Abstract
Bones and teeth represent a common finding in ancient DNA studies and in forensic casework, even after a long burial. Genetic typing is the gold standard for the personal identification of skeletal remains, but there are two main factors involved in the successful DNA typing of such samples: (1) the set-up of an efficient DNA extraction method; (2) the identification of the most suitable skeletal element for the downstream genetic analyses. In this paper, a protocol based on the processing of 0.5 g of bone powder decalcified using Na2EDTA proved to be suitable for a semi-automated DNA extraction workflow using the Maxwell® FSC DNA IQ™ Casework Kit (Promega, Madison, WI, USA). The performance of this method in terms of DNA recovery and quality was compared with a full demineralisation extraction protocol based on Qiagen technology and kits. No statistically significant differences were scored according to the DNA recovery and DNA degradation index (p-values ≥ 0.176; r ≥ 0.907). This new DNA extraction protocol was applied to 88 bone samples (41 femurs, 19 petrous bones, 12 metacarpals and 16 molars) allegedly belonging to 27 World War II Italian soldiers found in a mass grave on the isle of Cres (Croatia). The results of the qPCR performed by the Quantifiler Human DNA Quantification kit showed values above the lowest Limit of Quantification (lLOQ; 23 pg/µL) for all petrous bones, whereas other bone types showed, in most cases, lower amounts of DNA. Replicate STR-CE analyses showed successful typing (that is, >12 markers) in all tests on the petrous bones, followed by the metacarpals (83.3%), femurs (52.2%) and teeth (20.0%). Full profiles (22/22 autosomal markers) were achieved mainly in the petrous bones (84.2%), followed by the metacarpals (41.7%). Stochastic amplification artefacts such as drop-outs or drop-ins occurred with a frequency of 1.9% in the petrous bones, whereas they were higher when the DNA recovered from other bone elements was amplified (up to 13.9% in the femurs). Overall, the results of this study confirm that petrous bone outperforms other bone elements in terms of the quantity and quality of the recovered DNA; for this reason, if available, it should always be preferred for genetic testing. In addition, our results highlight the need for accurate planning of the DVI operation, which should be carried out by a multi-disciplinary team, and the tricky issue of identifying other suitable skeletal elements for genetic testing. Overall, the results presented in this paper support the need to adopt preanalytical strategies positively related to the successful genetic testing of aged skeletal remains in order to reduce costs and the time of analysis.
1. Introduction
Bones and teeth are the most resistant components in the human body, and they represent the usual finding even after a long burial. Genetic typing is the gold standard for personal identification of skeletal remains [1,2]. Furthermore, considering that genetic testing of skeletal elements is expensive and time consuming, it is strongly recommended to select suitable bone elements [3]. In fact, assuming the absence of PCR inhibitors such as humic acids [3] in DNA extracts, the success of DNA typing depends mainly on the amount and the quality of the genetic material available [3,4,5,6,7,8,9,10,11].
Bone taphonomy is a complex process in which both the type of skeletal elements chosen and environmental factors play key roles [3,12]. Although no strict correlation between bone tissue preservation and DNA preservation has been found so far, it is commonly known that skeletal elements from different anatomical regions or even different portions of the same bone element may produce different results [3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21]. The reasons underlying different outcomes have been evaluated in a few studies and indicate that bone density could be a protective factor in DNA integrity [22]. It is also well established that the bone element offering the highest DNA yields and quality is the temporal bone, particularly the inner ear part of the petrous bone [15,16,17,18,19,20,21,23,24]. When the petrous bone is not available, other skeletal elements, such as long bones (mainly femurs and tibias) and intact teeth, are to be preferred for genetic testing [3,19]. More recently, metacarpal bones [21] and vertebrae [13] were described to provide good chances of typing results in forensics. In order to confirm the usefulness of short bone elements, these findings should be evaluated in larger sets of bones buried in different environmental conditions. A standard forensic approach is to collect as many bones as possible from the same skeleton [4,5,6,7,8,9,10,11,14]. Eventually, case-to-case strategies need to be adopted in relation to the number of skeletons found, the post mortem interval, the feasibility of transporting all skeletal remains, the facilities available in the laboratory for storing the remains, etc.
Another crucial factor for successful typing is the DNA extraction protocol. As recently reviewed [14], several DNA extraction protocols were developed in the last decade with silica-based or magnetic silica-based bead procedures replacing the traditional phenol/chloroform organic purification. Moreover, even if several different approaches have been proposed [14,25], it is generally accepted in forensics that Na2EDTA-decalcified bone powder provides higher yields of DNA than the un-decalcified one [3,14]. Recently, commercial kits specifically designed for DNA extraction from 50 to 150 mg of bone powder have been available to scientists and used in forensic casework analyses [14,26,27,28,29,30]. However, when challenging bone samples are processed, larger amounts of bone powder may be required for a successful PCR-CE-based typing. In these cases, it may also be necessary to plan alternative strategies according to the laboratory’s instrumentations and the number of samples to be processed [3,14].
Recently, we were involved in a forensic investigation of the skeletal remains found in a Second World War (WWII) mass grave which, according to historical records, belonged to 27 male Italian soldiers. To process the request promptly, it was decided to move from the manual preparation of the samples [31] to the new semi-automated workflow described below. In addition, as this method was applied to a set (n = 88) of samples, the preliminary results of the DNA typing are illustrated.
2. Materials and Methods
This paper describes the modifications introduced to the standard protocol by increasing to 0.5 g the amount of bone powder processed using the Maxwell® FSC DNA IQ™ Casework Kit (Promega, Madison, WI, USA). The experiments were performed in one laboratory (Lab A), and the resulting DNA outcomes were compared to those made with a highly performing DNA extraction method [32] set up in another laboratory (Lab B) also involved in the present study. The protocol of Lab B was based on the DNA extraction from the same amount of bone powder (500 mg) using Qiagen technology and kits [13,21,33,34].
Details on the optimization of the proposed protocol are reported in Appendix A.
2.1. Bone Samples
The skeletons were exhumed in May 2019 from a mass grave on the island of Cres (Croatia). According to historical records, the skeletal remains belonged to 27 Italian soldiers. No clothes or uniforms were found, confirming the information on record which reported the soldiers to have been executed naked on April 21, 1945. The mass grave was located at sea level, and the remains were buried about 1–2 m under the ground surface. The island of Cres is located in the north-east Adriatic Sea (44°42′ N; 14°24′ E), and the calcareous soil facilitates the flow of rainwater. The climate is mild, with average temperatures ranging from 7.9 °C in winter to 23.1 °C in summer; the rainfall is 838 mm/year.
Twenty-seven metal boxes were handed over to the Italian government on 13 November 2019. The anthropological and medico-legal examination performed in 2022 by the University of Bari (Italy) showed that the skeletal remains belonged to more than 27 subjects because 29 right human femurs were scored. The skulls were always fractured with disarticulation of the temporal bones; in addition, in a few boxes, the bones appeared commingled, i.e., belonging to more than one subject (paper in preparation). For the genetic testing, bones and teeth were sampled for a total of 341 specimens. Strict precautions to prevent contamination [32,35,36,37] were adopted from this step only. The specimens were then delivered to the University of Trieste (Italy), where they were stored at room temperature, in the dark, until molecular analysis.
