Advances in and Perspectives on Transgenic Technology and CRISPR-Cas9 Gene Editing in Broccoli
Abstract
:1. Introduction
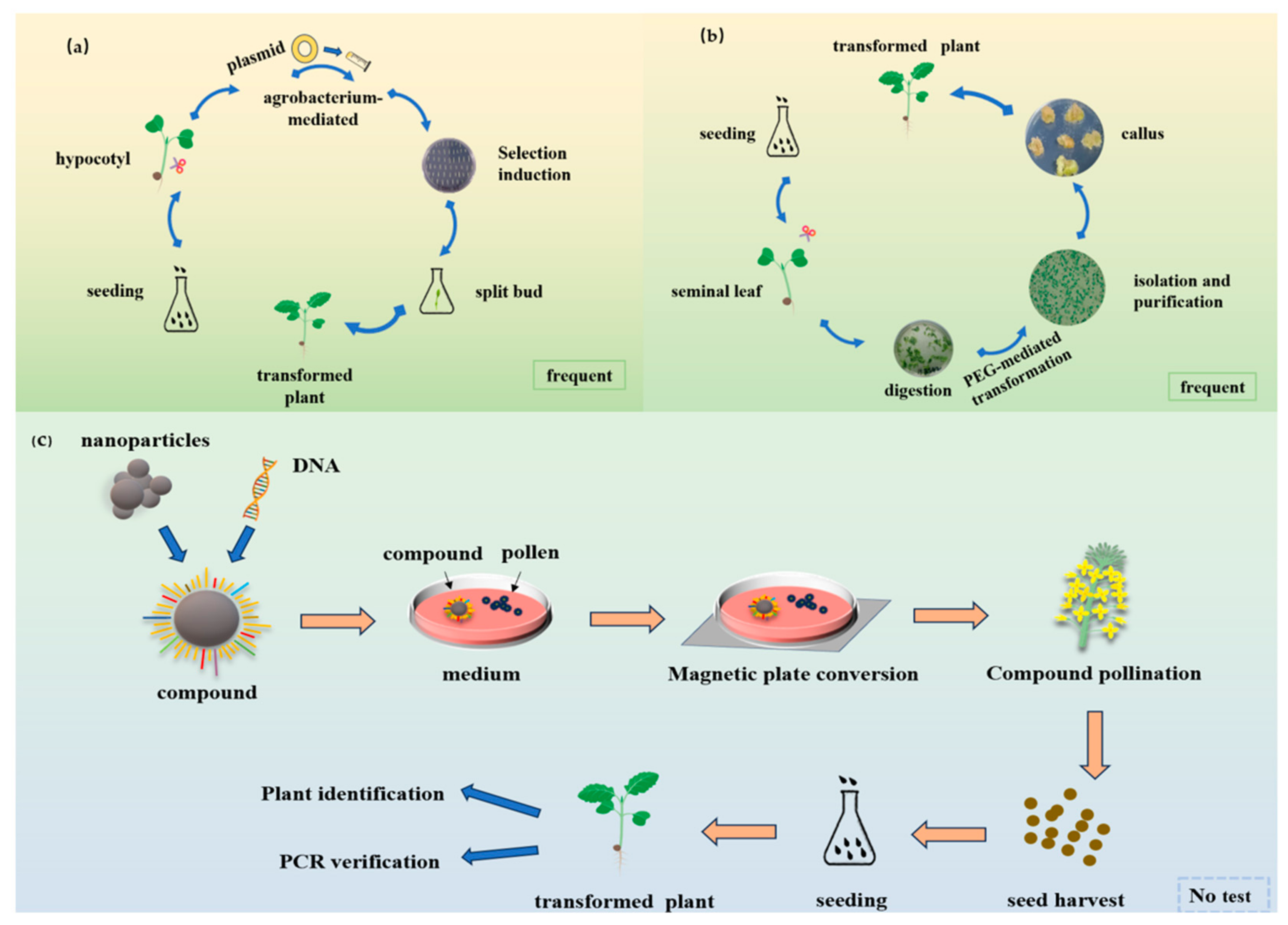
2. Construction and Optimization of the Genetic Transformation System for Broccoli
3. Research and Application of Genetic Transformation Technology in Broccoli
3.1. Agrobacterium Tumefaciens-Mediated Genetic Transformation System
3.1.1. Agronomic Traits
3.1.2. Quality Traits
3.1.3. Biotic and Abiotic Stress
3.1.4. Others
3.2. Application of the Protoplast-Mediated Instantaneous Transformation System
3.3. Others
4. Research and Application of Gene Editing Technology in Broccoli
5. Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gasmi, A.; Gasmi Benahmed, A.; Shanaida, M.; Chirumbolo, S.; Menzel, A.; Anzar, W.; Arshad, M.; Cruz-Martins, N.; Lysiuk, R.; Beley, N.; et al. Anticancer activity of broccoli, its organosulfur and polyphenolic compounds. Crit. Rev. Food Sci. Nutr. 2023, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.-H.; Lim, S.-B. Antioxidant and Anticancer Activities of Broccoli By-Products from Different Cultivars and Maturity Stages at Harvest. Prev. Nutr. Food Sci. 2015, 20, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Radošević, K.; Srček, V.G.; Bubalo, M.C.; Rimac Brnčić, S.; Takács, K.; Redovniković, I.R. Assessment of glucosinolates, antioxidative and antiproliferative activity of broccoli and collard extracts. J. Food Compos. Anal. 2017, 61, 59–66. [Google Scholar] [CrossRef]
- Tang, G.-Y.; Meng, X.; Li, Y.; Zhao, C.-N.; Liu, Q.; Li, H.-B. Effects of Vegetables on Cardiovascular Diseases and Related Mechanisms. Nutrients 2017, 9, 857. [Google Scholar] [CrossRef] [PubMed]
- Soares, A.; Carrascosa, C.; Raposo, A. Influence of Different Cooking Methods on the Concentration of Glucosinolates and Vitamin C in Broccoli. Food Bioprocess Technol. 2017, 10, 1387–1411. [Google Scholar] [CrossRef]
- Favela-González, K.M.; Hernández-Almanza, A.Y.; De la Fuente-Salcido, N.M. The value of bioactive compounds of cruciferous vegetables (Brassica) as antimicrobials and antioxidants: A review. J. Food Biochem. 2020, 44, e13414. [Google Scholar] [CrossRef]
- Barber, T.M.; Kabisch, S.; Pfeiffer, A.F.H.; Weickert, M.O. The Health Benefits of Dietary Fibre. Nutrients 2020, 12, 3209. [Google Scholar] [CrossRef] [PubMed]
- Mahn, A.; Castillo, A. Potential of Sulforaphane as a Natural Immune System Enhancer: A Review. Molecules 2021, 26, 752. [Google Scholar] [CrossRef] [PubMed]
- Houghton, C.A. Sulforaphane: Its “Coming of Age” as a Clinically Relevant Nutraceutical in the Prevention and Treatment of Chronic Disease. Oxidative Med. Cell. Longev. 2019, 2019, 2716870. [Google Scholar] [CrossRef]
- Wang, T.T.Y.; Schoene, N.W.; Milner, J.A.; Kim, Y.S. Broccoli-derived phytochemicals indole-3-carbinol and 3,3′-diindolylmethane exerts concentration-dependent pleiotropic effects on prostate cancer cells: Comparison with other cancer preventive phytochemicals. Mol. Carcinog. 2011, 51, 244–256. [Google Scholar] [CrossRef]
- Nandini, D.B.; Rao, R.; Deepak, B.S.; Reddy, P. Sulforaphane in broccoli: The green chemoprevention!! Role in cancer prevention and therapy. J. Oral Maxillofac. Pathol. 2020, 24, 405. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.V.; Srivastava, S.K.; Choi, S.; Lew, K.L.; Antosiewicz, J.; Xiao, D.; Zeng, Y.; Watkins, S.C.; Johnson, C.S.; Trump, D.L.; et al. Sulforaphane-induced Cell Death in Human Prostate Cancer Cells Is Initiated by Reactive Oxygen Species. J. Biol. Biol. Chem. Chem. 2005, 280, 19911–19924. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, H.; Guo, C.; Li, J.; Li, M.; Zhao, M.; Fu, Z.; Zhang, Z.; Li, F.; Zhao, X.; et al. Sulforaphane activates CD8+T cells antitumor response through IL-12RB2/MMP3/FasL-induced MDSCs apoptosis’. J. Immunother. Cancer 2024, 12, e007983. [Google Scholar] [CrossRef] [PubMed]
- Ishida, M.; Hara, M.; Fukino, N.; Kakizaki, T.; Morimitsu, Y. Glucosinolate metabolism, functionality and breeding for the improvement of Brassicaceae vegetables. Breed. Sci. 2014, 64, 48–59. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lu, Q.; Li, N.; Xu, M.; Miyamoto, T.; Liu, J. Sulforaphane suppresses metastasis of triple-negative breast cancer cells by targeting the RAF/MEK/ERK pathway. npj Breast Cancer 2022, 8, 40. [Google Scholar] [CrossRef] [PubMed]
- Guan, I.A.; Liu, J.S.T.; Sawyer, R.C.; Li, X.; Jiao, W.; Jiramongkol, Y.; White, M.D.; Hagimola, L.; Passam, F.H.; Tran, D.P.; et al. Integrating Phenotypic and Chemoproteomic Approaches to Identify Covalent Targets of Dietary Electrophiles in Platelets. ACS Cent. Sci. 2024, 10, 344–357. [Google Scholar] [CrossRef] [PubMed]
- Aranaz, P.; Navarro-Herrera, D.; Romo-Hualde, A.; Zabala, M.; López-Yoldi, M.; González-Ferrero, C.; Gil, A.G.; Alfredo Martinez, J.; Vizmanos, J.L.; Milagro, F.I.; et al. Broccoli extract improves high fat diet-induced obesity, hepatic steatosis and glucose intolerance in Wistar rats. J. Funct. Foods 2019, 59, 319–328. [Google Scholar] [CrossRef]
- Syed, R.U.; Moni, S.S.; Break, M.K.B.; Khojali, W.M.A.; Jafar, M.; Alshammari, M.D.; Abdelsalam, K.; Taymour, S.; Alreshidi, K.S.M.; Elhassan Taha, M.M.; et al. Broccoli: A Multi-Faceted Vegetable for Health: An In-Depth Review of Its Nutritional Attributes, Antimicrobial Abilities, and Anti-inflammatory Properties. Antibiotics 2023, 12, 1157. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Xia, Y.; Liu, H.-Y.; Guo, H.; He, X.-Q.; Liu, Y.; Wu, D.-T.; Mai, Y.-H.; Li, H.-B.; Zou, L.; et al. Nutritional values, beneficial effects, and food applications of broccoli (Brassica oleracea var. italica Plenck). Trends Food Sci. Technol. 2022, 119, 288–308. [Google Scholar] [CrossRef]
