Abstract
PIWI-interacting RNAs (piRNAs), a class of small non-coding RNAs (sncRNAs) with 24–32 nucleotides (nt), were initially identified in the reproductive system. Unlike microRNAs (miRNAs) or small interfering RNAs (siRNAs), piRNAs normally guide P-element-induced wimpy testis protein (PIWI) families to slice extensively complementary transposon transcripts without the seed pairing. Numerous studies have shown that piRNAs are abundantly expressed in the brain, and many of them are aberrantly regulated in central neural system (CNS) disorders. However, the role of piRNAs in the related developmental and pathological processes is unclear. The elucidation of piRNAs/PIWI would greatly improve the understanding of CNS development and ultimately lead to novel strategies to treat neural diseases. In this review, we summarized the relevant structure, properties, and databases of piRNAs and their functional roles in neural development and degenerative disorders. We hope that future studies of these piRNAs will facilitate the development of RNA-based therapeutics for CNS disorders.
1. Introduction
PiRNAs, the largest subclass of sncRNAs in the PIWI family [1], were initially identified in the reproductive system [2,3]. The mature piRNAs are generated from either an initial transcript originating from a piRNA cluster or the conversion of double-stranded RNA precursors by RNase type-III enzymes [4,5,6]. PiRNAs bind and guide PIWI proteins to target specific mRNAs through sequence complementarity, and play important roles in gene regulation, transposon element (TE) repression, and antiviral defense [7,8]. Compared to other small RNAs, piRNAs utilize a more adaptable approach to silencing target mRNA, albeit necessitating longer complementary sequences [7,9].
In germ cells, piRNAs normally maintain gene integrity by silencing TEs [10,11,12,13]. PiRNAs/PIWI are also essential for regulating spermatogenesis/oogenesis [10,14], such as PIWI1/2/3 deficiency causing male mouse sterility by promoting aberrant transposon and transcriptome activity [15,16,17,18].
An increasing number of studies have highlighted the potential roles of the piRNAs-genes/lncRNAs complex in regulating neuronal cell differentiation, migration, and development [19,20]. PiRNAs are also involved in various neurological processes, including cognition, learning, and memory-related behaviors [21,22,23]. What is more, dysregulation of the piRNAs/PIWI pathway is linked to neurodevelopmental and degenerative diseases [24,25,26,27]. For example, in two Parkinson’s disease Caenorhabditis elegans models, 21ur-10824, 21ur-11898, and 21ur-13215 are significantly up-regulated in both α-syn(A53T)Tg and AβTg and α-syn(A53T)Tg transgenic strains [28].
PiRNAs have been increasingly recognized for their role in CNS development, including neural stem cell (NSC) differentiation and neurocognitive recovery, as well as in neurodevelopmental and neurodegenerative activities [20,24,25,27]. To this end, this review aims to summarize the progress in elucidating the structure, properties, and regulatory roles of piRNAs, particularly in CNS development. It also describes their potential applications in neurodevelopmental and degenerative diseases, both in human and animal models, thereby providing valuable insights for future research and clinical practice in neural disorders.
2. The Basic Information of piRNAs
To gain a better understanding of the piRNAs/PIWI complex, we will review the biogenesis, classification, distribution, and relevant databases of piRNAs in this section.
2.1. piRNA Biogenesis
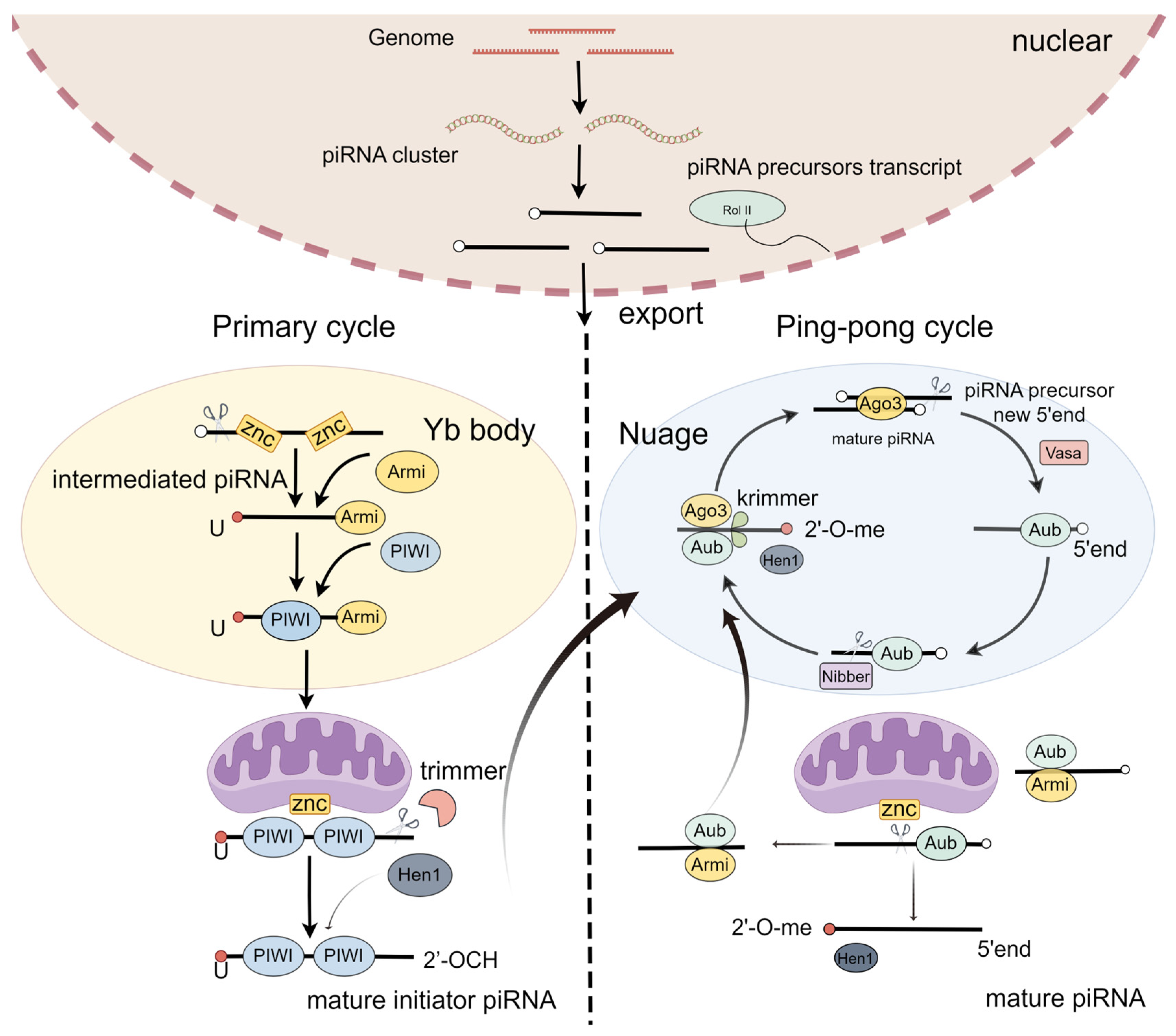
PiRNA biogenesis is a multistep process starting with transcription and the early processing of precursor RNA in the nucleus, export, and precursor cleavage, followed by processing to mature piRNAs and loading into PIWI proteins [29]. It includes two steps: the primary loop and the ping-pong loop (Figure 1) [30,31].
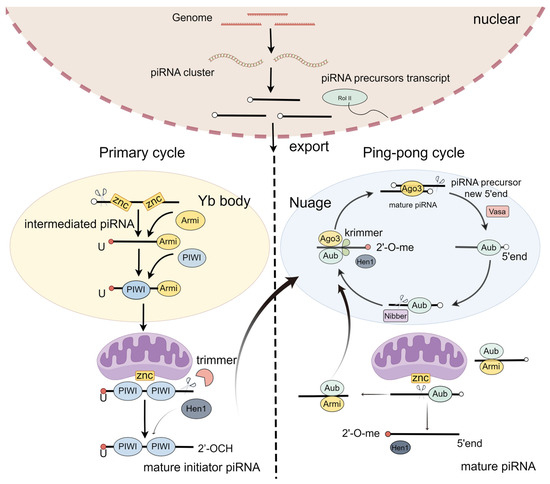
Figure 1.
PiRNA biogenesis.
In the primary loop, piRNA precursors originate from the piRNA cluster loci [29]. In the nucleus, the piRNA precursors are exposed to the Yb body and cleaved by the endonuclease Zuc, generating intermediate piRNAs, with the first nucleotide being uridine at the 5′ end [32,33]. These intermediate piRNAs further undergo trimming at their 3′ ends, eventually producing mature initiator piRNAs [34,35]. Finally, these mature initiator piRNAs are transported into the nucleus [32,36]. Thus, the primary loop initiates the downstream response of the ping-pong loop.
A secondary pathway, also called a ping-pong loop, involves the processing of pre-piRNAs initiated by the slicer activity of cytoplasmic PIWI proteins Aub or Ago3, loaded with complementary piRNAs [4,37]. To produce the responder pre-piRNAs in the ping-pong cycle, the initiator piRNAs bind PIWI proteins and are cleaved in a Zuc-dependent or independent manner [38,39]. To expand the diversity of piRNAs, the piRNA/AGO3 complex cuts the target RNA [40]. Taken together, the ping-pong pathway functions as an amplification cycle to increase the abundance and diversity of piRNAs.
2.2. The Structure, Classification, and Distribution of the piRNA/PIWI Complex
2.2.1. Structure of the piRNA/PIWI Complex
As a member of the Argonaute protein family, the structures of PIWI proteins are composed of PAZ in the N-terminal, RNase H-like, and Piwi/Argonaute/Zwille domain in the C-terminal [41]. The N-terminal PAZ can be attributed to PIWI protein–small RNA complex formation [42]. In the C-terminal, the RNase H-like domain shows RNA endonuclease activity in RNA-DNA hybrid molecules [43], and the Piwi/Argonaute/Zwille domain participates in the RNA interference process by binding piRNAs [44]. The PIWI proteins are highly conserved across multiple species [45,46,47], such as PIWI, Aub, and Ago3 in the fruit fly [48], MIWI/MWI2 (PIWIL1/4) and MILI (PIWIL2) in the mouse [49] and HIWI (PIWIL1), HILI (PIWIL2), HIWI3 (PIWIL3), and HIWI2 (PIWIL4) in humans [50].
Mature piRNAs are around 24–32 nt, with a preference for uridine at the 5′ end and 2′O-methylation at the 3′ end [35,45,51]. Compared to miRNAs, piRNAs usually lack sequence conservation and Dicer biogenesis [52]. Moreover, piRNAs are more selective than miRNAs when identifying a target through a weak seed region [53]. To ensure valid silencing TE activity, piRNAs closely monitor the sequence pairing through the seed gate [53]. Unlike other sncRNAs, piRNAs have been found only in animals and appear across different species [54].
2.2.2. PiRNAs in Different Genomic Areas and Cell Types
In most mammals, mature piRNAs are mainly divided into three major categories: piRNAs in TE, intergenic piRNAs, and 3′-UTR [55,56,57,58,59]. As the major class, TE piRNAs typically target the transcription and translation of TEs by RNAi in Drosophila and Danio rerio [60,61]. Intergenic piRNAs are mainly found in adult mammalian testes, such as in mice, rats, and humans [62,63,64]. In mice and chickens, the precursors of 3′UTR piRNAs are guided post-termination by 80S ribosomes, which harbor embedded TEs and produce piRNAs that cleave these TEs [65]. However, over 80% of piRNAs (including TE, 3′UTR, and intergenic piRNAs) lack obvious targets in mammals [65,66]. Overall, while piRNAs are associated with diverse genomic functions, their specific produced source in mammals still requires further exploration.