For the present study, all the right femurs (n = 29) and all the right petrous bones (n = 19) were selected, as well as 12 left femurs, 12 metacarpals and 16 molar teeth (with no sign of caries), for a total of 88 samples (see Table A4 of Appendix B). The criterion adopted was to analyse at least two samples for each skeleton. Following the indication of previous studies, we used the bone type tissues that offered the best chances of genetic typing, i.e., the compact portion of the diaphysis for the femurs [33], the compact portion of the epiphysis for metacarpals [21] and the inner ear part for petrous bones [20]. Lastly, given that there is no DNA in enamel, the latter was removed from the 12 molars selected, and the remaining entire teeth were then used for DNA extraction [14].
2.2. Bone Cleaning and Pulverisation
All the procedures described in this paragraph were performed in a room designed exclusively for processing old skeletal remains. To remove soil, the surfaces of the bones and teeth were cleaned mechanically using brushes and rotary sanding tools (for petrous bones, further mechanical cleaning was conducted after isolation of the inner part). To remove exogenous DNA contamination [3,14], the tooth or approximately 1.5 g of each bone sample was incubated in 0.5% sodium hypochlorite for 4 min with gentle agitation, washed in sterile bi-distilled water three times and dried at room temperature overnight. In addition, the surfaces of the samples were UV-radiated for 5 min before pulverisation.
Bones and teeth were pulverised at 30 Hz for 1–2 min [32] using a MM 400 Planetary Ball Mills (Retsch, Haan, Germany) equipped with metal grinding vials of 25 mL and metal balls with Ø of 16 mm (Verder, Castleford, UK). Liquid nitrogen was used to prevent the heating of the samples [32]. The 88 powder samples were stored in 50 mL Falcon tubes at room temperature in the dark until molecular analysis.
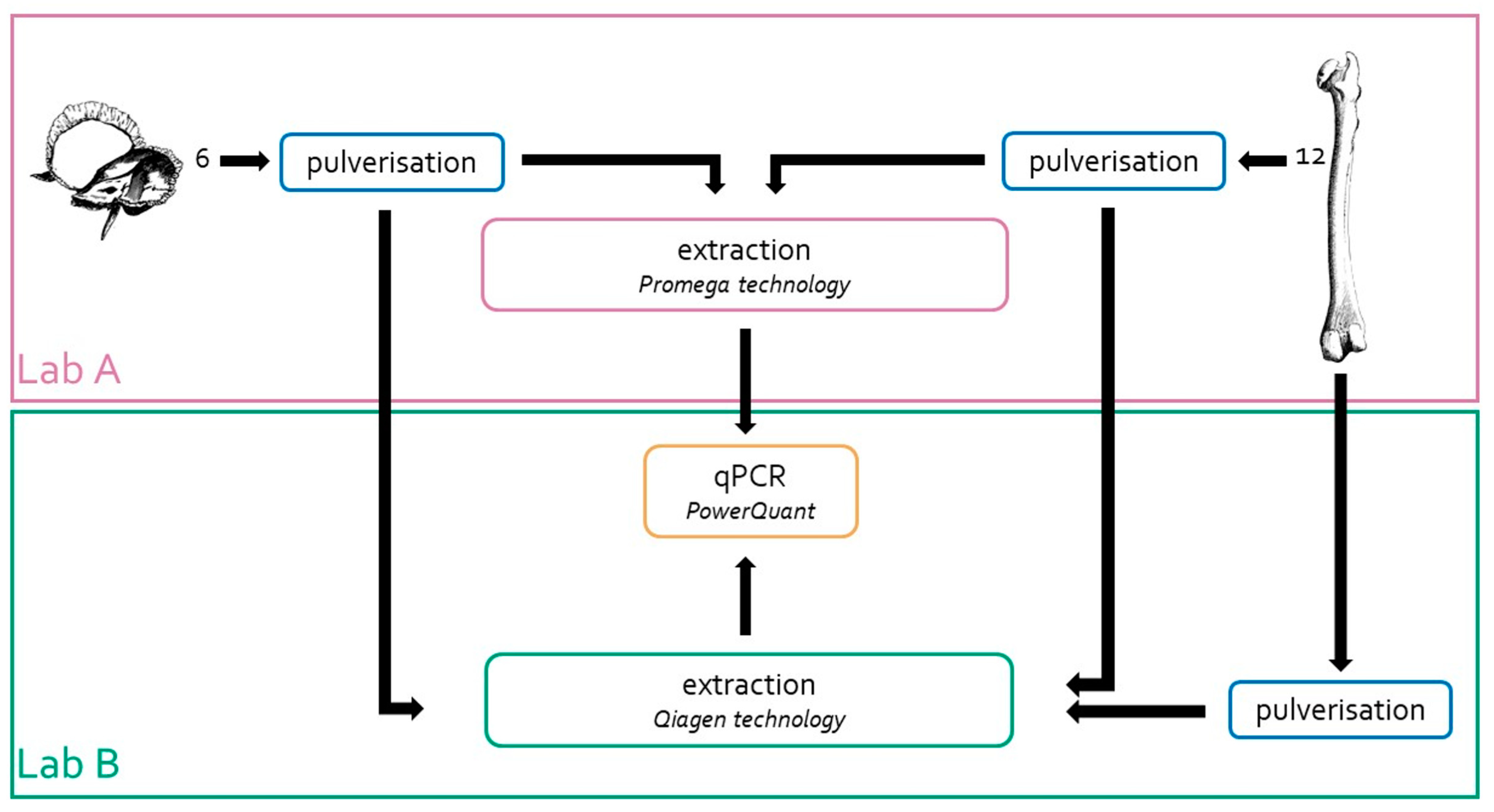
Bone fragments from the 12 left femurs were collected and sent to Lab B for bone pulverisation and DNA extraction, according to the workflow shown in Figure 1. For pulverisation, Lab B used a Bead Beater MillMix 20 (Tehtica-Domel, Železniki, Slovenia) equipped with metal grinding vials of 25 mL and metal balls with Ø of 20 mm (Tehtica-Domel, Livonia, MI, USA); 30 Hz for 1–2 min was applied [32]. After pulverisation, the samples were processed immediately.
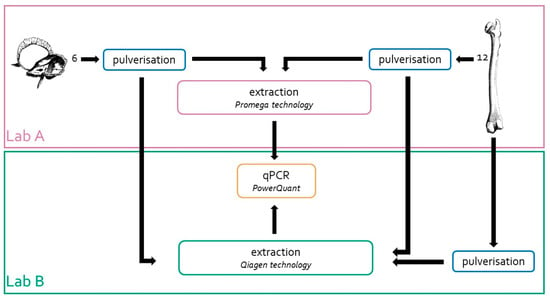
Figure 1.
Strategy adopted to assess the efficiency of the extraction method developed in Lab A. The same powders from eighteen bone samples (6 from petrous bone and 12 from femur) were extracted in two different laboratories (Lab A and Lab. B). In addition, 12 femur samples were pulverised and extracted in Lab B. DNA quantification of all these samples was carried out only in Lab B by using the PowerQuant kit.