- Bardelčíková, A.; Šoltys, J.; Mojžiš, J. Oxidative Stress, Inflammation and Colorectal Cancer: An Overview. Antioxidants 2023, 12, 901. [Google Scholar] [CrossRef]
- Tavassolifar, M.j.; Vodjgani, M.; Salehi, Z.; Izad, M. The Influence of Reactive Oxygen Species in the Immune System and Pathogenesis of Multiple Sclerosis. Autoimmune Dis. 2020, 2020, 5793817. [Google Scholar] [CrossRef] [PubMed]
- Bešlo, D.; Golubić, N.; Rastija, V.; Agić, D.; Karnaš, M.; Šubarić, D.; Lučić, B. Antioxidant Activity, Metabolism, and Bioavailability of Polyphenols in the Diet of Animals. Antioxidants 2023, 12, 1141. [Google Scholar] [CrossRef] [PubMed]
- Mei, G.; Chen, A.; Wang, Y.; Li, S.; Wu, M.; Hu, Y.; Liu, X.; Hou, X. A simple and efficient in planta transformation method based on the active regeneration capacity of plants. Plant Commun. 2024, 5, 100822. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Wang, K. Strategies for genotype-flexible plant transformation. Curr. Opin. Biotechnol. 2023, 79, 102848. [Google Scholar] [CrossRef] [PubMed]
- Hwang, H.-H.; Yu, M.; Lai, E.-M. Agrobacterium-Mediated Plant Transformation: Biology and Applications. Arab. Book 2017, 15, e0186. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Srivastava, D.K. Biotechnological advancement in genetic improvement of broccoli (Brassica oleracea L. var. italica), an important vegetable crop. Biotechnol. Lett. 2016, 38, 1049–1063. [Google Scholar] [CrossRef] [PubMed]
- Gao, C. Genome engineering for crop improvement and future agriculture. Cell 2021, 184, 1621–1635. [Google Scholar] [CrossRef] [PubMed]
- Su, W.; Xu, M.; Radani, Y.; Yang, L. Technological Development and Application of Plant Genetic Transformation. Int. J. Mol. Sci. 2023, 24, 10646. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Wang, H. Recent advances of genome editing and related technologies in China. Gene Ther. 2020, 27, 312–320. [Google Scholar] [CrossRef]
- Chamani Mohasses, F.; Mousavi Pakzad, S.M.; Moatamed, E.; Entesari, M.; Bidadi, H.; Molaahmad Nalousi, A.; Jamshidi, S.; Ghareyazie, B.; Mohsenpour, M. Efficient genetic transformation of rice using Agrobacterium with a codon-optimized chromoprotein reporter gene (ChromoP) and introducing an optimized iPCR method for transgene integration site detection. Plant Cell Tissue Organ Cult. (PCTOC) 2023, 156, 5. [Google Scholar] [CrossRef]
- Toki, S.; Hara, N.; Ono, K.; Onodera, H.; Tagiri, A.; Oka, S.; Tanaka, H. Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J. 2006, 47, 969–976. [Google Scholar] [CrossRef]
- Khan, H.; McDonald, M.C.; Williams, S.J.; Solomon, P.S. Assessing the efficacy of CRISPR/Cas9 genome editing in the wheat pathogen Parastagonspora nodorum. Fungal Biol. Biotechnol. 2020, 7, 4. [Google Scholar] [CrossRef] [PubMed]
- Wu, E.; Lenderts, B.; Glassman, K.; Berezowska-Kaniewska, M.; Christensen, H.; Asmus, T.; Zhen, S.; Chu, U.; Cho, M.-J.; Zhao, Z.-Y. Optimized Agrobacterium-mediated sorghum transformation protocol and molecular data of transgenic sorghum plants. Vitr. Cell. Dev. Biol.—Plant 2013, 50, 9–18. [Google Scholar] [CrossRef]
- Kumar, V.; Campbell, L.M.; Rathore, K.S. Rapid recovery- and characterization of transformants following Agrobacterium-mediated T-DNA transfer to sorghum. Plant Cell Tissue Organ Cult. (PCTOC) 2010, 104, 137–146. [Google Scholar] [CrossRef]
- Kumlehn, J.; Serazetdinova, L.; Hensel, G.; Becker, D.; Loerz, H. Genetic transformation of barley (Hordeum vulgare L.) via infection of androgenetic pollen cultures with Agrobacterium tumefaciens. Plant Biotechnol. J. 2006, 4, 251–261. [Google Scholar] [CrossRef]
- Zhangsun, D.; Luo, S.; Chen, R.; Tang, K. Improved Agrobacterium-mediated genetic transformation of GNA transgenic sugarcane. Biologia 2007, 62, 386–393. [Google Scholar] [CrossRef]
- Budeguer, F.; Enrique, R.; Perera, M.F.; Racedo, J.; Castagnaro, A.P.; Noguera, A.S.; Welin, B. Genetic Transformation of Sugarcane, Current Status and Future Prospects. Front. Plant Sci. 2021, 12, 768609. [Google Scholar] [CrossRef]
- Popelka, J.C.; Altpeter, F. Agrobacterium tumefaciens-mediated genetic transformation of rye (Secale cereale L.). Mol. Breed 2003, 11, 203–211. [Google Scholar] [CrossRef]
- Ramadevi, R.; Rao, K.V.; Reddy, V.D. Agrobacterium tumefaciens-mediated genetic transformation and production of stable transgenic pearl millet (Pennisetum glaucum [L.] R. Br.). Vitr. Cell. Dev. Biol.—Plant 2014, 50, 392–400. [Google Scholar] [CrossRef]
- Wang, H.; Zheng, Y.; Zhou, Q.; Li, Y.; Liu, T.; Hou, X. Fast, simple, efficient Agrobacterium rhizogenes-mediated transformation system to non-heading Chinese cabbage with transgenic roots. Hortic. Plant J. 2024, 10, 450–460. [Google Scholar] [CrossRef]
- Li, X.; Li, H.; Zhao, Y.; Zong, P.; Zhan, Z.; Piao, Z. Establishment of A Simple and Efficient Agrobacterium-mediated Genetic Transformation System to Chinese Cabbage (Brassica rapa L. ssp. pekinensis). Hortic. Plant J. 2021, 7, 117–128. [Google Scholar] [CrossRef]
- Huang, W.; Zheng, A.; Huang, H.; Chen, Z.; Ma, J.; Li, X.; Liang, Q.; Li, L.; Liu, R.; Huang, Z.; et al. Effects of sgRNAs, Promoters, and Explants on the Gene Editing Efficiency of the CRISPR/Cas9 System in Chinese Kale. Int. J. Mol. Sci. 2023, 24, 13241. [Google Scholar] [CrossRef]