PiRNAs and PIWI are also abundant in multiple somatic cells, such as in neuronal, liver, exosome, and cancer tissue cells [21,29,34,67]. The aberrant expression of ectopic piRNAs has been identified across a range of human diseases. For example, in the CNS, PIWI and piRNAs have been linked to neural differentiation [68,69], dendritic complexity [21,22], and neural crest specification [70]. These findings highlight the broad applicability of piRNAs as potential biomarkers and therapeutic targets in various diseases, especially in CNS disorders. Further research underlying piRNA dysregulation in different neural cell types is necessary to fully elucidate their therapeutic potential.
2.3. The Databases of piRNAs
Due to the lack of conserved sequences and structural features, distinguishing piRNAs from miRNAs and siRNAs is a challenge [71]. Numerous piRNA clusters have been annotated, and the relevant databases have been generated [7]. Here, we review the public and specific databases of piRNA research in Table 1.

Table 1.
The relevant piRNA databases.
Several public databases provide comprehensive information on piRNA analysis [73,75]. NONCODE and deepBase 3.0 provide basic information about the expression profile, mapping, potential function, and interaction function of piRNAs [72,73,74,75]. However, these databases cover only a limited number of species and rarely include the functions and target analyses of piRNAs.
PiRNAbank is the first piRNA database, including annotated information, a sequence homology-based search, clusters, corresponding genes, and repeat elements [77]. As the largest database, piRBase covers approximately 181 million sequences and includes 440 datasets from 44 species for piRNAs [81,82]. What is more, version 3.0 integrates piRNA annotation in all aspects, including the piRNA gold standard of sets, clusters, variants, splicing junction, and expression [76]. Nevertheless, piRBase v3.0 has collected data on diseases and visualizes the piRNAs–targets network [76]. Other databases, such as PiRNAclusters, piRTarBase, and piRNA-IPdb have also been used for cluster identification and target prediction [79,80].
These databases provide an excellent piRNA research platform for piRNA analysis in terms of functional analysis and multiple pathological processes [83,84,85]. Therefore, further data updating can be on improving naming conventions, experimental verification of the specific piRNA in diseases, data integration, annotation information, and the target prediction standard.
3. PiRNAs/PIWI in Neural Differentiation and Neurocognitive Behaviors
In the past two decades, piRNAs have been characterized in neuronal cells and are shown to have indispensable roles in brain function and neuronal diseases [22,24,86]. However, the precise mechanism remains unclear. To further enhance our understanding, we summarize the multiple functions of piRNAs/PIWI in neuronal differentiation, the blood–brain barrier (BBB) and neurocognitive behaviors (learning, memory, and anxiety), food intake, and neuron injury.
3.1. Neuronal Differentiation
3.1.1. PIWI Proteins
In PIWI families, PIWIL1 and PIWIL2 have been mostly proposed to control memory and anxiety [87,88]. The first evidence that the PIWI protein exists in the brain was found in the Aplysia hippocampus [21]. The author suggested that down-regulated MILI (PIWIL2) improves memory-relative behavior by demethylating CREB2 [21,22]. Similarly, in a rat cerebral ischemic injury model, the attenuation of PIWIL2 also increases CREB2 by promoting DNM3A methylation, ultimately preventing cerebral ischemic injury [89]. Recent research also showed that PIWIL2 is essential for proper neurogenesis in the postnatal mouse hippocampus. The depletion of PIWIL2 and piRNAs in the adult hippocampus impairs adult neural progenitor cells’ differentiation toward a neural fate, induces senescence, and generates reactive glia [88]. Overall, one of the possible neuronal regulatory pathways involving PIWIL2 is the passive demethylation of DNM3A.
In the E14.5 cerebral cortex of rats, PIWIL1 silencing leads to neuronal polarization and migration by modulating the microtubule-associated proteins (MAPs, MAP2, and MAP2B) [87]. In the chicken neural crest region, Chiwi (PIWIL1) is markedly higher at early neurulation stages, while Chili (PIWIL2) shows low transcript levels at all stages [70]. The loss of PIWIL1 impedes neural crest specification and emigration by targeting ERNI in the Sox2 pathway [70]. What is more, knockdown PIWIL1 inhibits food intake by targeting NPF1, a type of neuropeptide in the brain correlated with appetite, dietary behaviors, and energy metabolism [90].
Along with PIWIL1 and PIWIL2, PIWIL4 is essential for human embryonic mediated neuronal differentiation [68]. The knockdown of PIWIL4 suppresses the partial recovery of embryonic stem cell markers (MAP2 and TUB33) in NT2 cells. In contrast, overexpressed PIWIL4 decreases OCT4A and NANOG expression [68]. To sum up, PIWI proteins can act as modulators in multiple neural differentiation processes, influencing various pathways such as methylation, MAPs, and Sox2. Thus, more sophisticated studies are needed in the future to further address the potential redundant and synergistic functions of different PIWI family members in cortical development.
3.1.2. PiRNAs
In terms of the CNS, piRNAs were initially identified in E15.5 cultures and the 6-week mouse hippocampus [69]. Some well-designed reviews have summarized the function of piRNAs in neural heterogeneity, neurogenesis, and neural plasticity [91,92]. In this part, we review their regulatory roles in neurogenesis.
Among multiple piRNAs tested in the mouse hippocampus, five uniquely expressed piRNAs (DQ541777, 705026, 719597, 689086, and 540285) were co-immunoprecipitated with MIWI [69]. Lee et al. found that DQ541777 extended throughout the dendrites in a punctate pattern in both E15 cultured mouse and E18 rat hippocampal neurons. What is more, the inhibition of DQ541777 activity leads to a statistically significant decrease in dendrite spine area, suggesting its role as a modulator of dendritic spine development [69]. During the retinoic acid (RA)-induced neuronal differentiation of NT2 cells, piRNAs (DQ582359 and DQ596268) are up-regulated [68]. Overall, neuronal piRNAs are abundant during the critical period of neural differentiation and respond to changes in PIWI protein levels.
In adult neural stem cells (aNSCs), piRNAs are focused on synaptic plasticity and memory control in the hippocampus [88,93]. The hippocampus shows the highest number of unique piRNA transcripts (5494 piRNAs) in the adult brain [94]. Recently, it was demonstrated that the piRNA pathway has a crucial role in maintaining the fate of hippocampal NSCs [88]. Researchers identified 298 piRNA clusters that are differentially expressed in mouse undifferentiated aNPCs, among which piR-cluster 1 is homologous to the human piR-61648 [88]. Moreover, inhibiting the piRNA pathway in adult NPCs leads to senescence-associated inflammation and an increase in reactive astrogliogenesis [88]. However, these mechanisms of piRNAs in translation and epigenetics have been poorly investigated in the context of the CNS.
These results indicate that piRNAs/PIWI function as early and late neuronal marker regulators, enhancing neuronal differentiation. Future research on the piRNAs/PIWI process will contribute to understanding various CNS diseases, including neurodevelopmental and neurodegenerative diseases.
3.2. The Blood–Brain Barrier (BBB)
The BBB is a neuroprotective layer that maintains the homeostasis of the central nervous system and provides an appropriate environment in which neurons can execute their functions [95,96]. In addition, down-regulated HIWI2/PIWIL4 protein suppresses two key tight junctions (CLDN1 and TJP1) through the increased phosphorylation of the AKT/GSJ3 signaling pathway in the BBB [97,98]. Increased PIWIL4 leads to the increased infiltration of inflammatory molecules, potentially disrupting the BBB balance [97]. This evidence profoundly supports the novel concept that piRNAs/PIWI-like proteins may have a potential role in governing the BBB by altering the tight junctions and inflammatory factors.
As a potential marker in regulating the BBB balance and in relevant diseases, the role of piRNAs has not yet been investigated. Therefore, further studies are needed to enhance our understanding of how piRNAs regulate the BBB balance.
3.3. Neurocognitive Behaviors (Learning, Memory, Anxiety, and Food Intake)
PiRNAs can act as potential biomarkers in neurocognitive behaviors. In this section, we summarize the potential roles of piRNAs in learning, memory, anxiety, and food intake.
3.3.1. Learning and Memory
Learning-related neuronal plasticity and the formation of memory depend on tightly controlled gene and ncRNA patterns [99]. Increasing evidence implicates piRNAs in learning and memory [22,100]. In the 5-HT induction of Aplysia CNS, up-regulated aca-piR-4/15 enhances memory-related plasticity by inhibiting DNA methylation levels in the CREB2 promoter [21]. Similarly, in the sensory neurons of Aplysia, the piR-F in TE facilitates serotonin-dependent CREB2 methylation [100]. These studies reveal the dynamic mechanisms underlying epigenetic regulation in long-term memory formation. To sum up, in Aplysia long-term memory formation, piRNAs regulate CREB2 methylation through promotor and TE factors. Future research should focus on the potential role of the piRNAs/PIWI pathway in mammalian memory-like behavior.
3.3.2. Anxiety
Since the 20th century, anxiety has gradually been recognized as a psychiatric disorder affecting normal emotional behavior [101]. The relevant research has suggested that the piRNA/PIWI complex plays a fundamental role in the pathology of anxiety. In the dorsal hippocampus of mice, knocking down PIWIL1/2 enhances the long-term fear memory without affecting anxiety or locomotion [22]. In contrast, selectively knocking down PIWIL2 increases movement velocity and distance, which is indicative of anxiety-like behavior [22]. It is only downregulating PIWIL1 that shows no observable behavioral effect. Other research shows that knocking down PIWIL2 in the dorsal hippocampus of adult mice is sufficient to reproduce hyperactivity [102]. In a PIWIL2 KO mouse model, DNA is preferentially hypomethylated within the long intersoersed nuclear element1 (LINE1) promoter [102]. Moreover, large numbers of LINE1-derived piRNAs have been identified [102]. In these PIWI protein KO models of anxiety, specific piRNAs have not been implicated. The possible mechanism in terms of behavior regulation is that endogenous siRNAs may bind to piRNAs, thereby silencing brain-expressed retrotransposons. Thus, the precise roles of piRNAs in anxiety still need to be elucidated.
3.3.3. Food Intake
Food intake, as a complex physiological process, is regulated by the CNS through neuropeptides and neurotransmitters [103]. In regulating food intake, specific genetic elements such as genes and small non-coding RNAs, including piRNAs, have been highlighted for their potential [103,104,105,106]. In the locust brain, PIWI1 and Ago3 exhibited relatively higher expression levels, but no signals of PIWI2 were detected, suggesting that PIWI1 and Ago3 may have a fundamental function in the CNS [90]. The knockdown of PIWI1 leads to the suppression of body weight in locust by decreasing the expression of neuropeptide NPF1, but not after the knockdown of Ago3 [90]. In addition, the intronic piRs-3-I3 might enhance the RNA splicing of NPF1 by preventing hairpin formation at the branch point sites, indicating that the piRNAs/PIWI complex has a regulatory role in food intake [90]. Few studies have been conducted in this field. Therefore, ongoing research is exploring their exact roles in mammalian models and their therapeutic potential for conditions related to food intake.
3.3.4. Axon Injury
In the adult mammalian CNS, injured axons fail to regenerate [107]. In addition, piRNAs are associated with axon regeneration following neural injury [52].