2.3. Bone Decalcification and DNA Extraction
The protocol described below was applied to all 88 powder samples. In detail, 0.5 g of each powder sample was decalcified by mixing it with 15 mL of Na2EDTA pH 8.0 in 50 mL Falcon tubes at 40 °C with shaking at 850 rpm overnight. After centrifugation (2800 rpm for 15 min) and two washes with 10 mL of water, the pellet was resuspended by adding 460 µL of extraction buffer (1.2% SDS, 10 mM Tris pH 8.0, 10 mM Na2EDTA pH 8.0 and 100 mM NaCl), 40 µL of 1 M DTT and 30 µL Proteinase K (20 mg/mL). After incubation overnight at 37 °C at 850 rpm, the samples were added with 40 µL Proteinase K (20 mg/mL) and incubated at 56 °C for 4 h at 850 rpm. At the end of this second incubation, the samples were transferred into 1.5 mL Eppendorf tubes and centrifuged at 14,000 rpm for 3 min. An amount of 800 µL of the sample was mixed with 400 µL of the Lysis Buffer (LB) provided with the Maxwell® FSC DNA IQ™ Casework Kit (Promega) and vortexed for ten seconds. The whole sample (1.2 mL, twice the volume recommended in the protocol) was loaded into the cartridge and processed in a Maxwell RCS TM441 (Promega) apparatus following the recommended procedures. The DNA IQ Casework protocol, freely available at www.promega.com, was followed. DNA was eluted in a final volume of 60 microliters. Negative extraction controls (NECs) were processed every 6–12 bone samples. To assess the procedure repeatability, the bone powder from six random petrous bones was used for DNA extraction by a different operator three months after storage at room temperature in the dark.
DNA extractions were also carried out in Lab B using the same amount of bone powder (0.5 g) and the Qiagen technology and protocols described by Zupanič Pajnič [32]. At the end, 18 bone powders prepared in Lab A and 12 bone powders prepared in Lab B were used to compare the DNA extraction efficiencies, as shown in Figure 1.
2.4. DNA Quantification
The DNA samples were quantified in duplicate by qPCR. Lab A used the Quantifiler Human DNA Quantification kit (ThermoFischer Scientifics; TFS, Waltham, MA, USA) following the conditions detailed in ref. [31]. This kit allows the simultaneous detection of a 62 bp long single-copy target within the telomerase reverse transcriptase gene (hTERT) and of an IPC (Internal Positive Control). The LOQ (Limit of Quantification) ranged from 0.023 ng/µL to 50 ng/µL; the LOD (Limit of Detection) was set previously at 0.001 ng/µL [31]. Lab B used the PowerQuant kit (Promega) following the conditions detailed in ref. [33]. This kit allows the simultaneous amplification of four targets: three multi-copied targets, that is, the Auto (84 bp long), the Y-specific (134 bp long) and the Deg (294 bp long), and the IPC. The LOQ ranged from 0.0032 ng/µL to 50 ng/µL both for the Auto and the Y target, whereas the lowest LOQ for the Deg was 0.0005 ng/µL; the LOD was set previously at 0.0001 ng/µL for all quantification targets [33].
2.5. DNA Typing
Only samples with detectable amounts of DNA in at least one of the two assays with Quantifiler were used for genotyping. This resulted in the genotyping of 52 out of 88 samples by the employment of the PowerPlex Fusion (Promega) kit, which simultaneously amplifies 22 autosomal STR markers and 2 Y-specific targets (Amelogenin and DYS391). In detail, all 19 samples from the petrous bones, 11 samples from the right femurs, 6 from the left femurs, 11 from the metacarpals and 5 samples from the teeth were amplified in the recommended final volume of 25 µL. A total of 30 PCR cycles were run for the samples with 0.5–1.0 ng of template, whereas 32 cycles were run for the samples with less than 0.5 ng of DNA (in those cases, 15 µL of DNA extract, i.e., the maximum volume allowed was loaded into the PCR tube). Negative and positive PCR controls as well NECs were analysed. The samples were then run through a 310 ABI automatic DNA sequencer (Applied Biosystems, Waltham, MA, USA), and the resulting data were analysed with the GeneMapperID® ver 3.2.1 software (Applied Biosystem). Analytical and stochastic thresholds were set at 50 and 150 rfu, respectively. After the first round of amplification, only samples with at least twelve autosomal STR markers successfully amplified underwent duplicate tests (this procedure was not performed for profiles matching other profiles already loaded in the database). Thereafter, only samples for which it was possible to generate “consensus” profiles [38] for at least 12 autosomal STR markers were considered “suitable for personal identification” [8] and classified as full (22/22 autosomal markers) and partial profiles (≥12 autosomal markers), respectively. Stochastic amplification artefacts such as allele drop-out and/or allele drop-in were scored by cross-checking the results of the replicated amplifications.
2.6. Data Analysis
Statistical analyses were carried out with the Stata/SE 16.0 package (StataCorp, College Station, TX, USA) and a t-test or ANOVA (when indicated) was used for calculation. Significance was assumed for a p-value < 0.05.
2.7. Exclusion Database
All the individuals involved in any step of the molecular analysis were genotyped following standard procedure. Informed consent was acquired before saliva sampling.
3. Results
3.1. Evaluation of the Extraction Method
The new protocol includes sample extraction through two incubation steps (at 37 °C and 56 °C) followed by automated DNA purification using the Maxwell RCS TM441 apparatus. As reported in Appendix A, a volume double the one suggested in the kit user’s manual (www.promega.com) (accessed on 8 May 2022) (1.2 mL, which is 0.8 mL of sample added to 0.4 mL of Lysis Buffer provided in the kit) was loaded into the Maxwell® FSC DNA IQ™ Casework Kit cartridge. Even if this modification implies a lower % recovery (about 20–25%) than when following the recommended procedure, the total recovery of DNA is higher (up to 50–55%).