- Sun, B.; Zheng, A.; Jiang, M.; Xue, S.; Yuan, Q.; Jiang, L.; Chen, Q.; Li, M.; Wang, Y.; Zhang, Y.; et al. CRISPR/Cas9-mediated mutagenesis of homologous genes in Chinese kale. Sci. Rep. 2018, 8, 16786. [Google Scholar] [CrossRef]
- Sheng, X.; Yu, H.; Wang, J.; Shen, Y.; Gu, H. Establishment of a stable, effective and universal genetic transformation technique in the diverse species of Brassica oleracea. Front. Plant Sci. 2022, 13, 1669. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, L.; Li, C.; Yang, Y.; Duan, Y.; Yang, Y.; Sun, X. Establishment of Agrobacterium-mediated genetic transformation and application of CRISPR/Cas9 genome-editing system to Brassica rapa var. rapa. Plant Methods 2022, 18, 98. [Google Scholar] [CrossRef] [PubMed]
- Saini, D.K.; Kaushik, P. Visiting eggplant from a biotechnological perspective: A review. Sci. Hortic. 2019, 253, 327–340. [Google Scholar] [CrossRef]
- Mahto, B.K.; Sharma, P.; Rajam, M.V.; Reddy, P.M.; Dhar-Ray, S. An efficient method for Agrobacterium-mediated genetic transformation of chilli pepper (Capsicum annuum L.). Indian J. Plant Physiol. 2018, 23, 573–581. [Google Scholar] [CrossRef]
- Hashmi, M.H.; Saeed, F.; Demirel, U.; Bakhsh, A. Establishment of highly efficient and reproducible Agrobacterium-mediated transformation system for tomato (Solanum lycopersicum L.). Vitr. Cell. Dev. Biol.—Plant 2022, 58, 1066–1076. [Google Scholar] [CrossRef]
- Sun, S.; Kang, X.-P.; Xing, X.-J.; Xu, X.-Y.; Cheng, J.; Zheng, S.-W.; Xing, G.-M. Agrobacterium-mediated transformation of tomato (Lycopersicon esculentum L. cv. Hezuo 908) with improved efficiency. Biotechnol. Biotechnol. Equip. 2015, 29, 861–868. [Google Scholar] [CrossRef]
- Chen, W.; Punja, Z. Transgenic herbicide- and disease-tolerant carrot (Daucus carota L.) plants obtained through Agrobacterium-mediated transformation. Plant Cell Rep. 2002, 20, 929–935. [Google Scholar] [CrossRef]
- Que, F.; Hou, X.-L.; Wang, G.-L.; Xu, Z.-S.; Tan, G.-F.; Li, T.; Wang, Y.-H.; Khadr, A.; Xiong, A.-S. Advances in research on the carrot, an important root vegetable in the Apiaceae family. Hortic. Res. 2019, 6, 69. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Jiang, J.; Wang, Y. Protoplast fusion for crop improvement and breeding in China. Plant Cell Tissue Organ Cult. (PCTOC) 2012, 112, 131–142. [Google Scholar] [CrossRef]
- Furuta, H.; Shinoyama, H.; Nomura, Y.; Maeda, M.; Makara, K. Production of intergeneric somatic hybrids of chrysanthemum [Dendranthema × grandiflorum (Ramat.) Kitamura] and wormwood (Artemisia sieversiana J. F. Ehrh. ex. Willd) with rust (Puccinia horiana Henning) resistance by electrofusion of protoplasts. Plant Sci. 2004, 166, 695–702. [Google Scholar] [CrossRef]
- Gieniec, M.; Siwek, J.; Oleszkiewicz, T.; Maćkowska, K.; Klimek-Chodacka, M.; Grzebelus, E.; Baranski, R. Real-time detection of somatic hybrid cells during electrofusion of carrot protoplasts with stably labelled mitochondria. Sci. Rep. 2020, 10, 18811. [Google Scholar] [CrossRef] [PubMed]
- Davey, M.R.; Anthony, P.; Power, J.B.; Lowe, K.C. Plant protoplasts: Status and biotechnological perspectives. Biotechnol. Adv. 2005, 23, 131–171. [Google Scholar] [CrossRef] [PubMed]
- Eeckhaut, T.; Lakshmanan, P.S.; Deryckere, D.; Van Bockstaele, E.; Van Huylenbroeck, J. Progress in plant protoplast research. Planta 2013, 238, 991–1003. [Google Scholar] [CrossRef] [PubMed]
- Aviv, D.; Fluhr, R.; Edelman, M.; Galun, E. Progeny analysis of the interspecific somatic hybrids: Nicotiana tabacum (CMS) + Nicotiana sylvestris with respect to nuclear and chloroplast markers. Theor. Appl. Genet. 1980, 56, 145–150. [Google Scholar] [CrossRef]
- Bruznican, S.; Eeckhaut, T.; Van Huylenbroeck, J.; De Keyser, E.; De Clercq, H.; Geelen, D. An asymmetric protoplast fusion and screening method for generating celeriac cybrids. Sci. Rep. 2021, 11, 4553. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.H.; Landgren, M.; Glimelius, K. Transfer of the Brassica tournefortii cytoplasm to B. napus for the production of cytoplasmic male sterile B. napus. Physiol. Plant. 2006, 96, 123–129. [Google Scholar] [CrossRef]
- Liu, F.; Ryschka, U.; Marthe, F.; Klocke, E.; Schumann, G.; Zhao, H. Culture and fusion of pollen protoplasts of Brassica oleracea L. var. italica with haploid mesophyll protoplasts of B. rapa L. ssp. pekinensis. Protoplasma 2007, 231, 89–97. [Google Scholar] [CrossRef]
- Yarrow, S.A.; Burnett, L.A.; Wildeman, R.P.; Kemble, R.J. The transfer of ‘Polima’ cytoplasmic male sterility from oilseed rape (Brassica napus) to broccoli (B. oleracea) by protoplast fusion. Plant Cell Rep. 1990, 9, 185–188. [Google Scholar] [CrossRef]
- Melchers, G.; Mohri, Y.; Watanabe, K.; Wakabayashi, S.; Harada, K. One-step generation of cytoplasmic male sterility by fusion of mitochondrial-inactivated tomato protoplasts with nuclear-inactivated Solanum protoplasts. Proc. Natl. Acad. Sci. USA 1992, 89, 6832–6836. [Google Scholar] [CrossRef]
- Pelletier, G.; Primard, C.; Vedel, F.; Chetrit, P.; Remy, R.; Rousselle; Renard, M. Intergeneric Cytoplasmic Hybridization in Cruciferae by Protoplast Fusion. Mol. Gen. Genet. MGG 1983, 191, 244–250. [Google Scholar] [CrossRef]
- Kisaka, H.; Kisaka, M.; Kanno, A.; Kameya, T. Intergeneric somatic hybridization of rice (Oryza sativa L.) and barley (Hordeum vulgare L.) by protoplast fusion. Plant Cell Rep. 1998, 17, 362–367. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Cui, H.; Zhi, D.; Zhou, C.; Xia, G. Analysis of remote asymmetric somatic hybrids between common wheat and Arabidopsis thaliana. Plant Cell Rep. 2007, 26, 1233–1241. [Google Scholar] [CrossRef]