In the C. elegans sensory system, piRNA transcriptions (PRG-1, TOFU-7, and PRDE-1) are involved in suppressing axon regrowth by regulating parn-1 and henn-1 in an independent transcription manner [20]. In the adult rat sciatic nerve, piR-1199 and piR-5781 were up- and down-regulated, separately [108]. Additionally, piR-219 and piR-1200 were initially down-regulated, returned to the baseline levels, and then showed an up-regulation trend [108]. In a cultured sensory neuron, knocking down the MIWI/piRNAs complex increased axon length by 60% and decreased axon retraction [108]. This article highlights the roles of piRNAs/PIWI in regulating the multiple transcription factors involved in sensory regrowth. Interestingly, after peripheral nerve injury in a mouse model, the depletion of MIWI/PIWIL1 enhanced a series of piRNA expressions (piR-14384, -69959, -52953, and -52274) [19]. Compared to the healthy group, depleted MIWI/PIWIL1 inhibited the EGR2, a neural transcription factor localized with the Schwan cell marker S100 [19]. Overall, piRNA-MIWI has a critical function in axon regrowth and repair after nerve injury in mouse models.
Although the piRNAs/PIWI complex has been reported in various neuronal behaviors, most research has concentrated on identifying piRNAs. Therefore, future studies should focus on improving the specific amplification and synthesis pathways of piRNAs in the nervous system.
4. piRNAs/PIWI in Neurodevelopmental and Psychiatric Disorders
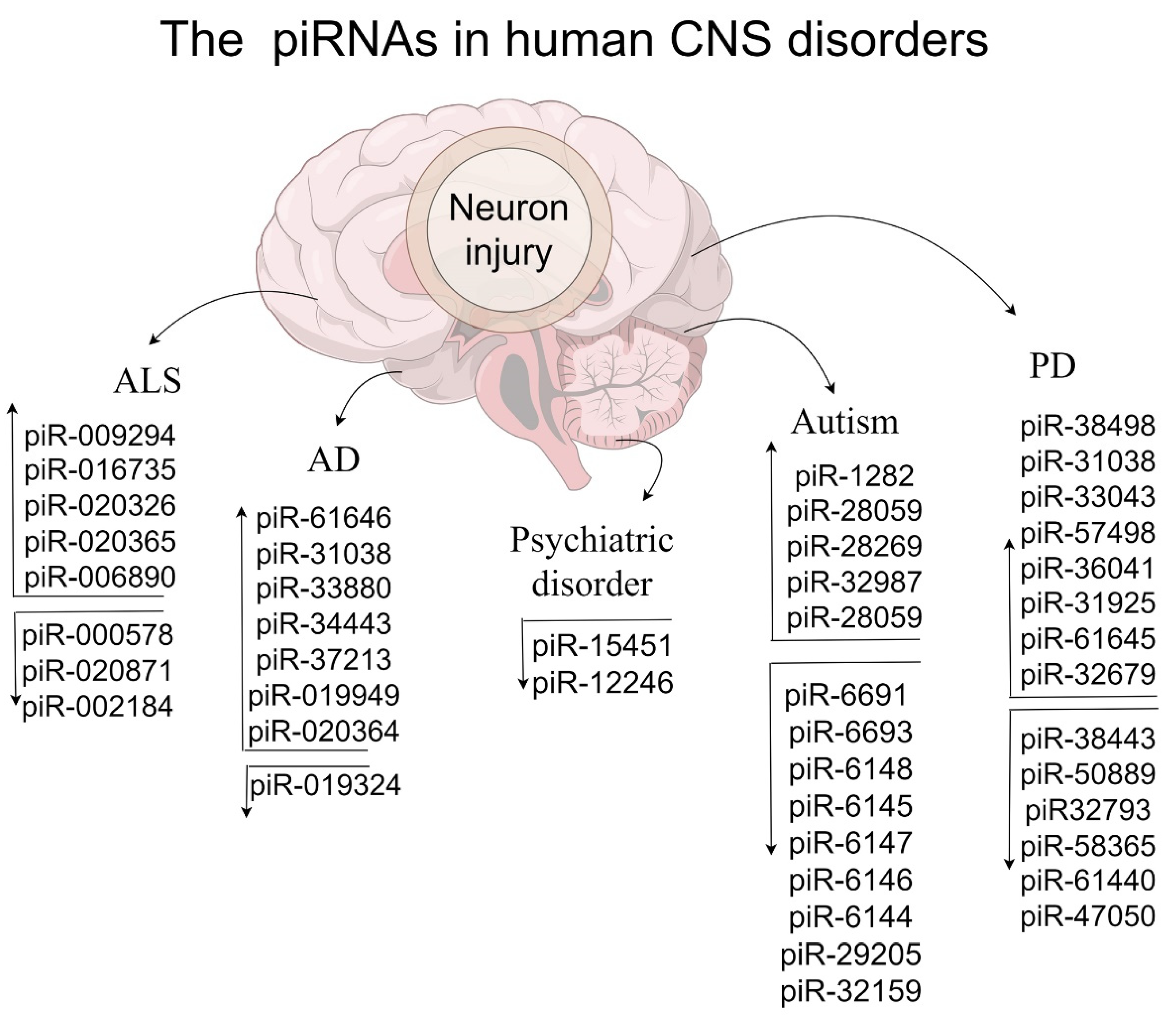
Multiple genes and ncRNAs lead to neurodevelopmental dysregulation, which gradually becomes a form of language disorders and brain dysfunction [109,110,111,112]. Unexpectedly, piRNAs show potential roles in neurodevelopmental disorders by directly or indirectly regulating gene/lncRNA [9,45,113]. Therefore, we summarize the underlying roles of piRNAs in stroke and psychiatric disorders (Table 2 and Figure 2), which is essential for improving the relevant diagnosis and treatment.

Table 2.
The piRNAs in CNS disorder models.
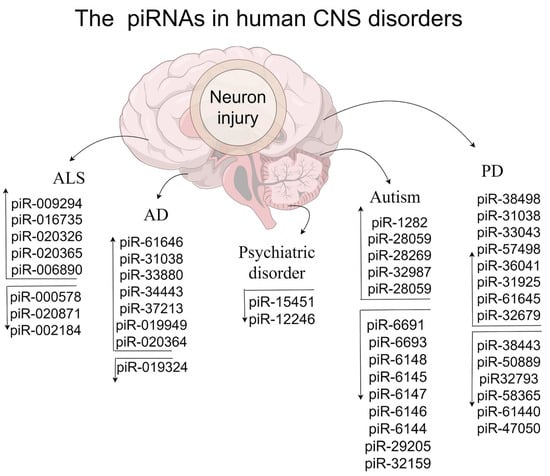
Figure 2.
The influence of human piRNAs in CNS disorders.
4.1. Stroke
Stroke is an acute cerebral vascular event with high mortality and morbidity. Recent studies have demonstrated that dysregulated piRNAs are strongly associated with stroke pathology by regulating the genes, proteins, and relevant signaling pathways [119,120,121].
In an ischemia rat brain, 54 up- and 51 down-regulated piRNAs were detected, with piR-177729 and piR-169523 showing the most significant alterations [114]. What is more, stroke-responsive piR-143106, -177729, -169523, and -70903 were predicted to target retrotransposon (RT) classes, as well as the zinc finger and Kruppel family [114]. These results suggest that abnormal piRNAs may lead to abnormal transposon expression, resulting in pathophysiologic changes after a stroke. In other research using an ischemia tolerance rat model, 5574 piRNAs were found, with rno-piR-000618, -017990, and -014971 up-regulated by targeting CREB2 [89]. Overall, stroke-responsive piRNAs restore the plasticity of damaged nerves and improve memory function after transient global cerebral ischemia.
Although mRNA and ncRNA therapies, including those with piRNAs, have great potential for treating neuronal diseases [121,122,123,124], only the identities of piRNAs have been explored so far. Understanding the piRNA/PIWI complex is likely to bring significant breakthroughs in understanding the complex interactions and molecular mechanisms activated after cerebral ischemia. Therefore, future research needs to discover potential piRNA targets and their associated proteins in terms of the pathophysiology of stroke.
4.2. Psychiatric Disorders
Psychiatric disorders are highly prevalent and cause an enormous burden of suffering, lost productivity, morbidity, and mortality [125]. Recent results also show the potential role of piRNAs in psychiatric behaviors, including schizophrenia and major depressive and bipolar disorders [126,127].
4.2.1. Schizophrenia (SCZ)
One study identified 37 piRNAs in the anterior cingulate cortex (ACC) of SCZ patients, with one piRNA being correlated with antipsychotic medication [127]. However, the authors did not demonstrate a change in level between the SCZ and control groups. In addition, piR-15451 and piR-12246 are down-regulated in the anterior cingulate cortex of schizophrenia subjects [127]. Conversely, piR-18252 has been correlated with antipsychotic medication issues [127]. However, the authors did not mention the changes seen between the schizophrenia and control subjects. Unlike with miRNAs, few studies have reported on correlations between piRNAs and major depression/bipolar disorder. One article mentioned that two circulated piRNAs (piR-007899 and piR-019162) might be related to symptoms following Gulf War Illness (GWI)-relevant exposure in rats [115]. CNS impairment issues, including memory dysregulation and depression, are among the most common symptoms reported in GWI.
However, only a few piRNAs and their relevant mechanisms have been identified in psychiatric processes. The possible reasons for this include the scarcity of clinical samples and the difficulty of constructing models. Therefore, detecting more specific piRNAs remains a major challenge.
4.2.2. Autism Spectrum Disorder (ASD)
ASD generally manifests in childhood and mainly presents with isolation, emotional dysregulation, and intellectual disability [128]. The pathological mechanism mainly involves multi-gene management [123,129]. The piRNAs/PIWI complex has also been identified as a potential regulator and diagnostic marker in ASD [130,131]. Knockdown PIWIL1 causes the delayed transition of newborn cortical neurons by enhancing MAPs, suggesting a relationship between the PIWI family and autism [87]. Compared to the healthy group, 21 up-regulated (most significant: hsa-piR-1282) and 16 down-regulated piRNAs (most significant: hsa-piR-32159) were identified in ASD samples [131]. Although 22 up-regulated piRNAs (most significant: hsa-piR-22380) and 7 down-regulated piRNAs (most significant: hsa-piR-27623) were found in the severe symptom vs. mild symptoms group, further validation experiments are needed to confirm their potential as diagnostic biomarkers. In addition, piRNAs in the digestive system and gut microbiota have been implicated in ASD [132]. Similarly, in one study of the saliva of children with autism, hsa-piR-6148, hsa-piR-6145, hsa-piR-6147, hsa-piR-6146, and hsa-piR-6144 were down-regulated and were associated with gastrointestinal (GI) disorders [132]. In ASD fecal samples, in one subject with ASD and a neurotypical control, hsa-piR-6691, -6693, and -29205 were significantly down-regulated, while hsa-piR-28269, -32987, and -28059 were up-regulated [130]. In the fecal samples of males with ASD, 66 up-regulated and 256 down-regulated piRNAs were identified [130]. In addition, the authors confirmed that piRNAs can be released into the gut lumen and may interact with the micro- and mycobiota, primarily targeting NACC1, SMAD4, TNRC6B, and TTN [130]. This not only provides a novel approach to the clinical diagnosis and analysis of ASD but also confirms the potential of piRNA as a biomarker.
Overall, the available evidence reflects a strong association between dysregulated piRNAs/PIWI and ASD [132,133]. From target prediction, over 50% of the targets of piRNAs may be lncRNAs, which are most abundant in the CNS. Therefore, further research will be focused on possible lncRNA targets and functional networks.
5. PiRNAs/PIWI in Neurodegenerative Disease
Exhibiting neuron degeneration in specific brain areas, neurodegenerative disorders are a class of pathological conditions that cause a lack of motor coordination and cognitive and memory impairment [134,135,136]. Several well-designed reviews have highlighted the dysregulation of the piRNA pathways in Alzheimer’s disease (AD), Parkinson’s disease (PD), and amyotrophic lateral sclerosis (ALS) [24,26,137]. To enhance understanding, we summarize the potential mechanisms of piRNAs/PIWI in both animal models and patients with neurodegenerative diseases (Table 2 and Figure 2).