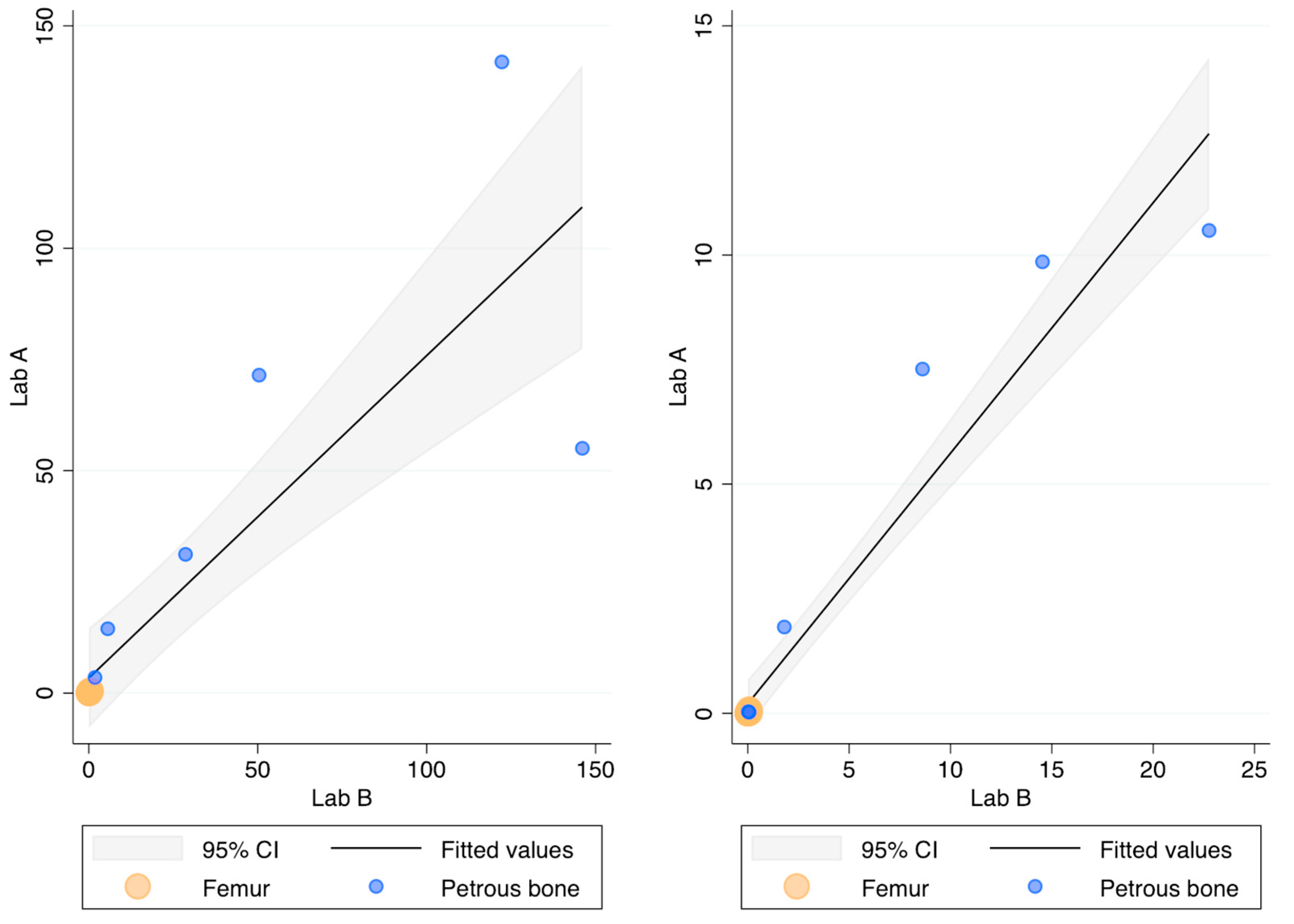
The recovery (ng DNA/g bone) of this DNA extraction method was compared with the protocol routinely employed in Lab B [32] by processing the same 18 bone powders. Figure 2 compares the recovery of the two methods as assessed by the 84 bp long Auto probe of the PowerQuant kit. No statistically significant difference was found (p-value = 0.709; r = 0.912). It has to be noted, however, that high amounts of DNA were not efficiently recovered in Lab A, likely due to the saturation of the capacity of the magnetic beads to bind the DNA (this outcome was observed even during the optimisation of the protocol; see Table A3).
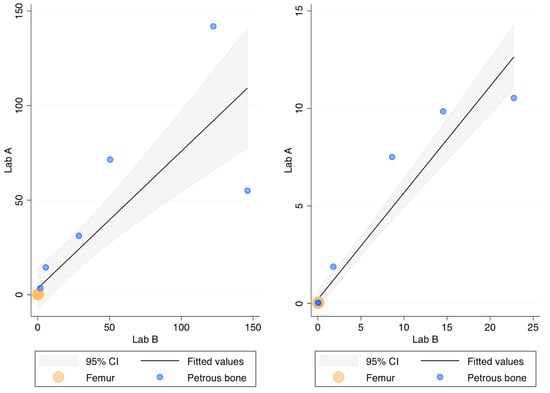
Figure 2.
DNA recovery as assessed by the Auto probe (left) and the Deg probe (right) of the PowerQuant kit; x-axis: ng DNA/g bone in Lab B; y-axis: ng DNA/g bone in Lab A.
The 294 bp long Deg probe showed no Cq values in two and five femur samples extracted in Lab A in Lab B, respectively. As shown in Figure 2, however, the quantification with the Deg probe did not show any difference (p-value = 0.176 r = 0.907) between the two laboratories. The Auto/Deg ratio, an indicator of the degradation extent, ranged from 5.5 to 418 and from 6.4 to 311 in Lab A and Lab B, respectively, but no statistically significant differences were observed between the samples extracted in the two laboratories (p-value = 0.548; r = 0.958). Finally, no differences were found for the IPC values (p-value = 0.754), showing that both protocols efficiently removed the PCR inhibitors.
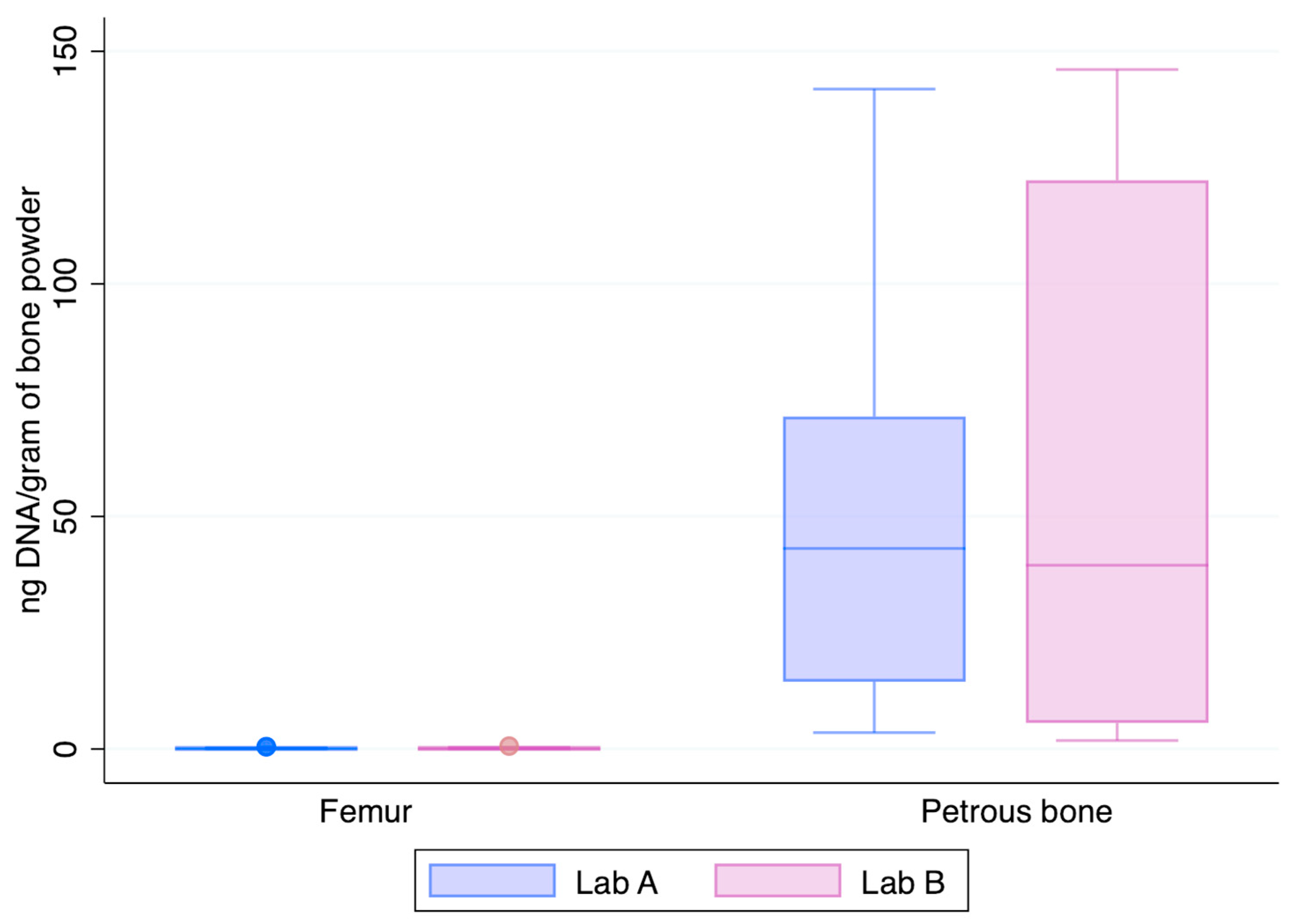
Figure 3 shows the normalised yields of DNA recovered from the two skeletal elements chosen at this step of the study, the femur and the petrous bone. The petrous bones yielded—on average—about 182-fold higher amounts of DNA than the femurs (median values: 52.9 ng vs. 0.143 ng and 59.1 ng vs. 0.150 ng in Lab A and Lab B, respectively). Higher recoveries from petrous bones are perfectly in agreement with previous data showing that those skeletal elements represent the best source of DNA from human bones [3,14], likely due to their intrinsic anatomical [23] and physiological features [22].