- Wang, Y.-P.; Sonntag, K.; Rudloff, E.; Chen, J.-M. Intergeneric Somatic Hybridization Between Brassica napus L. and Sinapis alba L. J. Integr. Plant Biol. 2005, 47, 84–91. [Google Scholar] [CrossRef]
- Wu, J.-H.; Ferguson, A.R.; Mooney, P.A. Allotetraploid hybrids produced by protoplast fusion for seedless triploid Citrus breeding. Euphytica 2005, 141, 229–235. [Google Scholar] [CrossRef]
- Cai, X.-D.; Fu, J.; Deng, X.-X.; Guo, W.-W. Production and molecular characterization of potential seedless cybrid plants between pollen sterile Satsuma mandarin and two seedy Citrus cultivars. Plant Cell Tissue Organ Cult. 2007, 90, 275–283. [Google Scholar] [CrossRef]
- Cheng, Y.; Guo, W.; Deng, X. Molecular characterization of cytoplasmic and nuclear genomes in phenotypically abnormal Valencia orange (Citrus sinensis) + Meiwa kumquat (Fortunella crassifolia) intergeneric somatic hybrids. Plant Cell Rep. 2003, 21, 445–451. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.-W.; Grosser, J.W. Somatic hybrid vigor in Citrus: Direct evidence from protoplast fusion of an embryogenic callus line with a transgenic mesophyll parent expressing the GFP gene. Plant Sci. 2005, 168, 1541–1545. [Google Scholar] [CrossRef]
- Li, A.; Jiang, J.; Zhang, Y.; Snowdon, R.J.; Liang, G.; Wang, Y. Molecular and cytological characterization of introgression lines in yellow seed derived from somatic hybrids between Brassica napus and Sinapis alba. Mol. Breed. 2010, 29, 209–219. [Google Scholar] [CrossRef]
- Wang, Y.P.; Sonntag, K.; Rudloff, E. Development of rapeseed with high erucic acid content by asymmetric somatic hybridization between Brassica napus and Crambe abyssinica. Theor. Appl. Genet. 2003, 106, 1147–1155. [Google Scholar] [CrossRef]
- Ananthakrishnan, G.; Ćalović, M.; Serrano, P.; Grosser, J.W. Production of additional allotetraploid somatic hybrids combining mandarings and sweet orange with pre-selected pummelos as potential candidates to replace sour orange rootstock. Vitr. Cell. Dev. Biol.—Plant 2006, 42, 367–371. [Google Scholar] [CrossRef]
- Yoo, S.-D.; Cho, Y.-H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Li, R.; Luo, H.; Wang, Z.; Li, M.-W.; Lam, H.-M.; Huang, C. Protoplasts: Small cells with big roles in plant biology. Trends Plant Sci. 2022, 27, 828–829. [Google Scholar] [CrossRef]
- Reed, K.M.; Bargmann, B.O.R. Protoplast Regeneration and Its Use in New Plant Breeding Technologies. Front. Genome Ed. 2021, 3, 734951. [Google Scholar] [CrossRef] [PubMed]
- Faraco, M.; Di Sansebastiano, G.P.; Spelt, K.; Koes, R.E.; Quattrocchio, F.M. One Protoplast Is Not the Other! Plant Physiol. 2011, 156, 474–478. [Google Scholar] [CrossRef] [PubMed]
- Takebe, I.; Otsuki, Y. Infection of tobacco mesophyll protoplasts by tobacco mosaic virus. Proc. Natl. Acad. Sci. USA 1969, 64, 843–848. [Google Scholar] [CrossRef]
- Jeong, Y.Y.; Lee, H.-Y.; Kim, S.W.; Noh, Y.-S.; Seo, P.J. Optimization of protoplast regeneration in the model plant Arabidopsis thaliana. Plant Methods 2021, 17, 21. [Google Scholar] [CrossRef]
- Abel, S.; Theologis, A. Transient transformation of Arabidopsis leaf protoplasts: A versatile experimental system to study gene expression. Plant J. 2004, 5, 421–427. [Google Scholar] [CrossRef]
- Wu, F.-H.; Shen, S.-C.; Lee, L.-Y.; Lee, S.-H.; Chan, M.-T.; Lin, C.-S. Tape-Arabidopsis Sandwich—A simpler Arabidopsis protoplast isolation method. Plant Methods 2009, 5, 16. [Google Scholar] [CrossRef]
- Kang, G.H.; Kang, B.-C.; Han, J.-S.; Lee, J.M. DNA-free genome editing in tomato protoplasts using CRISPR/Cas9 ribonucleoprotein delivery. Hortic. Environ. Biotechnol. 2023, 65, 131–142. [Google Scholar] [CrossRef]
- Liu, Y.; Andersson, M.; Granell, A.; Cardi, T.; Hofvander, P.; Nicolia, A. Establishment of a DNA-free genome editing and protoplast regeneration method in cultivated tomato (Solanum lycopersicum). Plant Cell Rep. 2022, 41, 1843–1852. [Google Scholar] [CrossRef] [PubMed]
- Stajič, E.; Kunej, U. Optimization of cabbage (Brassica oleracea var. capitata L.) protoplast transformation for genome editing using CRISPR/Cas9. Front. Plant Sci. 2023, 14, 1245433. [Google Scholar] [CrossRef]
- Sun, B.; Zhang, F.; Xiao, N.; Jiang, M.; Yuan, Q.; Xue, S.; Miao, H.; Chen, Q.; Li, M.; Wang, X.; et al. An efficient mesophyll protoplast isolation, purification and PEG-mediated transient gene expression for subcellular localization in Chinese kale. Sci. Hortic. 2018, 241, 187–193. [Google Scholar] [CrossRef]
- Yang, D.; Zhao, Y.; Liu, Y.; Han, F.; Li, Z. A high-efficiency PEG-Ca2+-mediated transient transformation system for broccoli protoplasts. Front. Plant Sci. 2022, 13, 1081321. [Google Scholar] [CrossRef] [PubMed]
- Meyer, C.M.; Goldman, I.L.; Grzebelus, E.; Krysan, P.J. Efficient production of transgene-free, gene-edited carrot plants via protoplast transformation. Plant Cell Rep. 2022, 41, 947–960. [Google Scholar] [CrossRef]
- Simpson, K.; Stange, C. Carrot protoplasts as a suitable method for protein subcellular localization. In Carotenoids: Carotenoid and Apocarotenoid Biosynthesis Metabolic Engineering and Synthetic Biology; Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2022; pp. 273–283. [Google Scholar]