5.1. Alzheimer’s Disease (AD)
AD is the most common neurodegenerative disease, characterized by progressive cognitive and functional decline [138,139]. Due to its complex pathology, a specific treatment strategy remains ambiguous. However, previous studies suggest that ncRNAs regulate the amyloid precursor protein (APP), tau, amyloid-β (Aβ) peptide, inflammation, and cell death in AD [140,141,142,143]. What is more, recent evidence highlights the potential roles of piRNAs in both AD models and patients [138,144].
5.1.1. AD Models
In tau-transgenic Drosophila, heterochromatin loss led to aberrant Ago3 expression [117]. When knocking down Ago3/PIWIL1 in tau-transgenic Drosophila brains, fewer apoptotic and proliferating cell nuclear antigen (PCNA)-positive cells were found, and the locomotor defect was reduced [27]. In the HASN539T C. elegans model, tdp-1 knock-out reduced the number of piRNAs from 112 to 0 [116]. Among them, 21ur-10488 and 21ur-2781 were predicted to target gsk-3 and tpi-1, which are related to the AD pathology [116]. These results show that the piRNAs/PIWI complex can serve as an ideal biomarker for therapy and diagnosis. However, few mammalian AD models have been reported.
5.1.2. AD Patients
In AD patients, Roy, Sarkar, et al. identified 146 up- and 3 down-regulated piRNAs [145]. Among them, the AD-associated genes, CYCS, LIN7C, KPNA6, and RAB11A, are regulated by four piRNAs (piR-38240, piR-34393, piR-40666, and piR-51810), which could lead to abnormal cell death, cellular homeostasis, and the transport of Aβ in the AD process [145]. In the human brain, 81 up-regulated and 22 down-regulated piRNAs were detected, of which 24 were specifically expressed [144]. Additionally, these AD-associated piRNAs were strongly correlated with AD-risk DNA variants (SNPs) through eQTL analysis [144]. Further research found associations of 17 variants across 8 loci, including APOE, APOJ, and SLC24A4, which were genome-wide significant [146]. Similarly, Mao, Fan et al. detected 9453 piRNAs from 4 AD patients, of which DQ570746 and DQ582201 accelerate aging by down-regulating CYP19A1 and CD33 in a TE-independent manner [147].
PiRNAs were first reported in the human cerebrospinal fluid exosomes of AD dementia patients [148]. Among them, down-regulated piR-019324 and up-regulated piR-019949/-020364 were identified in the exosomes, which may modulate the tau and Aβ42/40 in mild cognitive impairment [148]. Additionally, in AD plasma-derived extracellular vesicles, up-regulated piR-000552 and -020450 were identified [149]. These piRNAs may serve as candidate non-invasive biomarkers to distinguish between clinical and preclinical AD. However, the sensitivity and specificity of exosomal piRNAs still need to be improved.
Currently, three authors have summarized the potential role of piRNAs in AD [24,137,150]. Therefore, extensive studies should focus on seeking novel piRNAs and their targets, especially in lncRNAs and snRNAs. Additionally, the application of GWAS in piRNA research will greatly advance the understanding of Alzheimer’s disease pathology.
5.2. Parkinson’s Disease (PD)
PD is the second most common neurodegenerative disease, which is characterized by the gradual reduction of dopaminergic neurons in the substantia nigra pars compacta [151,152]. The heterochromatin decompensation and reduction of piRNAs/PIWI drive transposable element dysregulation in tauopathy, which is one of the pathological changes in PD [27,153].
5.2.1. PD Models
In a C. elegans model of PD, 79 up- (most significant: 21ur-5499) and 33 down-regulated (most significant: 21ur-10292) piRNAs have been found [116]. Among them, 21ur-1412 may target two PD-relevant genes (T08G11.1 and rab-39), while 21ur-10487, -839, and -2781 may target four PD-relevant genes (tpa-10, taf-1, dop-3, and rem-8) [116]. These targets act as effective signatures for PD: dysregulated Tpa-1 causes metabolic imbalances in a-synuclein [154]; defects in T08G11.1 that are related to membrane lipid homeostasis appear in VPS13C-PD cases [155]; the absence of taf-1 (TAFA in humans), dop-3 (DRD2 in humans), and rme-8 (DNAJC13 in humans) leads to dopaminergic dysfunction in PD brains [156,157,158]. In two other C. elegans PD models (α-syn(A53T)Tg and AβTg; α-syn(A53T)Tg transgenic strains), 21ur-10824, 21ur-11898, and 21ur-13215 were up-regulated [28], which is consistent with previous studies of PD [159]. What is more, in these two strains, the deletion of tofu-1 (a piRNA biogenesis gene) ameliorates behavioral phenotypes, improves thrashing, extends the lifespan, reduces α-synuclein expression, and alleviates dopaminergic neuron degeneration [28]. These results underscore the indispensable roles of neuronal piRNAs in the molecular basis of PD.
5.2.2. PD Patients
In PD patient studies, many dysregulated piRNAs were detected in fibroblasts (113 up- and 168 down-regulated), IPSCs/ESCs (193 up- and 62 down-regulated), and neurons (150 up- and 329 down-regulated) [159]. Among these, the fraction of memory piRNAs in neurons was deregulated and predominantly enriched in both SINE and LINE [159]. In one study of PD patients, 20 piRNAs were significantly altered in the prefrontal cortex, with 55 altered piRNAs in the amygdala, and hsa-piR-748391 was co-expressed in both regions [25]. In addition, hsa-piR-131693 levels decreased as the PD progressed. In the prefrontal cortex, transposon-derived piRNAs were massively detected [25], and their targets (pseudogenes HSP90AA1 and EEF1A1) have been reported to regulate the abnormal aggregation of α-synuclein and neuronal damage [160,161]. Because multiple piRNAs have a broad targeting capacity, the above piRNAs still need further verification or refinement.
The accumulating evidence suggests that piRNAs are critical regulators affecting the etiology of PD [8,110,162]. Future work should focus on exploring comprehensive piRNA databases across different species and cell types, as well as on identifying specific temporal and spatial expression patterns in tissues.
5.3. Amyotrophic Lateral Sclerosis (ALS)
ALS is a progressive motor neurodegenerative disease that causes selective motor neuron loss, progressive muscle wasting, and, ultimately, death [163]. Dysregulated TDP-43 and FUS proteins contribute to ALS pathogenesis by causing aberrant post-translational modification and subcellular mis-localization in neurons [163,164,165,166]. Recent research has highlighted the critical roles of piRNAs/PIWI in ALS [45].
5.3.1. ALS Models
After specifically knocking down the cabeza (Caz, a FUS homolog) in Drosophila, Aub was overexpressed, promoting crawling speed and climbing ability in both larvae and adults [118]. In the larvae CNS, knocking down Caz increased the level of pre-piRNAs and co-immunoprecipitation in the nuclears, while Aub overexpression had the opposite effect [118]. In addition, Caz knockdown decreased mature-side piRNA expression, while Aub overexpression or inhibition had no effect [118]. Overall, these results suggest that Caz negatively regulates the expression levels of pre-piRNAs and mature piRNAs, contributing to ALS.
5.3.2. ALS Patients
In ALS human brains, three down-regulated piRNAs (hsa-piR-000578, -020871, and -002184) and two up-regulated piRNAs (hsa-piR-009294 and -016735) were identified [133,167]. Meanwhile, protein-level analysis also showed a decrease in PIWIL1 and an increase in PIWIL4 [167]. These dysfunctional PIWIL1-directed piRNAs mislocate with TDP-43 in cytoplasmic inclusions, which may be an important determinant of TDP-43 accumulation in the cytoplasm and may contribute to ALS pathogenesis [167]. In the biofluids of ALS, up-regulated piRNAs in both CSF (hsa-piR-020326, -020365, and -006890) and serum (hsa-piR-006890, -008114, -000775, and -000765) have been identified by RNA-seq [168]. In addition, as diagnostic biomarkers, serum piRNAs may have more diagnostic value compared to cerebrospinal fluid (CSF). A comprehensive assessment of cell-free piRNAs/PIWI complexes in the context of ALS diagnosis is still needed.
Currently, the study of novel piRNAs in ALS relies on high-throughput, long-read, and extensive platforms. Simultaneously, their biological functions still warrant further clarification through a functional analysis of ALS pathophysiology.
6. Conclusions
Although many pieces of evidence indicate the fundamental function of piRNAs in diverse neural activities and their association with disease occurrence, elucidating the molecular mechanisms underlying neuronal piRNA-mediated gene regulation is just beginning. PiRNA research still faces major limitations, such as non-obvious targets, imperfect databases, multiple genomic locations, and lower abundance. Several neurodegenerative diseases have shown increased transposon expression, yet the roles of promoter and TE piRNAs in regulating neural development and degenerative diseases still need to be explored.