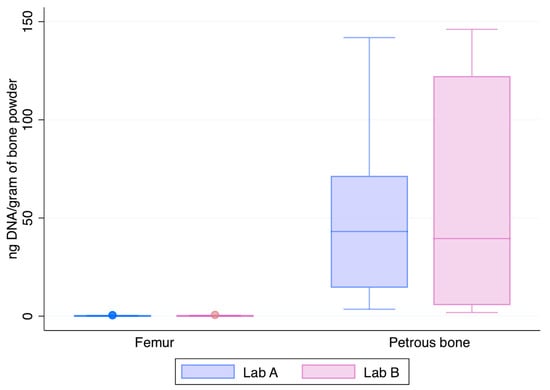
Figure 3.
DNA recovery as assessed by the Auto probe of the PowerQuant kit from the 12 femurs and 6 petrous bones (x-axis); y-axis: nanograms of DNA/gram of bone powder.
Since the pulverisation procedure, mainly the fineness of the bone powder [27] and the heat developed during the grinding procedure [35,39], can also affect DNA recovery, 12 femur fragments were pulverised and extracted in Lab B, as shown in Figure 1. The comparison of the DNA recovery among samples processed in different laboratories showed a p-value of 0.666, as assessed by the ANOVA test. This result rules out pulverisation in Lab A as a procedure capable of interfering significantly with efficient DNA recoveries, which leads to the conclusion that the femurs found in the mass grave actually contained very low amounts of DNA (on average, no more than 0.3 ng/g of bone powder). As reported in previous work, up to 3.2 nanograms of DNA/gram of bone powder was recovered, on average, from a set of 69 femurs found in a different WWII mass grave [33].
Lastly, two operators extracted the same six powders from petrous bones in Lab A, at different times. Although the number of samples is small, and the Quantifiler kit provided data only on quantifying a 62 bp long target, good repeatability (p-value = 0.291; r = 0.902) was found over time (three months of storage at room temperature in the dark). The results of the recoveries (nanograms of DNA/gram of bone powder) are reported in Figure A1.
PCR inhibition was never detected when the Cqs of the IPC probes of the two kits were evaluated.
3.2. DNA Quantification of the Bone Samples
The Quantifiler (TFS) kit was used to assess the DNA amount recovered from the 88 bone samples extracted with the method described in the present study (Lab A). The results are summarised in Table 1.

Table 1.
qPCR and STR typing results from different bone types. n: number of DNA samples; >LOD: percentage of qPCR tests above the Limit of Detection; lLOQ: percentage of qPCR tests above the lowest Limit of Quantification; pg/µL: average (±standard deviation) of DNA concentration as assessed by the Quantifiler kit; in brackets, the median (in bold), the minimum and the maximum values; STR typing: number of PCR tests with successful STR typing (that is, ≥12 markers) out of the total number of PCR tests (the percentage is in the bracket); markers: median number of markers scored in the partial profiles; S.A.: percentage of stochastic PCR artefacts (drop-outs and/or drop-ins) in the replicates; n.a.: not applicable.
The petrous bone always provided quantification values higher than the lowest LOQ (23 pg/µL), with a mean amount of 440 pg/µL. Out of the remaining bone types, only the metacarpal and the femur showed values within the range of the LOQ (in 16.6% and 3.4% of the tests, respectively). Detectable levels of DNA (that is, LOD ≥ 1 pg/µL) were, however, found in all bone types, with a frequency ranging from 15.6% (tooth) to 58.3% (metacarpals). No inhibition was detected, as shown by the Cq of the IPC [40,41].