- Gao, L.; Shen, G.; Zhang, L.; Qi, J.; Zhang, C.; Ma, C.; Li, J.; Wang, L.; Malook, S.U.; Wu, J. An efficient system composed of maize protoplast transfection and HPLC–MS for studying the biosynthesis and regulation of maize benzoxazinoids. Plant Methods 2019, 15, 144. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Song, D.; Gao, L.; Ajayo, B.S.; Wang, Y.; Huang, H.; Zhang, J.; Liu, H.; Liu, Y.; Yu, G.; et al. Optimization of isolation and transfection conditions of maize endosperm protoplasts. Plant Methods 2020, 16, 96. [Google Scholar] [CrossRef]
- Gomez-Cano, L.; Yang, F.; Grotewold, E. Isolation and Efficient Maize Protoplast Transformation. Bio-Protoc. 2019, 9, e3346. [Google Scholar] [CrossRef]
- Xiong, L.; Li, C.; Li, H.; Lyu, X.; Zhao, T.; Liu, J.; Zuo, Z.; Liu, B. A transient expression system in soybean mesophyll protoplasts reveals the formation of cytoplasmic GmCRY1 photobody-like structures. Sci. China Life Sci. 2019, 62, 1070–1077. [Google Scholar] [CrossRef] [PubMed]
- Subburaj, S.; Agapito-Tenfen, S.Z. Establishment of targeted mutagenesis in soybean protoplasts using CRISPR/Cas9 RNP delivery via electro–transfection. Front. Plant Sci. 2023, 14, 1255819. [Google Scholar] [CrossRef] [PubMed]
- Hanzawa, Y.; Wu, F. A Simple Method for Isolation of Soybean Protoplasts and Application to Transient Gene Expression Analyses. J. Vis. Exp. 2018, e57258. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Tao, L.; Zeng, L.; Vega-Sanchez, M.E.; Umemura, K.; Wang, G.L. A highly efficient transient protoplast system for analyzing defence gene expression and protein–protein interactions in rice. Mol. Plant Pathol. 2006, 7, 417–427. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Yu, G.; Chen, Z.; Han, J.; Hu, Y.; Wang, K. Optimization of protoplast isolation, transformation and its application in sugarcane (Saccharum spontaneum L). Crop J. 2021, 9, 133–142. [Google Scholar] [CrossRef]
- Wu, S.; Zhu, H.; Liu, J.; Yang, Q.; Shao, X.; Bi, F.; Hu, C.; Huo, H.; Chen, K.; Yi, G. Establishment of a PEG-mediated protoplast transformation system based on DNA and CRISPR/Cas9 ribonucleoprotein complexes for banana. BMC Plant Biol. 2020, 20, 425. [Google Scholar] [CrossRef] [PubMed]
- Barceló, M.; Wallin, A.; Medina, J.J.; Gil-Ariza, D.J.; López-Casado, G.; Juarez, J.; Sánchez-Sevilla, J.F.; López-Encina, C.; López-Aranda, J.M.; Mercado, J.A.; et al. Isolation and culture of strawberry protoplasts and field evaluation of regenerated plants. Sci. Hortic. 2019, 256, 108552. [Google Scholar] [CrossRef]
- Gou, Y.-J.; Li, Y.-L.; Bi, P.-P.; Wang, D.-J.; Ma, Y.-Y.; Hu, Y.; Zhou, H.-C.; Wen, Y.-Q.; Feng, J.-Y. Optimization of the protoplast transient expression system for gene functional studies in strawberry (Fragaria vesca). Plant Cell Tissue Organ Cult. (PCTOC) 2020, 141, 41–53. [Google Scholar] [CrossRef]
- Maddumage, R.; Fung, R.M.; Weir, I.; Ding, H.; Simons, J.L.; Allan, A.C. Efficient transient transformation of suspension culture-derived apple protoplasts. Plant Cell Tissue Organ Cult. 2002, 70, 77–82. [Google Scholar] [CrossRef]
- Altpeter, F.; Springer, N.M.; Bartley, L.E.; Blechl, A.; Brutnell, T.P.; Citovsky, V.; Conrad, L.; Gelvin, S.B.; Jackson, D.; Kausch, A.P.; et al. Advancing Crop Transformation in the Era of Genome Editing. Plant Cell 2016, 28, 1510–1520. [Google Scholar] [CrossRef]
- Bhowmik, P.; Ellison, E.; Polley, B.; Bollina, V.; Kulkarni, M.; Ghanbarnia, K.; Song, H.; Gao, C.; Voytas, D.F.; Kagale, S. Targeted mutagenesis in wheat microspores using CRISPR/Cas9. Sci. Rep. 2018, 8, 6502. [Google Scholar] [CrossRef] [PubMed]
- Yue, J.-J.; Yuan, J.-L.; Wu, F.-H.; Yuan, Y.-H.; Cheng, Q.-W.; Hsu, C.-T.; Lin, C.-S. Protoplasts: From Isolation to CRISPR/Cas Genome Editing Application. Front. Genome Ed. 2021, 3, 717017. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; Prasad, M. Advances in Agrobacterium tumefaciens-mediated genetic transformation of graminaceous crops. Protoplasma 2015, 253, 691–707. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Cong, Y.; Liu, Y.; Wang, T.; Shuai, Q.; Chen, N.; Gai, J.; Li, Y. Optimization of Agrobacterium-Mediated Transformation in Soybean. Front. Plant Sci. 2017, 8, 246. [Google Scholar] [CrossRef] [PubMed]
- Klimek-Chodacka, M.; Kadluczka, D.; Lukasiewicz, A.; Malec-Pala, A.; Baranski, R.; Grzebelus, E. Effective callus induction and plant regeneration in callus and protoplast cultures of Nigella damascena L. Plant Cell Tissue Organ Cult. (PCTOC) 2020, 143, 693–707. [Google Scholar] [CrossRef]
- Poddar, S.; Tanaka, J.; Cate, J.H.D.; Staskawicz, B.; Cho, M.-J. Efficient isolation of protoplasts from rice calli with pause points and its application in transient gene expression and genome editing assays. Plant Methods 2020, 16, 151. [Google Scholar] [CrossRef]
- Ma, C.; Liu, M.; Li, Q.; Si, J.; Ren, X.; Song, H. Efficient BoPDS Gene Editing in Cabbage by the CRISPR/Cas9 System. Hortic. Plant J. 2019, 5, 164–169. [Google Scholar] [CrossRef]
- Zhao, Y.; Yang, D.; Liu, Y.; Han, F.; Li, Z. A highly efficient genetic transformation system for broccoli and subcellular localization. Front. Plant Sci. 2023, 14, 1091588. [Google Scholar] [CrossRef] [PubMed]
- Han, F.; Liu, Y.; Fang, Z.; Yang, L.; Zhuang, M.; Zhang, Y.; Lv, H.; Wang, Y.; Ji, J.; Li, Z. Advances in Genetics and Molecular Breeding of Broccoli. Horticulturae 2021, 7, 280. [Google Scholar] [CrossRef]
- Henzi, M.X.; Christey, M.C.; Mcneil, D.L. Factors that influence Agrobacterium rhizogenes-mediated transformation of broccoli (Brassica oleracea L. var. italica). Plant Cell Rep. 2000, 19, 994–999. [Google Scholar] [CrossRef]