Author Contributions
X.P. and N.M. wrote the paper; X.P., N.M. and T.S. edited the paper. Figure design: X.P., Z.W., W.D. and S.L.; tables: Z.W., W.D. and S.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Fundamental Research Funds for the Central Universities (ZQN-1020, N.M.), the Scientific Research Funds of Huaqiao University (16BS815 (N.M.), 19BS303 (N.M.), and Z16Y0017 (T.S.), Innovation Awards of Quanzhou Talents (2018C057R), and the National Natural Science Foundation of China (32100775 (N.M.), 31771141 (T.S.)).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Li, D.; Taylor, D.H.; van Wolfswinkel, J.C. PIWI-mediated control of tissue-specific transposons is essential for somatic cell differentiation. Cell Rep. 2021, 37, 109776. [Google Scholar] [CrossRef] [PubMed]
- Girard, A.; Sachidanandam, R.; Hannon, G.J.; Carmell, M.A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 2006, 442, 199–202. [Google Scholar] [CrossRef] [PubMed]
- Aravin, A.; Gaidatzis, D.; Pfeffer, S.; Lagos-Quintana, M.; Landgraf, P.; Iovino, N.; Morris, P.; Brownstein, M.J.; Kuramochi-Miyagawa, S.; Nakano, T.; et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 2006, 442, 203–207. [Google Scholar] [CrossRef]
- Czech, B.; Hannon, G.J. One Loop to Rule Them All: The Ping-Pong Cycle and piRNA-Guided Silencing. Trends Biochem. Sci. 2016, 41, 324–337. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Dou, M.; Song, X.; Dong, Y.; Liu, S.; Liu, H.; Tao, J.; Li, W.; Yin, X.; Xu, W. The emerging role of the piRNA/piwi complex in cancer. Mol. Cancer 2019, 18, 123. [Google Scholar] [CrossRef]
- Jia, D.D.; Jiang, H.; Zhang, Y.F.; Zhang, Y.; Qian, L.L.; Zhang, Y.F. The regulatory function of piRNA/PIWI complex in cancer and other human diseases: The role of DNA methylation. Int. J. Biol. Sci. 2022, 18, 3358–3373. [Google Scholar] [CrossRef] [PubMed]
- Zuo, L.; Wang, Z.; Tan, Y.; Chen, X.; Luo, X. piRNAs and Their Functions in the Brain. Int. J. Hum. Genet. 2016, 16, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.Y.; Wen, Z.J.; Xu, H.M.; Zhang, Y.; Zhang, Y.F. Exosomal noncoding RNAs in central nervous system diseases: Biological functions and potential clinical applications. Front. Mol. Neurosci. 2022, 15, 1004221. [Google Scholar] [CrossRef]
- Gainetdinov, I.; Vega-Badillo, J.; Cecchini, K.; Bagci, A.; Colpan, C.; De, D.; Bailey, S.; Arif, A.; Wu, P.H.; MacRae, I.J.; et al. Relaxed targeting rules help PIWI proteins silence transposons. Nature 2023, 619, 394–402. [Google Scholar] [CrossRef]
- He, C.; Wang, K.; Gao, Y.; Wang, C.; Li, L.; Liao, Y.; Hu, K.; Liang, M. Roles of Noncoding RNA in Reproduction. Front. Genet. 2021, 12, 777510. [Google Scholar] [CrossRef]
- Saritas, G.; Main, A.M.; Winge, S.B.; Mørup, N.; Almstrup, K. PIWI-interacting RNAs and human testicular function. WIREs Mech. Dis. 2022, 14, e1572. [Google Scholar] [CrossRef] [PubMed]
- Siomi, M.C.; Sato, K.; Pezic, D.; Aravin, A.A. PIWI-interacting small RNAs: The vanguard of genome defence. Nat. Reviews. Mol. Cell Biol. 2011, 12, 246–258. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Siomi, M.C. The piRNA pathway in Drosophila ovarian germ and somatic cells. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2020, 96, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Onishi, R.; Yamanaka, S.; Siomi, M.C. piRNA- and siRNA-mediated transcriptional repression in Drosophila, mice, and yeast: New insights and biodiversity. EMBO Rep. 2021, 22, e53062. [Google Scholar] [CrossRef] [PubMed]
- Hasuwa, H.; Iwasaki, Y.W.; Au Yeung, W.K.; Ishino, K.; Masuda, H.; Sasaki, H.; Siomi, H. Production of functional oocytes requires maternally expressed PIWI genes and piRNAs in golden hamsters. Nat. Cell Biol. 2021, 23, 1002–1012. [Google Scholar] [CrossRef]
- Loubalova, Z.; Fulka, H.; Horvat, F.; Pasulka, J.; Malik, R.; Hirose, M.; Ogura, A.; Svoboda, P. Formation of spermatogonia and fertile oocytes in golden hamsters requires piRNAs. Nat. Cell Biol. 2021, 23, 992–1001. [Google Scholar] [CrossRef] [PubMed]
- Perillo, G.; Shibata, K.; Wu, P.H. piRNAs in sperm function and embryo viability. Reproduction 2023, 165, R91–R102. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, F.; Chen, Q.; Li, M.; Lv, X.; Xiao, Y.; Zhang, Z.; Hou, L.; Lai, Y.; Zhang, Y.; et al. The piRNA pathway is essential for generating functional oocytes in golden hamsters. Nat. Cell Biol. 2021, 23, 1013–1022. [Google Scholar] [CrossRef]
- Sohn, E.J.; Jo, Y.R.; Park, H.T. Downregulation MIWI-piRNA regulates the migration of Schwann cells in peripheral nerve injury. Biochem. Biophys. Res. Commun. 2019, 519, 605–612. [Google Scholar] [CrossRef]
- Kim, K.W.; Tang, N.H.; Andrusiak, M.G.; Wu, Z.; Chisholm, A.D.; Jin, Y. A Neuronal piRNA Pathway Inhibits Axon Regeneration in C. elegans. Neuron 2018, 97, 511–519.e6. [Google Scholar] [CrossRef]
- Rajasethupathy, P.; Antonov, I.; Sheridan, R.; Frey, S.; Sander, C.; Tuschl, T.; Kandel, E.R. A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. Cell 2012, 149, 693–707. [Google Scholar] [CrossRef] [PubMed]
- Leighton, L.J.; Wei, W.; Marshall, P.R.; Ratnu, V.S.; Li, X.; Zajaczkowski, E.L.; Spadaro, P.A.; Khandelwal, N.; Kumar, A.; Bredy, T.W. Disrupting the hippocampal Piwi pathway enhances contextual fear memory in mice. Neurobiol. Learn. Mem. 2019, 161, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Landry, C.D.; Kandel, E.R.; Rajasethupathy, P. New mechanisms in memory storage: piRNAs and epigenetics. Trends Neurosci. 2013, 36, 535–542. [Google Scholar] [CrossRef] [PubMed]
- Pierouli, K.; Papakonstantinou, E.; Papageorgiou, L.; Diakou, I.; Mitsis, T.; Dragoumani, K.; Spandidos, D.A.; Bacopoulou, F.; Chrousos, G.P.; Goulielmos, G.; et al. Role of non-coding RNAs as biomarkers and the application of omics technologies in Alzheimer’s disease (Review). Int. J. Mol. Med. 2023, 51, 5. [Google Scholar] [CrossRef]
- Zhang, T.; Wong, G. Dysregulation of Human Somatic piRNA Expression in Parkinson’s Disease Subtypes and Stages. Int. J. Mol. Sci. 2022, 23, 2469. [Google Scholar] [CrossRef] [PubMed]
- Wakisaka1, K.T.; Ima, a.Y. The dawn of piRNA research in various neuronal disorders. Front. Biosci. 2019, 24, 1440–1451. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Samimi, H.; Gamez, M.; Zare, H.; Frost, B. Pathogenic tau-induced piRNA depletion promotes neuronal death through transposable element dysregulation in neurodegenerative tauopathies. Nat. Neurosci. 2018, 21, 1038–1048. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Wang, C.; Zhang, T.; Li, R.; Chen, L.; Leung, K.L.; Lakso, M.; Zhou, Q.; Zhang, H.; Wong, G. PIWI-interacting RNA expression regulates pathogenesis in a Caenorhabditis elegans model of Lewy body disease. Nat. Commun. 2023, 14, 6137. [Google Scholar] [CrossRef]
- Chen, S.; Ben, S.; Xin, J.; Li, S.; Zheng, R.; Wang, H.; Fan, L.; Du, M.; Zhang, Z.; Wang, M. The biogenesis and biological function of PIWI-interacting RNA in cancer. J. Hematol. Oncol. 2021, 14, 93. [Google Scholar] [CrossRef]
- Mohn, F.; Handler, D.; Brennecke, J. Noncoding RNA. piRNA-guided slicing specifies transcripts for Zucchini-dependent, phased piRNA biogenesis. Science 2015, 348, 812–817. [Google Scholar] [CrossRef]
- Wu, Z.; Yu, X.; Zhang, S.; He, Y.; Guo, W. Novel roles of PIWI proteins and PIWI-interacting RNAs in human health and diseases. Cell Commun. Signal. 2023, 21, 343. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, S.; Liu, K. Structural insights into piRNA biogenesis. Biochim. Biophys. Acta Gene Regul. Mech. 2022, 1865, 194799. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Yang, J.; Cho, W.C.; Zheng, Y. Argonaute proteins: Structural features, functions and emerging roles. J. Adv. Res. 2020, 24, 317–324. [Google Scholar] [CrossRef]
- Iwasaki, Y.W.; Siomi, M.C.; Siomi, H. PIWI-Interacting RNA: Its Biogenesis and Functions. Annu. Rev. Biochem. 2015, 84, 405–433. [Google Scholar] [CrossRef]
- Hombach, S.; Kretz, M. Non-coding RNAs: Classification, Biology and Functioning. Adv. Exp. Med. Biol. 2016, 937, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Gainetdinov, I.; Colpan, C.; Cecchini, K.; Arif, A.; Jouravleva, K.; Albosta, P.; Vega-Badillo, J.; Lee, Y.; Ozata, D.M.; Zamore, P.D. Terminal modification, sequence, length, and PIWI-protein identity determine piRNA stability. Mol. Cell 2021, 81, 4826–4842.e8. [Google Scholar] [CrossRef]
- Hirakata, S.; Siomi, M.C. Assembly and Function of Gonad-Specific Non-Membranous Organelles in Drosophila piRNA Biogenesis. Noncoding RNA 2019, 5, 52. [Google Scholar] [CrossRef]
- Munafo, M.; Manelli, V.; Falconio, F.A.; Sawle, A.; Kneuss, E.; Eastwood, E.L.; Seah, J.W.E.; Czech, B.; Hannon, G.J. Daedalus and Gasz recruit Armitage to mitochondria, bringing piRNA precursors to the biogenesis machinery. Genes. Dev. 2019, 33, 844–856. [Google Scholar] [CrossRef]
- Wang, X.; Zeng, C.; Liao, S.; Zhu, Z.; Zhang, J.; Tu, X.; Yao, X.; Feng, X.; Guang, S.; Xu, C. Molecular basis for PICS-mediated piRNA biogenesis and cell division. Nat. Commun. 2021, 12, 5595. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Zhou, T.; Chen, Q. Exploring the expanding universe of small RNAs. Nat. Cell Biol. 2022, 24, 415–423. [Google Scholar] [CrossRef]
- Cerutti, L.; Mian, N.; Bateman, A. Domains in gene silencing and cell differentiation proteins: The novel PAZ domain and redefinition of the Piwi domain. Trends Biochem. Sci. 2000, 25, 481–482. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, W.; Liu, Y.; Pi, M.; Jiang, Y.; Ainiwaer, A.; Mao, S.; Chen, H.; Ran, Y.; Sun, S.; et al. Emerging roles and potential application of PIWI-interacting RNA in urological tumors. Front. Endocrinol. 2022, 13, 1054216. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sheng, G.; Juranek, S.; Tuschl, T.; Patel, D.J. Structure of the guide-strand-containing argonaute silencing complex. Nature 2008, 456, 209–213. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Wang, T.; Gao, X.Q.; Chen, X.Z.; Wang, F.; Zhou, L.Y. Emerging functions of piwi-interacting RNAs in diseases. J. Cell Mol. Med. 2021, 25, 4893–4901. [Google Scholar] [CrossRef] [PubMed]
