approach. J. Agric. Food Chem. 2020, 68, 4051–4061. [Google Scholar] [CrossRef] [PubMed]
- Hershberger, J.; Tanaka, R.; Wood, J.C.; Kaczmar, N.; Wu, D.; Hamilton, J.P.; DellaPenna, D.; Buell, C.R.; Gore, M.A. Transcriptome-wide association and prediction for carotenoids and tocochromanols in fresh sweet corn kernels. Plant Genome 2022, 15, e20197. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, P.; Yun, P.; Wang, L.; Gao, G.; Zhang, Q.; Luo, L.; Zhang, C.; He, Y. Genetic dissection of vitamin E content in rice (Oryza sativa) grains using recombinant inbred lines derived from a cross between’Zhenshan97B’ and ‘Nanyangzhan’. Plant Breeding 2019, 138, 820–829. [Google Scholar] [CrossRef]
- Zhan, W.; Liu, J.; Pan, Q.; Wang, H.; Yan, S.; Li, K.; Deng, M.; Li, W.; Liu, N.; Kong, Q.; et al. An allele of Zm PORB 2 encoding a protochlorophyllide oxidoreductase promotes tocopherol accumulation in both leaves and kernels of maize. Plant J. 2019, 100, 114–127. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Fernie, A.R.; Yan, J. The Past, Present, and Future of Maize Improvement: Domestication, Genomics, and Functional Genomic Routes toward Crop Enhancement. Plant Commun. 2020, 1, 100010. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Liu, B.; Zhang, L.; Zhang, W.; Chen, R.; Wang, L. Overexpression of the maize γ-tocopherol methyltransferase gene (ZmTMT) increases α-tocopherol content in transgenic Arabidopsis and maize seeds. Transgenic Res. 2020, 29, 95–104. [Google Scholar]
- Dai, D.; Ma, Z.; Song, R. Maize kernel development. Mol. Breed. 2021, 41, 2. [Google Scholar] [CrossRef]
- Kaur, S.; Rakshit, S.; Choudhary, M.; Das, A.K.; Kumar, R.R. Meta-analysis of QTLs associated with popping traits in maize (Zea mays L.). PLoS ONE 2021, 16, e0256389. [Google Scholar]
- Fritsche, S.; Wang, X.; Jung, C. Recent Advances in our Understanding of Tocopherol Biosynthesis in Plants: An Overview of Key Genes, Functions, and Breeding of Vitamin E Improved Crops. Antioxidants 2017, 6, 99. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Han, N.; Liu, C.; Buerte, B.; Zhou, C.; Chen, J.; Wang, M.; Zhang, Y.; Tang, Y.; Zhu, M.; et al. Functional dissection of HGGT and HPT in barley vitamin E biosynthesis via CRISPR/Cas9-enabled genome editing. Ann. Bot. 2020, 126, 929–942. [Google Scholar] [CrossRef] [PubMed]
- Woodhouse, M.R.; Cannon, E.K.; Portwood, J.L.; Harper, L.C.; Gardiner, J.M.; Schaeffer, M.L.; Andorf, C.M. A pan-genomic approach to genome databases using maize as a model system. BMC Plant Biol. 2021, 21, 385. [Google Scholar] [CrossRef] [PubMed]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef] [PubMed]