- Metz, T.D.; Dixit, R.; Earle, E.D. Agrobacterium tumefaciens-mediated transformation of broccoli (Brassica oleracea var. italica) and cabbage (B. oleracea var. capitata). Plant Cell Rep 1995, 15, 287–292. [Google Scholar] [PubMed]
- Desneux, N.; Kim, Y.-J.; Lee, J.-H.; Harn, C.H.; Kim, C.-G. Transgenic Cabbage Expressing Cry1Ac1 Does Not Affect the Survival and Growth of the Wolf Spider, Pardosa astrigera L. Koch (Araneae: Lycosidae). Plos ONE 2016, 11, e0153395. [Google Scholar] [CrossRef]
- Chen, G.C.B.; Xu, F.; Lei, J. Development of Adjustable Male Sterile Plant in Chinese Flowering Cabbageby Antisense DAD1 Fragment Transformation. Afr. J. Biotechnol. 2010, 9, 4534–4541. [Google Scholar]
- Li, H.; Zhang, Q.; Li, L.; Yuan, J.; Wang, Y.; Wu, M.; Han, Z.; Liu, M.; Chen, C.; Song, W.; et al. Ectopic Overexpression of bol-miR171b Increases Chlorophyll Content and Results in Sterility in Broccoli (Brassica oleracea L var. italica). J. Agric. Food Chem. 2018, 66, 9588–9597. [Google Scholar] [CrossRef]
- Cao, H.; Liu, R.; Zhang, J.; Liu, Z.; Fan, S.; Yang, G.; Jin, Z.; Pei, Y. Improving sulforaphane content in transgenic broccoli plants by overexpressing MAM1, FMOGS–OX2, and Myrosinase. Plant Cell Tissue Organ Cult. (PCTOC) 2021, 146, 461–471. [Google Scholar] [CrossRef]
- Kim, Y.-C.; Cha, A.; Hussain, M.; Lee, K.; Lee, S. Impact of Agrobacterium-infiltration and transient overexpression of BroMYB28 on glucoraphanin biosynthesis in broccoli leaves. Plant Biotechnol. Rep. 2019, 14, 373–380. [Google Scholar] [CrossRef]
- Li, S.; Zhang, L.; Wang, Y.; Xu, F.; Liu, M.; Lin, P.; Ren, S.; Ma, R.; Guo, Y.-D. Knockdown of a cellulose synthase gene BoiCesA affects the leaf anatomy, cellulose content and salt tolerance in broccoli. Sci. Rep. 2017, 7, 41397. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Jiang, J.-J.; Miao, L.-X.; He, C.-M. Over-expression of a C3H-type zinc finger gene contributes to salt stress tolerance in transgenic broccoli plants. Plant Cell Tissue Organ Cult. (PCTOC) 2017, 130, 239–254. [Google Scholar] [CrossRef]
- Jiang, M.; Miao, L.; Zhang, H.; Zhu, X. Over-Expression of a Transcription Factor Gene BoC3H4 Enhances Salt Stress Tolerance but Reduces Sclerotinia Stem Rot Disease Resistance in Broccoli. J. Plant Growth Regul. 2019, 39, 1162–1176. [Google Scholar] [CrossRef]
- Jiang, M.; Ye, Z.-H.; Zhang, H.-J.; Miao, L.-X. Broccoli Plants Over-expressing an ERF Transcription Factor Gene BoERF1 Facilitates Both Salt Stress and Sclerotinia Stem Rot Resistance. J. Plant Growth Regul. 2018, 38, 1–13. [Google Scholar] [CrossRef]
- Jiang, M.; He, C.-M.; Miao, L.-X.; Zhang, Y.-C. Overexpression of a Broccoli Defensin Gene BoDFN Enhances Downy Mildew Resistance. J. Integr. Agric. 2012, 11, 1137–1144. [Google Scholar] [CrossRef]
- Jiang, M.; Liu, Q.-E.; Liu, Z.-N.; Li, J.-Z.; He, C.-M. Over-expression of a WRKY transcription factor gene BoWRKY6 enhances resistance to downy mildew in transgenic broccoli plants. Australas. Plant Pathol. 2016, 45, 327–334. [Google Scholar] [CrossRef]
- Jiang, M.; Miao, L.-X.; He, C. Overexpression of an Oil Radish Superoxide Dismutase Gene in Broccoli Confers Resistance to Downy Mildew. Plant Mol. Biol. Report. 2012, 30, 966–972. [Google Scholar] [CrossRef]
- Jiang, M.; Jiang, J.J.; He, C.M.; Guan, M. Broccoli plants over-expressing a cytosolic ascorbate peroxidase gene increase resistance to downy mildew and heat stress. J. Plant Pathol. 2016, 98, 413–420. [Google Scholar] [CrossRef]
- Liu, Y.; Wei, M.; Liu, Y.; Fang, Z.; Zhang, Y.; Yang, L.; Lv, H.; Wang, Y.; Ji, J.; Zhang, X.; et al. Functional characterization of BoGL5 by an efficient CRISPR/Cas9 genome editing system in broccoli. Sci. Hortic. 2023, 319, 112136. [Google Scholar] [CrossRef]
- Li, Z.; Song, L.; Liu, Y.; Han, F.; Liu, W. Electrophysiological, Morphologic, and Transcriptomic Profiling of the Ogura-CMS, DGMS and Maintainer Broccoli Lines. Plants 2022, 11, 561. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-Q.; Li, G.-Q.; Yao, X.-Q.; Huang, L.; Wu, X.-Y.; Xie, Z.-J. Characterization of Ogura CMS fertility-restored interspecific hybrids and backcross progenies from crosses between broccoli and rapeseed. Euphytica 2020, 216, 194. [Google Scholar] [CrossRef]
- Huang, J.L.W.; Liu, Y.; Han, F.; Fang, Z.; Yang, L.; Zhuang, M.; Zhang, Y.; Lv, H.; Wang, Y. Creation of Fertility Restorer Materials for Ogura CMS Broccoli and the Study of Its Genetic Background. Acta Hortic. Sin 2022, 49, 533–547. [Google Scholar] [CrossRef]
- Li, Z.; Mei, Y.; Liu, Y.; Fang, Z.; Yang, L.; Zhuang, M.; Zhang, Y.; Lv, H. The evolution of genetic diversity of broccoli cultivars in China since 1980. Sci. Hortic. 2019, 250, 69–80. [Google Scholar] [CrossRef]
- Wu, J.; Guo, W.; Cui, S.; Tang, X.; Zhang, Q.; Lu, W.; Jin, Y.; Zhao, J.; Mao, B.; Chen, W. Broccoli seed extract rich in polysaccharides and glucoraphanin ameliorates DSS-induced colitis via intestinal barrier protection and gut microbiota modulation in mice. J. Sci. Food Agric. 2022, 103, 1749–1760. [Google Scholar] [CrossRef]
- Chen, Z.; Debernardi, J.M.; Dubcovsky, J.; Gallavotti, A. Recent advances in crop transformation technologies. Nat. Plants 2022, 8, 1343–1351. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.P.; Li, Z.; Liu, Y.; Li, L.; Fang, Z.; Yang, L.; Zhuang, M.; Zhang, Y.; Lv, H. Transcriptome reveals the gene expression patterns of sulforaphane metabolism in broccoli florets. Plos ONE 2019, 14, e0213902. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liu, Y.; Yuan, S.; Han, F.; Fang, Z.; Yang, L.; Zhuang, M.; Zhang, Y.; Lv, H.; Wang, Y.; et al. Fine mapping of the major QTLs for biochemical variation of sulforaphane in broccoli florets using a DH population. Sci. Rep. 2021, 11, 9004. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Liu, Y.; Han, F.; Fang, Z.; Yang, L.; Zhuang, M.; Zhang, Y.; Lv, H.; Wang, Y.; Ji, J.; et al. Genetic diversity and population structure analysis of 161 broccoli cultivars based on SNP markers. Hortic. Plant J. 2021, 7, 423–433. [Google Scholar] [CrossRef]