- Copley, K.E.; Shorter, J. Repetitive elements in aging and neurodegeneration. Trends Genet. 2023, 39, 381–400. [Google Scholar] [CrossRef] [PubMed]
- Suleiman, M.; Kounosu, A.; Murcott, B.; Dayi, M.; Pawluk, R.; Yoshida, A.; Viney, M.; Kikuchi, T.; Hunt, V.L. piRNA-like small RNAs target transposable elements in a Clade IV parasitic nematode. Sci. Rep. 2022, 12, 10156. [Google Scholar] [CrossRef] [PubMed]
- Houwing, S.; Kamminga, L.M.; Berezikov, E.; Cronembold, D.; Girard, A.; van den Elst, H.; Filippov, D.V.; Blaser, H.; Raz, E.; Moens, C.B.; et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell 2007, 129, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Siomi, M.C.; Miyoshi, T.; Siomi, H. piRNA-mediated silencing in Drosophila germlines. Semin. Cell Dev. Biol. 2010, 21, 754–759. [Google Scholar] [CrossRef] [PubMed]
- Chuma, S.; Nakano, T. piRNA and spermatogenesis in mice. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013, 368, 20110338. [Google Scholar] [CrossRef] [PubMed]
- Ghildiyal, M.; Seitz, H.; Horwich, M.D.; Li, C.; Du, T.; Lee, S.; Xu, J.; Kittler, E.L.; Zapp, M.L.; Weng, Z.; et al. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science 2008, 320, 1077–1081. [Google Scholar] [CrossRef] [PubMed]
- Aravin, A.A.; Naumova, N.M.; Tulin, A.V.; Vagin, V.V.; Rozovsky, Y.M.; Gvozdev, V.A. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 2001, 11, 1017–1027. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.W. PIWI Proteins and piRNAs in the Nervous System. Mol. Cells 2019, 42, 828–835. [Google Scholar] [CrossRef] [PubMed]
- Anzelon, T.A.; Chowdhury, S.; Hughes, S.M.; Xiao, Y.; Lander, G.C.; MacRae, I.J. Structural basis for piRNA targeting. Nature 2021, 597, 285–289. [Google Scholar] [CrossRef]
- Wang, X.; Ramat, A.; Simonelig, M.; Liu, M.F. Emerging roles and functional mechanisms of PIWI-interacting RNAs. Nat. Rev. Mol. Cell Biol. 2023, 24, 123–141. [Google Scholar] [CrossRef] [PubMed]
- Keam, S.P.; Young, P.E.; McCorkindale, A.L.; Dang, T.H.; Clancy, J.L.; Humphreys, D.T.; Preiss, T.; Hutvagner, G.; Martin, D.I.; Cropley, J.E.; et al. The human Piwi protein Hiwi2 associates with tRNA-derived piRNAs in somatic cells. Nucleic Acids Res. 2014, 42, 8984–8995. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, P.; Ghosh, S.; Wang, B.; Heyns, M.; Li, D.; Mackey, J.R.; Kovalchuk, O.; Damaraju, S. Genome-wide profiling of transfer RNAs and their role as novel prognostic markers for breast cancer. Sci. Rep. 2016, 6, 32843. [Google Scholar] [CrossRef] [PubMed]
- Honda, S.; Kawamura, T.; Loher, P.; Morichika, K.; Rigoutsos, I.; Kirino, Y. The biogenesis pathway of tRNA-derived piRNAs in Bombyx germ cells. Nucleic Acids Res. 2017, 45, 9108–9120. [Google Scholar] [CrossRef] [PubMed]
- Jensen, S.; Brasset, E.; Parey, E.; Roest Crollius, H.; Sharakhov, I.V.; Vaury, C. Conserved Small Nucleotidic Elements at the Origin of Concerted piRNA Biogenesis from Genes and lncRNAs. Cells 2020, 9, 1491. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.H.; Lee, B.; Li, X.Z. The birth of piRNAs: How mammalian piRNAs are produced, originated, and evolved. Mamm. Genome 2022, 33, 293–311. [Google Scholar] [CrossRef] [PubMed]
- Feltzin, V.L.; Khaladkar, M.; Abe, M.; Parisi, M.; Hendriks, G.J.; Kim, J.; Bonini, N.M. The exonuclease Nibbler regulates age-associated traits and modulates piRNA length in Drosophila. Aging Cell 2015, 14, 443–452. [Google Scholar] [CrossRef]
- Cacchione, S.; Cenci, G.; Raffa, G.D. Silence at the End: How Drosophila Regulates Expression and Transposition of Telomeric Retroelements. J. Mol. Biol. 2020, 432, 4305–4321. [Google Scholar] [CrossRef] [PubMed]
- Gou, L.T.; Dai, P.; Yang, J.H.; Xue, Y.; Hu, Y.P.; Zhou, Y.; Kang, J.Y.; Wang, X.; Li, H.; Hua, M.M.; et al. Pachytene piRNAs instruct massive mRNA elimination during late spermiogenesis. Cell Res. 2014, 24, 680–700. [Google Scholar] [CrossRef] [PubMed]
- Goh, W.S.; Falciatori, I.; Tam, O.H.; Burgess, R.; Meikar, O.; Kotaja, N.; Hammell, M.; Hannon, G.J. piRNA-directed cleavage of meiotic transcripts regulates spermatogenesis. Genes. Dev. 2015, 29, 1032–1044. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Kang, J.Y.; Gou, L.T.; Wang, J.; Xue, Y.; Skogerboe, G.; Dai, P.; Huang, D.W.; Chen, R.; Fu, X.D.; et al. MIWI and piRNA-mediated cleavage of messenger RNAs in mouse testes. Cell Res. 2015, 25, 193–207. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.H.; Wang, R.H.; Du, K.; Zhu, J.; Zheng, J.; Xie, L.H.; Pereira, A.A.; Zhang, C.; Ricci, E.P.; Li, X.Z. Coupled protein synthesis and ribosome-guided piRNA processing on mRNAs. Nat. Commun. 2021, 12, 5970. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.H.; Cui, H.; Song, C.; Shen, J.T.; Zhuo, X.; Wang, R.H.; Yu, X.; Ndamba, R.; Mu, Q.; Gu, H.; et al. Amniotes co-opt intrinsic genetic instability to protect germ-line genome integrity. Nat. Commun. 2023, 14, 812. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Huang, Y.; Ding, Y.; Shi, L.; Zhong, X.; Kang, M.; Li, C. Analysis of piRNA expression spectra in a non-alcoholic fatty liver disease mouse model induced by a methionine- and choline-deficient diet. Exp. Ther. Med. 2020, 19, 3829–3839. [Google Scholar] [CrossRef] [PubMed]
- Subhramanyam, C.S.; Cao, Q.; Wang, C.; Heng, Z.S.L.; Zhou, Z.; Hu, Q. Role of PIWI-like 4 in modulating neuronal differentiation from human embryonal carcinoma cells. RNA Biol. 2020, 17, 1613–1624. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.J.; Banerjee, S.; Zhou, H.; Jammalamadaka, A.; Arcila, M.; Manjunath, B.S.; Kosik, K.S. Identification of piRNAs in the central nervous system. Rna 2011, 17, 1090–1099. [Google Scholar] [CrossRef] [PubMed]
- Galton, R.; Fejes-Toth, K.; Bronner, M.E. Co-option of the piRNA pathway to regulate neural crest specification. Sci. Adv. 2022, 8, eabn1441. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Tu, S.; Stubna, M.; Wu, W.S.; Huang, W.C.; Weng, Z.; Lee, H.C. The piRNA targeting rules and the resistance to piRNA silencing in endogenous genes. Science 2018, 359, 587–592. [Google Scholar] [CrossRef] [PubMed]
- Bu, D.; Yu, K.; Sun, S.; Xie, C.; Skogerbo, G.; Miao, R.; Xiao, H.; Liao, Q.; Luo, H.; Zhao, G.; et al. NONCODE v3.0: Integrative annotation of long noncoding RNAs. Nucleic Acids Res. 2012, 40, D210–D215. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Yuan, J.; Li, H.; Li, M.; Zhao, G.; Bu, D.; Zhu, W.; Wu, W.; Chen, R.; Zhao, Y. NONCODEv4: Exploring the world of long non-coding RNA genes. Nucleic Acids Res. 2013, 42, D98–D103. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Liu, S.; Wang, J.; Xuan, J.; Zhang, X.; Qu, L.; Zheng, L.; Yang, J. deepBase v3.0: Expression atlas and interactive analysis of ncRNAs from thousands of deep-sequencing data. Nucleic Acids Res. 2021, 49, D877–D883. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.H.; Shao, P.; Zhou, H.; Chen, Y.Q.; Qu, L.H. deepBase: A database for deeply annotating and mining deep sequencing data. Nucleic Acids Res. 2010, 38, D123–D130. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Shi, Y.; Zhou, H.; Zhang, P.; Song, T.; Ying, Z.; Yu, H.; Li, Y.; Zhao, Y.; Zeng, X.; et al. piRBase: Integrating piRNA annotation in all aspects. Nucleic Acids Res. 2022, 50, D265–D272. [Google Scholar] [CrossRef] [PubMed]
- Sai Lakshmi, S.; Agrawal, S. piRNABank: A web resource on classified and clustered Piwi-interacting RNAs. Nucleic Acids Res. 2008, 36, D173–D177. [Google Scholar] [CrossRef] [PubMed]
- Rosenkranz, D.; Zischler, H.; Gebert, D. piRNAclusterDB 2.0: Update and expansion of the piRNA cluster database. Nucleic Acids Res. 2022, 50, D259–D264. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.S.; Brown, J.S.; Chen, T.T.; Chu, Y.H.; Huang, W.C.; Tu, S.; Lee, H.C. piRTarBase: A database of piRNA targeting sites and their roles in gene regulation. Nucleic Acids Res. 2019, 47, D181–D187. [Google Scholar] [CrossRef] [PubMed]
- Barrenada, O.; Larriba, E.; Brieno-Enriquez, M.A.; Mazo, J.D. piRNA-IPdb: A PIWI-bound piRNAs database to mining NGS sncRNA data and beyond. BMC Genom. 2021, 22, 765. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Si, X.; Skogerbo, G.; Wang, J.; Cui, D.; Li, Y.; Sun, X.; Liu, L.; Sun, B.; Chen, R.; et al. piRBase: A web resource assisting piRNA functional study. Database 2014, 2014, bau110. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, P.; Lu, Y.; Li, Y.; Zheng, Y.; Kan, Y.; Chen, R.; He, S. piRBase: A comprehensive database of piRNA sequences. Nucleic Acids Res. 2019, 47, D175–D180. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.D.; Alam, W.; Tayara, H.; Chong, K.T. Identification of Functional piRNAs Using a Convolutional Neural Network. IEEE/ACM Trans. Comput. Biol. Bioinform. 2022, 19, 1661–1669. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Hoeksema, J.; Liang, C. piRNN: Deep learning algorithm for piRNA prediction. PeerJ 2018, 6, e5429. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Chen, L.; Li, R.; Liu, N.; Huang, X.; Wong, G. PIWI-interacting RNAs in human diseases: Databases and computational models. Brief. Bioinform. 2022, 23, bbac217. [Google Scholar] [CrossRef]
- Wang, H.; Shi, B.; Zhang, X.; Shen, P.; He, Q.; Yin, M.; Pan, Y.; Ma, J. Exosomal hsa-piR1089 promotes proliferation and migration in neuroblastoma via targeting KEAP1. Pathol. Res. Pract. 2023, 241, 154240. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.P.; Yao, M.J.; Chang, S.Y.; Gou, L.T.; Liu, M.F.; Qiu, Z.L.; Yuan, X.B. Novel function of PIWIL1 in neuronal polarization and migration via regulation of microtubule-associated proteins. Mol. Brain 2015, 8, 39. [Google Scholar] [CrossRef] [PubMed]