3.3. Genetic Typing of the Challenging Bone Samples
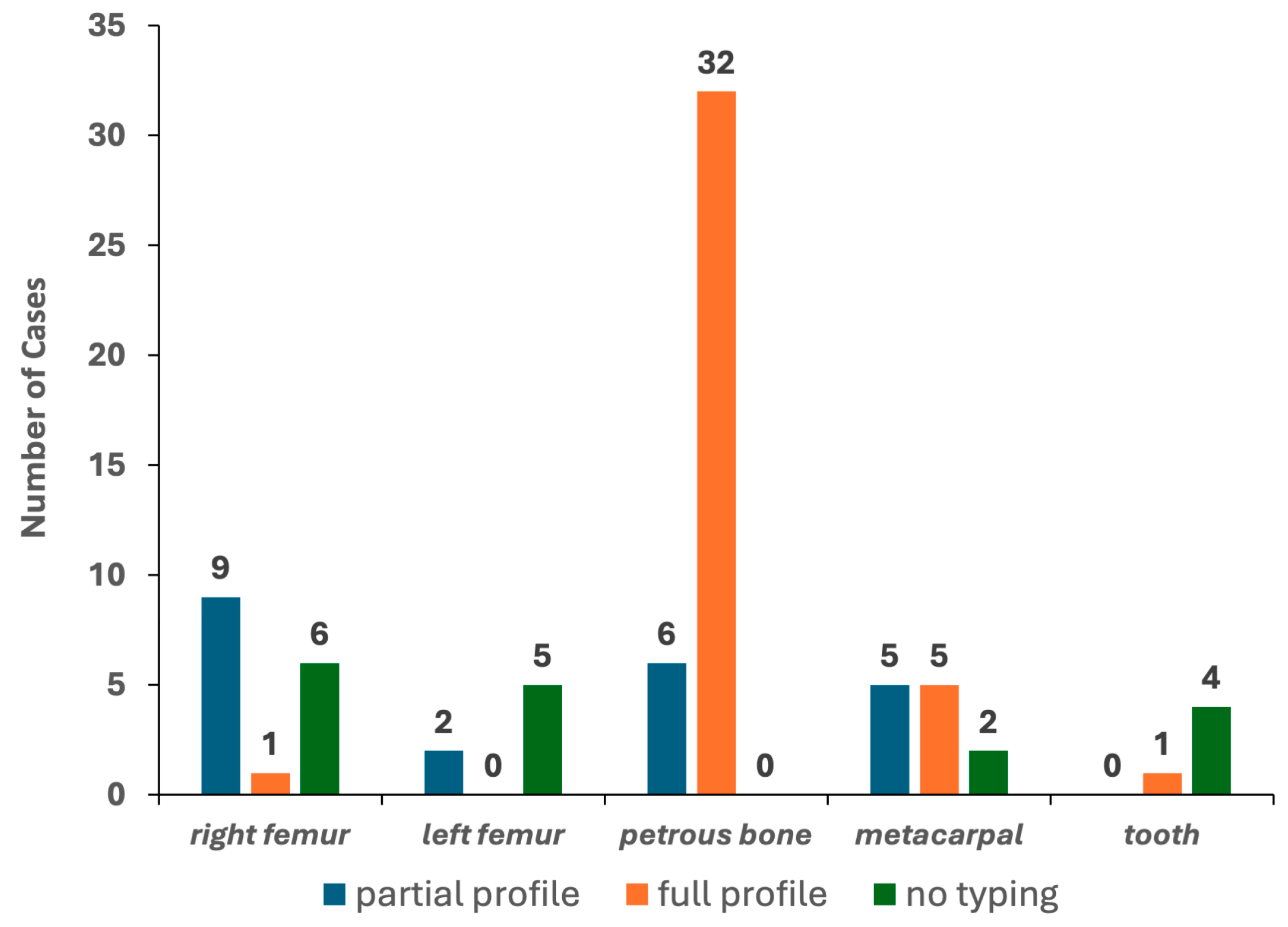
To this aim, only bone elements with detectable levels of DNA in at least one of the two qPCR assays were selected. The results yielded from 78 PCR tests (excluding the DNA control samples) are reported in Table 1. Genetic typing from the petrous bone samples was always achieved, with full profiles (22 out of 22 autosomal STR markers) in 84.2% of the tests (see Figure 4). Six PCR tests gave partial profiles characterised by 16 markers/electropherograms (median value). As shown in Figure A2, a typical ski-slope profile was obtained from these challenging samples, in agreement with the scored degradation levels [35].
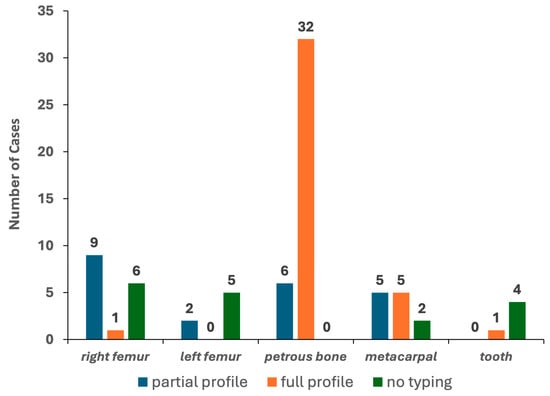
Figure 4.
Results of the 78 PCR tests performed with the PowerPlex Fusion kit.
The other bone elements were successfully typed with different rates (from 20% to 83.3% for teeth and metacarpals, respectively). Only five, one and one full electropherogram was achieved for the metacarpal, tooth and femur, respectively. Partial profiles characterised by 18–20 markers/electropherograms (median values) were obtained in 16 tests, whereas the remaining 17 tests showed 5 markers/electropherograms (median value; min = 0; max = 9) and were then classified, such as “no typing” (see Figure 4). Remarkably, the petrous bone provided the same genotypes in the two PCR replicates for 98.1% of the markers, whereas the other bone types yielded profiles affected by stochastic artefacts in up to 13.9% of the replicates likely originated by high degradation levels and/or low amounts of template used for the PCR amplification [35].
As discussed above, consensus profiles [38] for at least twelve autosomal STR markers [8] in duplicated experiments were considered “suitable for personal identification” [8]. The consensus [38] approach allowed the identification of 22 different STR profiles suitable for comparisons (see Table A4 of Appendix B). Out of them, 17 were full profiles, whereas the remaining 5 were partial, with at least 16 scored markers. Importantly, bones found in the same metal box did not match in three cases. However, all results of the genetic typing can be considered useful for comparison with reference samples (relatives of the alleged soldiers) we are still trying to identify and collect.
No amplicons were scored in the PCR and extraction negative controls. All the profiles were compared with the ones stored in the laboratory exclusion database, and no matches were identified.
4. Discussion
When the skeletal remains collected in the mass grave in the isle of Cres were delivered to our lab, there were two main goals to achieve: the first was to set up a reliable protocol for DNA extraction in order to maximise the DNA extraction efficiency from these very challenging skeletal remains, and the second was to select the best-performing bone elements for individual genetic identification.
To this aim, we first used the Bone DNA extraction kit (Promega) on 150 mg of bone powder from femurs. As this method allowed no or very poor qPCR quantification values, we decided to increase the amount of starting bone powder to 0.5 g and set up a protocol based on the employment of the Maxwell® FSC DNA IQ™ Casework Kit (Promega). The main advantage of this semi-automated DNA extraction protocol compared to other protocols, some of which are based on organic purification [3,14,31,42,43], is the decrease in the risk of human error and cross-contamination of the samples due to the automation of the DNA purification steps, especially by reducing physical manipulations of the specimens. The bone lysates, in fact, are transferred from 50 mL Falcon tubes into the cartridge after three simple and quick steps: (1) short centrifugation (in an Eppendorf tube) to pellet the undigested bone powder (if any); (2) transferring the sample to a second Eppendorf tube and mixing it with the buffer provided in the kit; (3) loading the sample into the prefilled cartridge.
The performances of the above-described protocol in terms of DNA recovery and the level of DNA degradation were compared with those of an optimised DNA extraction protocol from bone samples based on the Qiagen technology and kits [32]. The comparison did not show any statistically significant difference between the two protocols; for this reason, the one described in this paper can be efficiently performed in any laboratory equipped with a Maxwell automated DNA extraction apparatus. Although it is not a real issue, one limit to the present method is the low repeatability when the yields of DNA are higher than 30–50 nanograms, likely due to the saturation of the magnetic beads.
The DNA extraction protocol was applied to 88 specimens from different bone elements (72) and 16 teeth (molars) selected for this study. The results of this preliminary report confirm that petrous bone is the bone element providing the best chances for successful DNA typing. The high amounts of DNA recovered from this bone element is the simplest but most convincing explanation for the successful typing scored in all 19 petrous bone samples considered in this study [41].