- Huang, X.; Cheng, B.; Wang, Y.; Liu, G.; Hu, L.; Yu, X.; He, H. Effects of fresh-cut and storage on glucosinolates profile using broccoli as a case study. Hortic. Plant J. 2023, 9, 285–292. [Google Scholar] [CrossRef]
- Li, Z.; Liu, Y.; Fang, Z.; Yang, L.; Zhuang, M.; Zhang, Y. Full Length Cloning, Expression and Correlation Analysis of P450 CYP79F1 Gene with Sulforaphane Content in Different Broccoli Organs. Sci. Agric. Sin. 2018, 51, 2357–2367. [Google Scholar] [CrossRef]
- Zhu, B.; Liang, Z.; Zang, Y.; Zhu, Z.; Yang, J. Diversity of glucosinolates among common Brassicaceae vegetables in China. Hortic. Plant J. 2023, 9, 365–380. [Google Scholar] [CrossRef]
- Baskar, V.; Park, S.W. Molecular characterization of BrMYB28 and BrMYB29 paralogous transcription factors involved in the regulation of aliphatic glucosinolate profiles in Brassica rapa ssp. pekinensis. Comptes Rendus Biol. 2015, 338, 434–442. [Google Scholar] [CrossRef]
- Guo, H.; Huang, Z.; Li, M.; Hou, Z. Growth, ionic homeostasis, and physiological responses of cotton under different salt and alkali stresses. Sci. Rep. 2020, 10, 21844. [Google Scholar] [CrossRef]
- Vicente, J.G.; Gunn, N.D.; Bailey, L.; Pink, D.A.C.; Holub, E.B. Genetics of resistance to downy mildew in Brassica oleracea and breeding towards durable disease control for UK vegetable production. Plant Pathol. 2011, 61, 600–609. [Google Scholar] [CrossRef]
- Gupta, M.; Vikram, A.; Bharat, N. Black rot—A devastating disease of crucifers: A review. Agric. Rev. 2013, 34, 269–278. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, Y.; Fang, Z.; Li, Z.; Yang, L.; Zhuang, M.; Zhang, Y.; Lv, H. Comparative Transcriptome Analysis between Broccoli (Brassica oleracea var. italica) and Wild Cabbage (Brassica macrocarpa Guss.) in Response to Plasmodiophora brassicae during Different Infection Stages. Front. Plant Sci. 2016, 7, 1929. [Google Scholar] [CrossRef]
- Ludwig-Müller, J.; Prinsen, E.; Rolfe, S.A.; Scholes, J.D. Metabolism and Plant Hormone Action During Clubroot Disease. J. Plant Growth Regul. 2009, 28, 229–244. [Google Scholar] [CrossRef]
- Piao, Z.; Ramchiary, N.; Lim, Y.P. Genetics of Clubroot Resistance in Brassica Species. J. Plant Growth Regul. 2009, 28, 252–264. [Google Scholar] [CrossRef]
- Ueno, H.; Matsumoto, E.; Aruga, D.; Kitagawa, S.; Matsumura, H.; Hayashida, N. Molecular characterization of the CRa gene conferring clubroot resistance in Brassica rapa. Plant Mol. Biol. 2012, 80, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Zhao, J.Z.; Tang, J.; Shelton, A.; Earle, E. Broccoli plants with pyramided cry1Ac and cry1C Bt genes control diamondback moths resistant to Cry1A and Cry1C proteins. Theor. Appl. Genet. 2002, 105, 258–264. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.-Z.; Cao, J.; Li, Y.; Collins, H.L.; Roush, R.T.; Earle, E.D.; Shelton, A.M. Transgenic plants expressing two Bacillus thuringiensis toxins delay insect resistance evolution. Nat. Biotechnol. 2003, 21, 1493–1497. [Google Scholar] [CrossRef] [PubMed]
- Viswakarma, N.; Bhattacharya, R.C.; Chakrabarty, R.; Bhat, S.R.; Kirti, P.B.; Shastri, N.V.; Chopra, V.L. Insect resistance of transgenic broccoli (‘Pusa Broccoli KTS-1’) expressing a syntheticcryIA(b)gene. J. Hortic. Sci. Biotechnol. 2015, 79, 182–188. [Google Scholar] [CrossRef]
- Kumar, P.; Gambhir, G.; Gaur, A.; Sharma, K.C.; Thakur, A.K.; Srivastava, D.K. Development of transgenic broccoli with cryIAa gene for resistance against diamondback moth (Plutella xylostella). 3 Biotech 2018, 8, 299. [Google Scholar] [CrossRef]
- Lv, Z.; Sun, H.; Du, W.; Li, R.; Mao, H.; Kopittke, P.M. Interaction of different-sized ZnO nanoparticles with maize (Zea mays): Accumulation, biotransformation and phytotoxicity. Sci. Total Environ. 2021, 796, 148927. [Google Scholar] [CrossRef]
- Pagano, L.; Rossi, R.; White, J.C.; Marmiroli, N.; Marmiroli, M. Nanomaterials biotransformation: In planta mechanisms of action. Environ. Pollut. 2023, 318, 120834. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Ying, Y.; Ping, J. Recent Advances in Plant Nanoscience. Adv. Sci. 2021, 9, 2103414. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Meng, Z.; Wang, Y.; Chen, W.; Sun, C.; Cui, B.; Cui, J.; Yu, M.; Zeng, Z.; Guo, S.; et al. Pollen magnetofection for genetic modification with magnetic nanoparticles as gene carriers. Nat. Plants 2017, 3, 956–964. [Google Scholar] [CrossRef] [PubMed]
- Sheng, X.; Zhao, Z.; Yu, H.; Wang, J.; Gu, H. Rapid alterations of DNA sequence and cytosine methylation induced by somatic hybridization between Brassica oleracea L. var. italica and Brassica nigra (L.) Koch. Plant Cell Tissue Organ Cult. (PCTOC) 2013, 115, 395–405. [Google Scholar] [CrossRef]
- Lian, Y.-J.; Lin, G.-Z.; Zhao, X.-M.; Lim, H.-T. Production and genetic characterization of somatic hybrids between leaf mustard (Brassica juncea) and broccoli (Brassica oleracea). Vitr. Cell. Dev. Biol.—Plant 2011, 47, 289–296. [Google Scholar] [CrossRef]