- Gasperini, C.; Tuntevski, K.; Beatini, S.; Pelizzoli, R.; Lo Van, A.; Mangoni, D.; Cossu, R.M.; Pascarella, G.; Bianchini, P.; Bielefeld, P.; et al. Piwil2 (Mili) sustains neurogenesis and prevents cellular senescence in the postnatal hippocampus. EMBO Rep. 2023, 24, e53801. [Google Scholar] [CrossRef] [PubMed]
- Zhan, L.; Chen, M.; Pang, T.; Li, X.; Long, L.; Liang, D.; Peng, L.; Sun, W.; Xu, E. Attenuation of Piwil2 induced by hypoxic postconditioning prevents cerebral ischemic injury by inhibiting CREB2 promoter methylation. Brain Pathol. 2023, 33, e13109. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Jiang, F.; Liu, X.; Liu, Q.; Fu, Y.; Li, R.; Hou, L.; Zhang, J.; He, J.; Kang, L. Piwi/piRNAs control food intake by promoting neuropeptide F expression in locusts. EMBO Rep. 2022, 23, e50851. [Google Scholar] [CrossRef] [PubMed]
- Chavda, V.; Madhwani, K.; Chaurasia, B. PiWi RNA in Neurodevelopment and Neurodegenerative Disorders. Curr. Mol. Pharmacol. 2022, 15, 517–531. [Google Scholar] [CrossRef] [PubMed]
- Ozata, D.M.; Gainetdinov, I.; Zoch, A.; O’Carroll, D.; Zamore, P.D. PIWI-interacting RNAs: Small RNAs with big functions. Nat. Rev. Genet. 2019, 20, 89–108. [Google Scholar] [CrossRef] [PubMed]
- Penning, A.; Tosoni, G.; Abiega, O.; Bielefeld, P.; Gasperini, C.; De Pietri Tonelli, D.; Fitzsimons, C.P.; Salta, E. Adult Neural Stem Cell Regulation by Small Non-coding RNAs: Physiological Significance and Pathological Implications. Front. Cell Neurosci. 2021, 15, 781434. [Google Scholar] [CrossRef] [PubMed]
- Perera, B.P.U.; Tsai, Z.T.; Colwell, M.L.; Jones, T.R.; Goodrich, J.M.; Wang, K.; Sartor, M.A.; Faulk, C.; Dolinoy, D.C. Somatic expression of piRNA and associated machinery in the mouse identifies short, tissue-specific piRNA. Epigenetics 2019, 14, 504–521. [Google Scholar] [CrossRef] [PubMed]
- Nishihara, H.; Perriot, S.; Gastfriend, B.D.; Steinfort, M.; Cibien, C.; Soldati, S.; Matsuo, K.; Guimbal, S.; Mathias, A.; Palecek, S.P.; et al. Intrinsic blood-brain barrier dysfunction contributes to multiple sclerosis pathogenesis. Brain 2022, 145, 4334–4348. [Google Scholar] [CrossRef] [PubMed]
- Hang, Z.; Zhou, L.; Xing, C.; Wen, Y.; Du, H. The blood-brain barrier, a key bridge to treat neurodegenerative diseases. Ageing Res. Rev. 2023, 91, 102070. [Google Scholar] [CrossRef] [PubMed]
- Roy, R.; Pattnaik, S.; Sivagurunathan, S.; Chidambaram, S. Small ncRNA binding protein, PIWI: A potential molecular bridge between blood brain barrier and neuropathological conditions. Med. Hypotheses 2020, 138, 109609. [Google Scholar] [CrossRef] [PubMed]
- Sivagurunathan, S.; Palanisamy, K.; Arunachalam, J.P.; Chidambaram, S. Possible role of HIWI2 in modulating tight junction proteins in retinal pigment epithelial cells through Akt signaling pathway. Mol. Cell. Biochem. 2016, 427, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Bartsch, D.; Ghirardi, M.; Skehel, P.A.; Karl, K.A.; Herder, S.P.; Chen, M.; Bailey, C.H.; Kandel, E.R. Aplysia CREB2 represses long-term facilitation: Relief of repression converts transient facilitation into long-term functional and structural change. Cell 1995, 83, 979–992. [Google Scholar] [CrossRef] [PubMed]
- Hao, L.; Yang, Z. Dynamical Mechanisms for Gene Regulation Mediated by Two Noncoding RNAs in Long-Term Memory Formation. Neural Plast. 2021, 2021, 6668389. [Google Scholar] [CrossRef]
- Craske, M.G.; Rauch, S.L.; Ursano, R.; Prenoveau, J.; Pine, D.S.; Zinbarg, R.E. What is an anxiety disorder? Depress. Anxiety 2009, 26, 1066–1085. [Google Scholar] [CrossRef] [PubMed]
- Nandi, S.; Chandramohan, D.; Fioriti, L.; Melnick, A.M.; Hebert, J.M.; Mason, C.E.; Rajasethupathy, P.; Kandel, E.R. Roles for small noncoding RNAs in silencing of retrotransposons in the mammalian brain. Proc. Natl. Acad. Sci. USA 2016, 113, 12697–12702. [Google Scholar] [CrossRef] [PubMed]
- Donkin, I.; Versteyhe, S.; Ingerslev, L.R.; Qian, K.; Mechta, M.; Nordkap, L.; Mortensen, B.; Appel, E.V.; Jorgensen, N.; Kristiansen, V.B.; et al. Obesity and Bariatric Surgery Drive Epigenetic Variation of Spermatozoa in Humans. Cell Metab. 2016, 23, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Sinnott-Armstrong, N.; Wagschal, A.; Wark, A.R.; Camporez, J.P.; Perry, R.J.; Ji, F.; Sohn, Y.; Oh, J.; Wu, S.; et al. A MicroRNA Linking Human Positive Selection and Metabolic Disorders. Cell 2020, 183, 684–701.e14. [Google Scholar] [CrossRef] [PubMed]
- Vinnikov, I.A.; Hajdukiewicz, K.; Reymann, J.; Beneke, J.; Czajkowski, R.; Roth, L.C.; Novak, M.; Roller, A.; Dörner, N.; Starkuviene, V.; et al. Hypothalamic miR-103 protects from hyperphagic obesity in mice. J. Neurosci. 2014, 34, 10659–10674. [Google Scholar] [CrossRef] [PubMed]
- Falibene, A.; Rossler, W.; Josens, R. Serotonin depresses feeding behaviour in ants. J. Insect Physiol. 2012, 58, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Curcio, M.; Bradke, F. Axon Regeneration in the Central Nervous System: Facing the Challenges from the Inside. Annu. Rev. Cell Dev. Biol. 2018, 34, 495–521. [Google Scholar] [CrossRef] [PubMed]
- Phay, M.; Kim, H.H.; Yoo, S. Analysis of piRNA-Like Small Non-coding RNAs Present in Axons of Adult Sensory Neurons. Mol. Neurobiol. 2018, 55, 483–494. [Google Scholar] [CrossRef] [PubMed]
- Parenti, I.; Rabaneda, L.G.; Schoen, H.; Novarino, G. Neurodevelopmental Disorders: From Genetics to Functional Pathways. Trends Neurosci. 2020, 43, 608–621. [Google Scholar] [CrossRef]
- Kuo, M.C.; Liu, S.C.; Hsu, Y.F.; Wu, R.M. The role of noncoding RNAs in Parkinson’s disease: Biomarkers and associations with pathogenic pathways. J. Biomed. Sci. 2021, 28, 78. [Google Scholar] [CrossRef]
- Kim, Y.S.; Choi, J.; Yoon, B.E. Neuron-Glia Interactions in Neurodevelopmental Disorders. Cells 2020, 9, 2176. [Google Scholar] [CrossRef] [PubMed]
- Kwon, H.C.; Bae, Y.; Lee, S.V. The Role of mRNA Quality Control in the Aging of Caenorhabditis elegans. Mol. Cells 2023, 46, 664–671. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Wong, G. An old weapon with a new function: PIWI-interacting RNAs in neurodegenerative diseases. Transl. Neurodegener. 2021, 10, 9. [Google Scholar] [CrossRef] [PubMed]
- Dharap, A.; Nakka, V.P.; Vemuganti, R. Altered expression of PIWI RNA in the rat brain after transient focal ischemia. Stroke 2011, 42, 1105–1109. [Google Scholar] [CrossRef] [PubMed]
- Pierce, L.M.; Kurata, W.E.; Matsumoto, K.W.; Clark, M.E.; Farmer, D.M. Long-term epigenetic alterations in a rat model of Gulf War Illness. Neurotoxicology 2016, 55, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Wang, C.; Chen, L.; Wong, G. Dysregulation of MicroRNAs and PIWI-Interacting RNAs in a Caenorhabditis elegans Parkinson’s Disease Model Overexpressing Human alpha-Synuclein and Influence of tdp-1. Front. Neurosci. 2021, 15, 600462. [Google Scholar] [CrossRef] [PubMed]
- Frost, B.; Hemberg, M.; Lewis, J.; Feany, M.B. Tau promotes neurodegeneration through global chromatin relaxation. Nat. Neurosci. 2014, 17, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Wakisaka, K.T.; Tanaka, R.; Hirashima, T.; Muraoka, Y.; Azuma, Y.; Yoshida, H.; Tokuda, T.; Asada, S.; Suda, K.; Ichiyanagi, K.; et al. Novel roles of Drosophila FUS and Aub responsible for piRNA biogenesis in neuronal disorders. Brain Res. 2019, 1708, 207–219. [Google Scholar] [CrossRef] [PubMed]
- Kaur, H.; Sarmah, D.; Saraf, J.; Vats, K.; Kalia, K.; Borah, A.; Yavagal, D.R.; Dave, K.R.; Ghosh, Z.; Bhattacharya, P. Noncoding RNAs in ischemic stroke: Time to translate. Ann. N.Y Acad. Sci. 2018, 1421, 19–36. [Google Scholar] [CrossRef] [PubMed]
- Saugstad, J.A. Non-Coding RNAs in Stroke and Neuroprotection. Front. Neurol. 2015, 6, 50. [Google Scholar] [CrossRef] [PubMed]
- Potemkin, N.; Clarkson, A.N. Non-coding RNAs in stroke pathology, diagnostics, and therapeutics. Neurochem. Int. 2023, 162, 105467. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Shirvani-Farsani, Z.; Hussen, B.M.; Taheri, M.; Arefian, N. Emerging Impact of Non-coding RNAs in the Pathology of Stroke. Front. Aging Neurosci. 2021, 13, 780489. [Google Scholar] [CrossRef] [PubMed]
- Chavda, V.; Madhwani, K. Coding and non-coding nucleotides’: The future of stroke gene therapeutics. Genomics 2021, 113, 1291–1307. [Google Scholar] [CrossRef] [PubMed]
- Carroll, D. RNA in Therapeutics: CRISPR in the Clinic. Mol. Cells 2023, 46, 4–9. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, S.F.; Shore, M.F. Prevention of psychiatric disorders. Harv. Rev. Psychiatry 1995, 3, 115–129. [Google Scholar] [CrossRef] [PubMed]
- Yoshino, Y.; Dwivedi, Y. Non-Coding RNAs in Psychiatric Disorders and Suicidal Behavior. Front. Psychiatry 2020, 11, 543893. [Google Scholar] [CrossRef] [PubMed]
- Ragan, C.; Patel, K.; Edson, J.; Zhang, Z.H.; Gratten, J.; Mowry, B. Small non-coding RNA expression from anterior cingulate cortex in schizophrenia shows sex specific regulation. Schizophr. Res. 2017, 183, 82–87. [Google Scholar] [CrossRef] [PubMed]
- McDaniel Peters, B.C.; Wood, W. Autism and Equine-Assisted Interventions: A Systematic Mapping Review. J. Autism Dev. Disord. 2017, 47, 3220–3242. [Google Scholar] [CrossRef] [PubMed]
- de Giambattista, C.; Ventura, P.; Trerotoli, P.; Margari, M.; Palumbi, R.; Margari, L. Subtyping the Autism Spectrum Disorder: Comparison of Children with High Functioning Autism and Asperger Syndrome. J. Autism Dev. Disord. 2019, 49, 138–150. [Google Scholar] [CrossRef] [PubMed]