The other bone samples seemed to be more compromised, as assessed by a lower amount of DNA recovered and a lower quality of the resulting genetic profiles. In fact, according to the sensitivity of the qPCR assay used, detectable amounts of DNA were found in approximately 25% of the qPCR tests performed on femur samples, in 58% of tests performed on metatarsal samples and in 16% of the tests performed on tooth samples (see Table 1). In forensic genetics, qPCR data are used mainly to address the operator on the strategies to be adopted in downstream typing approaches (volume of sample to be added to the PCR reaction and number of PCR cycles) [1,35]. It is well known that the use of different quantification kits can result in different quantification values for many reasons (length and molecular sequence of the targets, chemistry, calibration standards, etc.) [40]. In addition, each kit has its own sensitivity (for example, the LOD of the PowerQuant is about 10-fold lower than that of the Quantifiler kit). Thus, even if other commercial kits could likely provide quantification values for part of the bone samples analysed in this study which showed no Cq in the Quantifiler assay, it is also true that sub-cellular amounts of DNA are of scarce utility for a reliable STR-CE typing. However, as recently pointed out for historical bone samples aged from 80 to 800 years [41], it is clear that the number of scored markers is positively linked to the amount of the template used for the genetic testing, both for traditional PCR-CE and next-generation sequencing approaches.
In agreement with the limited amounts of template available for STR amplification, the genetic typing showed low percentages of successful typing. Overall, our results partially agree with previous data highlighting that metatarsals are ideal bone elements for genetic typing [21], even if a possible explanation for our finding could be ascribed to the peculiar burial conditions of the mass grave of the isle of Cres [3,14,44,45,46,47]. In addition, the low-quality genetic results obtained for molar teeth in our study do not support the positive outcome described for these elements recovered from some mass graves [14,38,42]. Again, as reported in several studies [3,12,14,44,45,46,47], environmental burial conditions (i.e., temperature and soil properties such as pH and ionic composition, hydrology, etc.) are key factors for biomolecule preservation.
The availability of genetic profiles based on at least 12 STR markers is recommended for successful individual identification in DVI (Disaster Victim Identification) casework [8]. However, it is an obvious consideration that the availability of more genotyping data can increase the chances of successful identification, in particular if the reference samples (ante mortem) are not first-degree relatives. Figure A1 provides an overview of the results achieved up to now. Seventeen full profiles (based on 22 autosomal STR markers) and five partial profiles with at least 16 markers were uploaded into our post mortem databases. Thus, the results presented in this paper seem to be promising and suggest that the analysis of other bone specimens collected from other skeletal elements could allow the recovery of all genetic profiles belonging to the 27 Italian soldiers whose bodies are likely to have been buried in that mass grave.
The anthropological and medico-legal examination of the 27 metal boxes handed over to the Italian government showed that the skeletal remains belonged to at least 29 subjects, and the bones were also likely commingled; still, only 19 right petrous bones were found within the metal boxes. DNA typing provided definitive evidence of commingled remains in three cases for which a comparison was performable. Therefore, our results highlight the need for accurate planning of the DVI operation, which should be carried out by a multi-disciplinary team. In particular, the added value of a forensic anthropologist should allow for collecting all the bone elements/fragments and for dealing with commingled scenarios [45], therefore reducing the effort for genetic typing.
A final consideration is that, while it is well known that an accurate selection of bone elements for DNA analysis is fundamental for successful genetic typing [3,14], the selection needs to be customised for each specific DVI scenario. In other words, while it is true that petrous bone usually outperforms other bone elements [15,16,17,18,19,20,21,46,48], the identification of other suitable skeletal elements seems to be a case-to-case tricky matter, depending on the features of the mass grave and the environmental conditions where the skeletons were buried [3,12,14,41,44,45,46,47]. This implies that an expensive and time-consuming strategy based on multiple samplings of different bone elements remains the only way to obtain genetic profiles from challenging skeletal remains [4,11,12,13,14,15,16,17,18,19,20,21], especially in DVI casework, as recommended by the DNA commission of the International Society for Forensic Genetics [8]. Thus, even the results presented in this paper highlight the need to search preanalytical parameters (such as bone density, bone composition and micro-computed tomography) positively related to DNA profiling, as performed in a few pilot studies performed on bones of archaeological [22,46,49,50] and forensic interest [51]. In conclusion, a multi-disciplinary approach is irreplaceable in the genetic analysis of DVI.
5. Conclusions
In this work, we described the set-up of a new protocol based on the processing of 0.5 g of decalcified bone powder whose DNA recovery was similar to a well-established protocol [32]; therefore, it was applied to a selected sample of bones found in a WWII mass grave. Although the total number of samples is limited (n = 88) and the qPCR quantification was conducted with a kit having a (relatively) low sensitivity, the results confirm that petrous bone should be preferred for genetic typing. Our results showed that petrous bone—likely due to its intrinsic properties—outperforms other bone elements (femurs, metacarpals and teeth) in quantity and quality. Overall, the results of this study support that burial conditions play a fundamental role in biomolecule preservation, and no generalisation is allowed. Femurs, which usually provide satisfying results [3,14,38,42], yielded approximately ten-fold less DNA than other femurs found in a different WWII mass grave studied by us [33]. Similarly, teeth found in the mass grave of the isle of Cres yielded low levels of DNA. Teeth are the hardest organs in the human body, with the enamel protecting the crown; still, in spite of their anatomical features and the absence of dental caries, they did not escape degradation processes. More encouraging, instead, were the results from metacarpals. However, further studies are needed to understand the role of environmental factors in DNA preservation within different types of bone remains.
Author Contributions
Conceptualization, B.D.S., P.F. and C.P.; formal analysis, M.C., M.G.C., R.V., F.I. and B.D.S.; validation, I.Z.P., S.S.C., P.G. and Y.A.; funding, P.F.; software, S.B. and B.B.; technical support, A.B.; writing—review and editing, P.F., C.P. and S.B. All authors have read and agreed to the published version of the manuscript.
Funding
This study was partially financially supported by Comunità di Lussino, Trieste, Italy.
Institutional Review Board Statement
The present study was approved by the Ethics Committee of the University of Trieste (prot. 129/29.03.2023), Trieste, Italy.
Informed Consent Statement
Informed consent was obtained from all living subjects involved in this study.
Data Availability Statement
Data are contained within the article. Other data presented in this study are available on request from the corresponding author.
Acknowledgments
The Authors wish to thank Licia Giadrossi and Cap. Federico Scopinich (Comunità di Lussino, Trieste, Italy) for promoting the fundraising that partially supported this study. Special thanks to Susanna Gerolami for the English revision of the manuscript.