- Kim, Y.-C.; Ahn, W.S.; Cha, A.; Jie, E.Y.; Kim, S.W.; Hwang, B.-H.; Lee, S. Development of glucoraphanin-rich broccoli (Brassica oleracea var. italica) by CRISPR/Cas9-mediated DNA-free BolMYB28 editing. Plant Biotechnol. Rep. 2022, 16, 123–132. [Google Scholar] [CrossRef]
- Wiktorek-Smagur, A.; Hnatuszko-Konka, K.; Kononowicz, A.K. Flower bud dipping or vacuum infiltration—two methods of Arabidopsis thaliana transformation. Russ. J. Plant Physiol. 2009, 56, 560–568. [Google Scholar] [CrossRef]
- Christian, M.; Cermak, T.; Doyle, E.L.; Schmidt, C.; Zhang, F.; Hummel, A.; Bogdanove, A.J.; Voytas, D.F. Targeting DNA Double-Strand Breaks with TAL Effector Nucleases. Genetics 2010, 186, 757–761. [Google Scholar] [CrossRef]
- Gaj, T.; Gersbach, C.A.; Barbas, C.F. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013, 31, 397–405. [Google Scholar] [CrossRef]
- Ahmar, S.; Hensel, G.; Gruszka, D. CRISPR/Cas9-mediated genome editing techniques and new breeding strategies in cereals—Current status, improvements, and perspectives. Biotechnol. Adv. 2023, 69, 108248. [Google Scholar] [CrossRef]
- Cai, Y.; Chen, L.; Hou, W. Genome Editing Technologies Accelerate Innovation in Soybean Breeding. Agronomy 2023, 13, 2045. [Google Scholar] [CrossRef]
- Erdoğan, İ.; Cevher-Keskin, B.; Bilir, Ö.; Hong, Y.; Tör, M. Recent Developments in CRISPR/Cas9 Genome-Editing Technology Related to Plant Disease Resistance and Abiotic Stress Tolerance. Biology 2023, 12, 1037. [Google Scholar] [CrossRef]
- Bansal, K.C.; Roy, S.; Ghoshal, B. Genome Editing Technologies for Efficient Use of Plant Genetic Resources. Indian J. Plant Genet. Resour. 2022, 35, 95–99. [Google Scholar] [CrossRef]
- Mali, P.; Esvelt, K.M.; Church, G.M. Cas9 as a versatile tool for engineering biology. Nat. Methods 2013, 10, 957–963. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, X.; Liu, H.; Li, Z. Highly efficient DNA-free plant genome editing using virally delivered CRISPR–Cas9. Nat. Plants 2020, 6, 773–779. [Google Scholar] [CrossRef] [PubMed]
- Neequaye, M.; Stavnstrup, S.; Harwood, W.; Lawrenson, T.; Hundleby, P.; Irwin, J.; Troncoso-Rey, P.; Saha, S.; Traka, M.H.; Mithen, R.; et al. CRISPR-Cas9-Mediated Gene Editing of MYB28 Genes Impair Glucoraphanin Accumulation of Brassica oleracea in the Field. CRISPR J. 2021, 4, 416–426. [Google Scholar] [CrossRef]
- Lv, Z.; Jiang, R.; Chen, J.; Chen, W. Nanoparticle-mediated gene transformation strategies for plant genetic engineering. Plant J. 2020, 104, 880–891. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Xu, C.; Li, T.; Ma, H.; Gong, J.; Li, X.; Sun, X.; Hu, X. Application of Nanotechnology in Plant Genetic Engineering. Int. J. Mol. Sci. 2023, 24, 14836. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Q.; Chen, Q.-J. Agrobacterium-mediated delivery of CRISPR/Cas reagents for genome editing in plants enters an era of ternary vector systems. Sci. China Life Sci. 2020, 63, 1491–1498. [Google Scholar] [CrossRef]
- Anand, A.; Bass, S.H.; Wu, E.; Wang, N.; McBride, K.E.; Annaluru, N.; Miller, M.; Hua, M.; Jones, T.J. An improved ternary vector system for Agrobacterium-mediated rapid maize transformation. Plant Mol. Biol. 2018, 97, 187–200. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, Y.; Lu, M.-H.; Chai, Y.-P.; Jiang, Y.-Y.; Zhou, Y.; Wang, X.-C.; Chen, Q.-J. A Novel Ternary Vector System United with Morphogenic Genes Enhances CRISPR/Cas Delivery in Maize. Plant Physiol. 2019, 181, 1441–1448. [Google Scholar] [CrossRef] [PubMed]
| Gene Transferred | Exosome Type | Expression Vector Types | Functional Information | References | |||
|---|---|---|---|---|---|---|---|
| Seminal leaf | Hypocotyls | CRISPR- Cas9 | Over- expressing | RNAi | |||
| BoiDAD1F | √ | √ | recoverable male sterility | [114] | |||
| Bol- miR171b | √ | √ | nearly completely male sterile and increased the chlorophyll content | [115] | |||
| BoMAM1 | √ | √ | led to an increase in SF content | [116] | |||
| BoFMOGS-OX2 | √ | √ | led to an increase in SF content | [116] | |||
| BoMyrosinase | √ | √ | led to an increase in SF content | [116] | |||
| BroMYB28 | √ | √ | increased glucoraphanin content | [117] | |||
| BoiCesA | √ | √ | enhanced salt tolerance; dwarf and smaller leaves | [118] | |||
| BoC3H | √ | √ | enhanced salt stress tolerance | [119] | |||
| BoC3H4 | √ | √ | enhanced salt stress tolerance | [120] | |||
| BoERF1 | √ | √ | enhanced salt stress tolerance; enhanced resistance to Sclerotinia stem rot | [121] | |||
| BoDFN | √ | √ | downy mildew resistance | [122] | |||
| BoWRKY6 | √ | √ | downy mildew resistance | [123] | |||
| RsrSOD | √ | √ | downy mildew resistance | [124] | |||
| BoAPX | √ | √ | enhanced resistance to downy mildew enhanced tolerance to heat stress | [125] | |||
| BoGL5 | √ | √ | mutants lacked cuticular waxes | [126] | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Meng, S.; Liu, Y.; Han, F.; Xu, T.; Zhao, Z.; Li, Z. Advances in and Perspectives on Transgenic Technology and CRISPR-Cas9 Gene Editing in Broccoli. Genes 2024, 15, 668. https://doi.org/10.3390/genes15060668
Zhang L, Meng S, Liu Y, Han F, Xu T, Zhao Z, Li Z. Advances in and Perspectives on Transgenic Technology and CRISPR-Cas9 Gene Editing in Broccoli. Genes. 2024; 15(6):668. https://doi.org/10.3390/genes15060668
Chicago/Turabian StyleZhang, Li, Sufang Meng, Yumei Liu, Fengqing Han, Tiemin Xu, Zhiwei Zhao, and Zhansheng Li. 2024. "Advances in and Perspectives on Transgenic Technology and CRISPR-Cas9 Gene Editing in Broccoli" Genes 15, no. 6: 668. https://doi.org/10.3390/genes15060668