- Chiappori, F.; Cupaioli, F.A.; Consiglio, A.; Di Nanni, N.; Mosca, E.; Licciulli, V.F.; Mezzelani, A. Analysis of Faecal Microbiota and Small ncRNAs in Autism: Detection of miRNAs and piRNAs with Possible Implications in Host-Gut Microbiota Cross-Talk. Nutrients 2022, 14, 1340. [Google Scholar] [CrossRef] [PubMed]
- Salloum-Asfar, S.; Elsayed, A.K.; Elhag, S.F.; Abdulla, S.A. Circulating Non-Coding RNAs as a Signature of Autism Spectrum Disorder Symptomatology. Int. J. Mol. Sci. 2021, 22, 6549. [Google Scholar] [CrossRef]
- Beversdorf, D.Q.; Sohl, K.; Levitskiy, D.; Tennant, P.; Goin-Kochel, R.P.; Shaffer, R.C.; Confair, A.; Middleton, F.A.; Hicks, S.D. Saliva RNA Biomarkers of Gastrointestinal Dysfunction in Children with Autism and Neurodevelopmental Disorders: Potential Implications for Precision Medicine. Front. Psychiatry 2021, 12, 824933. [Google Scholar] [CrossRef]
- Sato, K.; Takayama, K.I.; Inoue, S. Role of piRNA biogenesis and its neuronal function in the development of neurodegenerative diseases. Front. Aging Neurosci. 2023, 15, 1157818. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Kundu, S.; Singh, A.; Singh, S. Understanding the Role of Histone Deacetylase and their Inhibitors in Neurodegenerative Disorders: Current Targets and Future Perspective. Curr. Neuropharmacol. 2022, 20, 158–178. [Google Scholar] [CrossRef] [PubMed]
- Jomova, K.; Vondrakova, D.; Lawson, M.; Valko, M. Metals, oxidative stress and neurodegenerative disorders. Mol. Cell Biochem. 2010, 345, 91–104. [Google Scholar] [CrossRef]
- Cooke, P.; Janowitz, H.; Dougherty, S.E. Neuronal Redevelopment and the Regeneration of Neuromodulatory Axons in the Adult Mammalian Central Nervous System. Front. Cell Neurosci. 2022, 16, 872501. [Google Scholar] [CrossRef] [PubMed]
- Lauretti, E.; Dabrowski, K.; Pratico, D. The neurobiology of non-coding RNAs and Alzheimer’s disease pathogenesis: Pathways, mechanisms and translational opportunities. Ageing Res. Rev. 2021, 71, 101425. [Google Scholar] [CrossRef]
- Tan, L.; Yu, J.T.; Hu, N.; Tan, L. Non-coding RNAs in Alzheimer’s disease. Mol. Neurobiol. 2013, 47, 382–393. [Google Scholar] [CrossRef] [PubMed]
- Scheltens, P.; De Strooper, B.; Kivipelto, M.; Holstege, H.; Chetelat, G.; Teunissen, C.E.; Cummings, J.; van der Flier, W.M. Alzheimer’s disease. Lancet 2021, 397, 1577–1590. [Google Scholar] [CrossRef]
- Wang, J.Z.; Grundke-Iqbal, I.; Iqbal, K. Kinases and phosphatases and tau sites involved in Alzheimer neurofibrillary degeneration. Eur. J. Neurosci. 2007, 25, 59–68. [Google Scholar] [CrossRef]
- Saba, R.; Goodman, C.D.; Huzarewich, R.L.; Robertson, C.; Booth, S.A. A miRNA signature of prion induced neurodegeneration. PLoS ONE 2008, 3, e3652. [Google Scholar] [CrossRef] [PubMed]
- Hébert, S.S.; Horré, K.; Nicolaï, L.; Papadopoulou, A.S.; Mandemakers, W.; Silahtaroglu, A.N.; Kauppinen, S.; Delacourte, A.; De Strooper, B. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/beta-secretase expression. Proc. Natl. Acad. Sci. USA 2008, 105, 6415–6420. [Google Scholar] [CrossRef] [PubMed]
- Salminen, A.; Kauppinen, A.; Suuronen, T.; Kaarniranta, K.; Ojala, J. ER stress in Alzheimer’s disease: A novel neuronal trigger for inflammation and Alzheimer’s pathology. J. Neuroinflammation 2009, 6, 41. [Google Scholar] [CrossRef]
- Qiu, W.; Guo, X.; Lin, X.; Yang, Q.; Zhang, W.; Zhang, Y.; Zuo, L.; Zhu, Y.; Li, C.R.; Ma, C.; et al. Transcriptome-wide piRNA profiling in human brains of Alzheimer’s disease. Neurobiol. Aging 2017, 57, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Roy, J.; Sarkar, A.; Parida, S.; Ghosh, Z.; Mallick, B. Small RNA sequencing revealed dysregulated piRNAs in Alzheimer’s disease and their probable role in pathogenesis. Mol. Biosyst. 2017, 13, 565–576. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Qiu, W.; Garcia-Milian, R.; Lin, X.; Zhang, Y.; Cao, Y.; Tan, Y.; Wang, Z.; Shi, J.; Wang, J.; et al. Genome-wide significant, replicated and functional risk variants for Alzheimer’s disease. J. Neural Transm. 2017, 124, 1455–1471. [Google Scholar] [CrossRef] [PubMed]
- Mao, Q.; Fan, L.; Wang, X.; Lin, X.; Cao, Y.; Zheng, C.; Zhang, Y.; Zhang, H.; Garcia-Milian, R.; Kang, L.; et al. Transcriptome-wide piRNA profiling in human brains for aging genetic factors. Jacobs J. Genet. 2019, 4, 14. [Google Scholar]
- Jain, G.; Stuendl, A.; Rao, P.; Berulava, T.; Pena Centeno, T.; Kaurani, L.; Burkhardt, S.; Delalle, I.; Kornhuber, J.; Hull, M.; et al. A combined miRNA-piRNA signature to detect Alzheimer’s disease. Transl. Psychiatry 2019, 9, 250. [Google Scholar] [CrossRef]
- Fitz, N.F.; Wang, J.; Kamboh, M.I.; Koldamova, R.; Lefterov, I. Small nucleolar RNAs in plasma extracellular vesicles and their discriminatory power as diagnostic biomarkers of Alzheimer’s disease. Neurobiol. Dis. 2021, 159, 105481. [Google Scholar] [CrossRef]
- Olufunmilayo, E.O.; Holsinger, R.M.D. Roles of Non-Coding RNA in Alzheimer’s Disease Pathophysiology. Int. J. Mol. Sci. 2023, 24, 12498. [Google Scholar] [CrossRef]
- Costa, H.N.; Esteves, A.R.; Empadinhas, N.; Cardoso, S.M. Parkinson’s Disease: A Multisystem Disorder. Neurosci. Bull. 2023, 39, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Paccosi, E.; Proietti-De-Santis, L. Parkinson’s Disease: From Genetics and Epigenetics to Treatment, a miRNA-Based Strategy. Int. J. Mol. Sci. 2023, 24, 9547. [Google Scholar] [CrossRef] [PubMed]
- Fakhri, S.; Darvish, E.; Narimani, F.; Moradi, S.Z.; Abbaszadeh, F.; Khan, H. The regulatory role of non-coding RNAs and their interactions with phytochemicals in neurodegenerative diseases: A systematic review. Brief. Funct. Genom. 2023, 22, 143–160. [Google Scholar] [CrossRef] [PubMed]
- Galvagnion, C.; Marlet, F.R.; Cerri, S.; Schapira, A.H.V.; Blandini, F.; Di Monte, D.A. Sphingolipid changes in Parkinson L444P GBA mutation fibroblasts promote α-synuclein aggregation. Brain 2022, 145, 1038–1051. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, O.; Gana-Weisz, M.; Banfi, S.; Nigro, V.; Bar-Shira, A.; Thaler, A.; Gurevich, T.; Mirelman, A.; Giladi, N.; Alcalay, R.N.; et al. Novel variants in genes related to vesicle-mediated-transport modify Parkinson’s disease risk. Mol. Genet. Metab. 2023, 139, 107608. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Q.; Pan, H.; Zhao, Y.; Wang, Y.; Xu, Q.; Tan, J.; Yan, X.; Li, J.; Tang, B.; Guo, J. Association Study of TAF1 Variants in Parkinson’s Disease. Front. Neurosci. 2022, 16, 846095. [Google Scholar] [CrossRef] [PubMed]
- Ohira, K.; Yokota, H.; Hirano, S.; Nishimura, M.; Mukai, H.; Horikoshi, T.; Sawai, S.; Yamanaka, Y.; Yamamoto, T.; Kakeda, S.; et al. DRD2 Taq1A Polymorphism-Related Brain Volume Changes in Parkinson’s Disease: Voxel-Based Morphometry. Park. Dis. 2022, 2022, 8649195. [Google Scholar] [CrossRef] [PubMed]
- Gagliardi, M.; Annesi, G.; Procopio, R.; Morelli, M.; Iannello, G.; Bonapace, G.; Mancini, M.; Nicoletti, G.; Quattrone, A. DNAJC13 mutation screening in patients with Parkinson’s disease from South Italy. Park. Relat. Disord. 2018, 55, 134–137. [Google Scholar] [CrossRef] [PubMed]
- Schulze, M.; Sommer, A.; Plotz, S.; Farrell, M.; Winner, B.; Grosch, J.; Winkler, J.; Riemenschneider, M.J. Sporadic Parkinson’s disease derived neuronal cells show disease-specific mRNA and small RNA signatures with abundant deregulation of piRNAs. Acta Neuropathol. Commun. 2018, 6, 58. [Google Scholar] [CrossRef] [PubMed]
- Nataf, S.; Guillen, M.; Pays, L. Common Neurodegeneration-Associated Proteins Are Physiologically Expressed by Human B Lymphocytes and Are Interconnected via the Inflammation/Autophagy-Related Proteins TRAF6 and SQSTM1. Front. Immunol. 2019, 10, 2704. [Google Scholar] [CrossRef] [PubMed]
- Aisha, Z.; Lei, J.; Zhang, Y.; Ma, J. EEF1A1 is Involved the Regulating Neuroinflammatory Processes in Parkinson’s Disease. J. Integr. Neurosci. 2023, 22, 122. [Google Scholar] [CrossRef] [PubMed]
- Simoes, F.A.; Joilin, G.; Peters, O.; Schneider, L.S.; Priller, J.; Spruth, E.J.; Vogt, I.; Kimmich, O.; Spottke, A.; Hoffmann, D.C.; et al. Potential of Non-Coding RNA as Biomarkers for Progressive Supranuclear Palsy. Int. J. Mol. Sci. 2022, 23, 14554. [Google Scholar] [CrossRef] [PubMed]
- Rutherford, N.J.; Zhang, Y.J.; Baker, M.; Gass, J.M.; Finch, N.A.; Xu, Y.F.; Stewart, H.; Kelley, B.J.; Kuntz, K.; Crook, R.J.; et al. Novel mutations in TARDBP (TDP-43) in patients with familial amyotrophic lateral sclerosis. PLoS Genet. 2008, 4, e1000193. [Google Scholar] [CrossRef] [PubMed]
- Sreedharan, J.; Blair, I.P.; Tripathi, V.B.; Hu, X.; Vance, C.; Rogelj, B.; Ackerley, S.; Durnall, J.C.; Williams, K.L.; Buratti, E.; et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science 2008, 319, 1668–1672. [Google Scholar] [CrossRef] [PubMed]
- Couthouis, J.; Hart, M.P.; Erion, R.; King, O.D.; Diaz, Z.; Nakaya, T.; Ibrahim, F.; Kim, H.J.; Mojsilovic-Petrovic, J.; Panossian, S.; et al. Evaluating the role of the FUS/TLS-related gene EWSR1 in amyotrophic lateral sclerosis. Hum. Mol. Genet. 2012, 21, 2899–2911. [Google Scholar] [CrossRef] [PubMed]
- Vance, C.; Rogelj, B.; Hortobagyi, T.; De Vos, K.J.; Nishimura, A.L.; Sreedharan, J.; Hu, X.; Smith, B.; Ruddy, D.; Wright, P.; et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 2009, 323, 1208–1211. [Google Scholar] [CrossRef] [PubMed]
- Abdelhamid, R.F.; Ogawa, K.; Beck, G.; Ikenaka, K.; Takeuchi, E.; Yasumizu, Y.; Jinno, J.; Kimura, Y.; Baba, K.; Nagai, Y.; et al. piRNA/PIWI Protein Complex as a Potential Biomarker in Sporadic Amyotrophic Lateral Sclerosis. Mol. Neurobiol. 2022, 59, 1693–1705. [Google Scholar] [CrossRef] [PubMed]
- Joilin, G.; Gray, E.; Thompson, A.G.; Talbot, K.; Leigh, P.N.; Newbury, S.F.; Turner, M.R.; Hafezparast, M. Profiling non-coding RNA expression in cerebrospinal fluid of amyotrophic lateral sclerosis patients. Ann. Med. 2022, 54, 3069–3078. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).