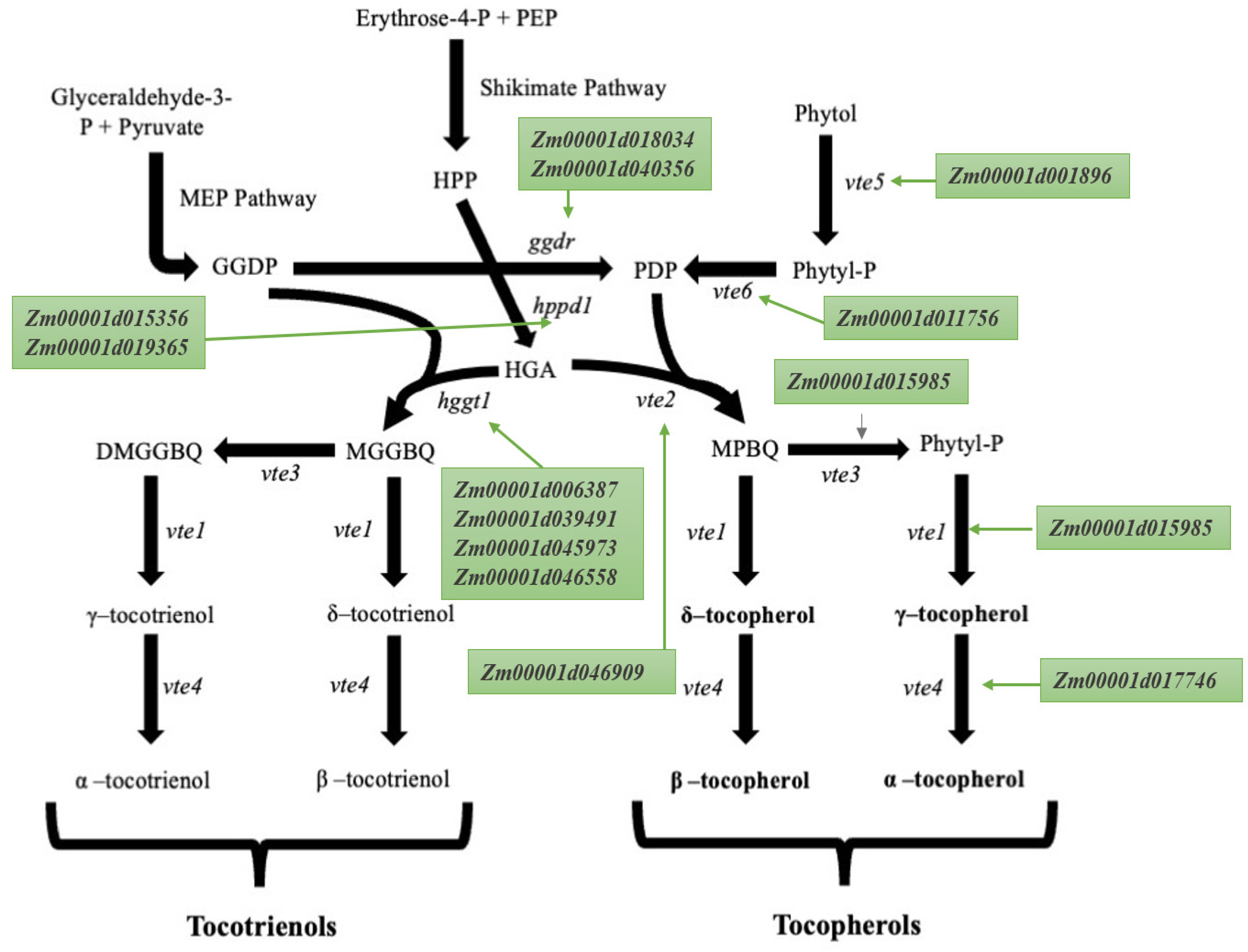

| Gene | v4 Gene Model ID | v2 Gene Model ID | Full Name | Chromosome | Start | End |
|---|---|---|---|---|---|---|
| ZmVTE4 | Zm00001d017746 | GRMZM2G035213 | vitamin E synthesis 4 | 5 | 205825585 | 205829216 |
| ZmVTE2 | Zm00001d046909 | GRMZM2G048472 | homogentisate phytyltransferase 1 | 9 | 110047718 | 110058786 |
| ZmVTE1 | Zm00001d015985 | GRMZM2G009785 | Tocopherol cyclase/sucrose export defective 1 (SXD1) | 5 | 136805707 | 136822194 |
| ZmVTE3 | Zm00001d031071 | GRMZM2G082998 | 2-methyl-6-phytyl-1,4-hydroquinone methyltransferase/MPBQ/MSBQ methyltransferase | 1 | 175969708 | 175971563 |
| ZmHGGT2 | Zm00001d006387 | GRMZM2G410644 | Homogentisate geranylgeranyltransferase/HGGT | 2 | 206364248 | 206367453 |
| ZmHGGT3 | Zm00001d039491 | GRMZM5G848876 | Homogentisate geranylgeranyltransferase/HGGT | 3 | 5766073 | 5768360 |
| ZmHGGT4 | Zm00001d045973 | GRMZM2G398628 | Homogentisate geranylgeranyltransferase/HGGT | 9 | 51047616 | 51049885 |
| ZmHGGT1 | Zm00001d046558 | GRMZM2G173358 | Homogentisate geranylgeranyltransferase/HGGT | 9 | 95895574 | 95899061 |
| ZmVTE5 | Zm00001d001896 | GRMZM2G104538 | vte5—vitamin E synthesis 5 | 2 | 2509566 | 2511414 |
| ZmVTE6 | Zm00001d011756 | GRMZM2G166383 | KOG4491—Predicted membrane protein | |||
| ZmGGDR | Zm00001d018034 | GRMZM2G105741 | Geranylgeranyl diphosphate reductase/Geranylgeranyl reductase | 5 | 212543655 | 212546035 |
| Zm00001d040356 | GRMZM2G419111 | Geranylgeranyl diphosphate reductase/Geranylgeranyl reductase | 3 | 39647288 | 39648626 | |
| ZmHPPD1 | Zm00001d015356 | GRMZM2G088396 | hppd1—4-hydroxyphenylpyruvate dioxygenase 1 | 5 | 86084654 | 86086755 |
| Zm00001d019365 | GRMZM2G374213 | 4-hydroxyphenylpyruvate dioxygenase/p-hydroxyphenylpyruvate oxidase | 7 | 29973891 | 29976259 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kassem, M.A.; Knizia, D.; Meksem, K. A Summary of Two Decades of QTL and Candidate Genes That Control Seed Tocopherol Contents in Maize (Zea mays L.). Genes 2024, 15, 472. https://doi.org/10.3390/genes15040472
Kassem MA, Knizia D, Meksem K. A Summary of Two Decades of QTL and Candidate Genes That Control Seed Tocopherol Contents in Maize (Zea mays L.). Genes. 2024; 15(4):472. https://doi.org/10.3390/genes15040472
Chicago/Turabian StyleKassem, My Abdelmajid, Dounya Knizia, and Khalid Meksem. 2024. "A Summary of Two Decades of QTL and Candidate Genes That Control Seed Tocopherol Contents in Maize (Zea mays L.)" Genes 15, no. 4: 472. https://doi.org/10.3390/genes15040472