Conflicts of Interest
Alessandro Bosetti was employed by the company Promega Italia, Milano, Italy. The remaining Authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
Appendix A
Appendix A.1. Optimisation of the Extraction Protocol with Maxwell Technology
Considering the high number of bone samples to analyse, we decided to move from a manual [31] to a semi-automatic protocol, based on the use of the Maxwell RCS extractor available in Lab A. First, the Bone DNA extraction kit (Promega) was used on 150 mg of bone powder from femurs. These bone elements were selected for this preliminary test because they usually provide suitable yields of DNA [3,14]; in addition, they represented the biggest specimens available. No or very poor quantification values from 150 mg of powder prompted us to increase to 0.5 g the starting amount of the sample. Therefore, a protocol based on the use of the Maxwell® FSC DNA IQ™ Casework Kit (Promega), designed for forensic but not bone samples, was set up. In addition, the kit’s brochure recommends using 400 µL of sample (extracted with commercial buffers) mixed with 200 µL of Lysis Buffer (LB) provided in the kit. Thereafter, the most significant experiments performed for optimising the extraction protocol are reported.
Appendix A.1.1. Assessment of the DNA Recovery from the Home-Made Extraction Buffer (Standard Volumes)
The following home-made extraction buffer (h-m EB) was prepared: 1.2% SDS, 10 mM Tris pH 8.0, 10 mM Na2EDTA pH 8.0, 100 mM NaCl, 80 mM DTT and Proteinase K (1 mg/mL). This buffer is described for DNA extraction from bone samples, followed by organic purification [31].
Liquid samples were spiked with human DNA (DNA K562, Promega) to assess the recovery. In detail, 5 and 30 ng of DNA were used to spike the h-m EB at the final concentrations of 25 pg/µL and 150 pg/µL, respectively. The eluted samples (50 µL) were then quantified with the Quantifiler kit in duplicate tests (see main text). The protocol and the results are shown below in Table A1.

Table A1.
Assessment of DNA recovery from the home-made extraction buffer with recommended volumes.
Table A1.
Assessment of DNA recovery from the home-made extraction buffer with recommended volumes.
| Liquid Medium | h-m EB | Water | h-m EB | Water |
|---|---|---|---|---|
| Number of tests | 3 | 3 | 3 | 3 |
| Volume | 400 µL | 400 µL | 400 µL | 400 µL |
| K562 DNA (final concentration) | 25 pg/µL | 25 pg/µL | 150 pg/µL | 150 pg/µL |
| LB | 200 µL | 200 µL | 200 µL | 200 µL |
| Total volume | 600 µL | 600 µL | 600 µL | 600 µL |
| % recovery (average ± st. dev.) | 91.7 ± 4 | 92.7 ± 6 | 94.5 ±5 | 92.2 ± 2 |
| p-value | 0.372 | 0.153 | ||
The results showed that the h-m EB does not interfere with the recovery of the spiking DNA (recovery of about 92–95%). In addition, no differences were found with water (p-value > 0.153) when using the recommended volumes (total volume 600 µL).
Appendix A.1.2. Assessment of the DNA Recovery from Increased Volumes
Since a volume of 400 µL is not suitable for processing 0.5 g of decalcified bone powder in a single purification step, the DNA recovery was evaluated by doubling the recommended volumes. In addition, for this test, the samples were spiked with human DNA, extracted and finally quantified by qPCR (Quantifiler kit) in duplicate tests (see Table A2).

Table A2.
Assessment of DNA recovery by doubling the recommended volumes.
Table A2.
Assessment of DNA recovery by doubling the recommended volumes.
| Liquid Medium | h-m EB | h-m EB | h-m EB | h-m EB |
|---|---|---|---|---|
| Number of tests | 3 | 3 | 3 | 3 |
| Volume | 400 µL | 800 µL | 400 µL | 800 µL |
| K562 DNA (final concentration) | 25 pg/µL | 25 pg/µL | 150 pg/µL | 150 pg/µL |
| LB | 200 µL | 400 µL | 200 µL | 400 µL |
| Total volume | 600 µL | 1.200 µL | 600 µL | 1.200 µL |
| % recovery (average ± st. dev.) | 94.3 ± 4 | 72.4 ± 3 | 93.9 ± 7 | 71.2 ± 4 |
| p-value | 5.8 × 10−7 | 9.2 × 10−5 | ||
The results showed that by doubling the recommended working volumes, the recovery decreased by about 21.2–22.6% (p-value: <9.2 × 10−5). Nevertheless, the total yield of recovered DNA was higher (about 52–54%).
Appendix A.1.3. Assessment of Bacterial Contamination
It is well known that bacterial contamination can represent the major source of DNA in skeletal remains [3,14]. Therefore, we evaluated if massive bacterial contamination interfered with the recovery of the human DNA. To this aim, 800 µL of h-m EB was spiked with human and Escherichia coli DNA (strain DH5α), extracted and finally quantified by the Quantifiler kit (in duplicate tests) following the above-reported protocol.The results are shown in Table A3.

Table A3.
Assessment of DNA recovery in samples containing 200 ng of E. coli DNA.
Table A3.
Assessment of DNA recovery in samples containing 200 ng of E. coli DNA.
| Liquid Medium | h-m EB | h-m EB | h-m EB | h-m EB |
|---|---|---|---|---|
| Number of tests | 3 | 3 | 3 | 3 |
| Volume | 800 µL | 800 µL | 800 µL | 800 µL |
| K562 DNA (final concentration) | 25 pg/µL | 25 pg/µL | 150 pg/µL | 150 pg/µL |
| E. coli DNA (final concentration) | - | 250 pg/µL | - | 250 pg/µL |
| LB | 400 µL | 400 µL | 400 µL | 400 µL |
| Total volume | 1.200 µL | 1.200 µL | 1.200 µL | 1.200 µL |
| % recovery (average ± st. dev.) | 71.1 ± 2 | 54.7 ± 9 | 70.2 ± 6 | 43.0 ± 14 |
| p-value | 0.005 | 0.002 | ||
These results showed that bacterial DNA indeed interfered with the recovery of the human DNA by scaling down the recovery by up to 38.6%. This finding is likely to be ascribed to a magnetic bead saturation.
In conclusion, the above-described approach seems to be a good compromise between suitable buffer volumes needed for efficient sample extraction and the features of the DNA extraction kit components.
Appendix B

Table A4.
List of the 88 samples analysed in this study. The numbers refer to sample codes; * indicates samples typed by a single PCR test (see text for details). The outcomes are shown as follows:
Table A4.
List of the 88 samples analysed in this study. The numbers refer to sample codes; * indicates samples typed by a single PCR test (see text for details). The outcomes are shown as follows:
| full-consensus STR profile | partial-consensus STR profile | unsuccessful STR typing | no Cq at qPCR | ||
| skeleton | rigth femur | left femur | petrous bone | metacarpal | molar teeth |
| #1 | 1.15bis * | 1.1 | 1.17 | ||
| #2 | 2.3bis | 2.1 | 2.5 | ||
| #3 | 3.2.1 | 3.2.2 * | 3.2 | 3.10 | |
| #4 | 3.3.1 | 3.3.2 | |||
| #5 | 4.16 | 4.1 | 4.12 | ||
| #6 | 4.2.1 * | 4.3.4 * | |||
| #7 | 5.7 | 5.1 | 5.3 * | ||
| #8 | 5.2.1 | 5.2.2 * | |||
| #9 | 6.12 * | 6.1 | 6.8 | ||
| #10 | 7.9 | 8.14 | 7.3 | ||
| #11 | 8.2.1 | 8.2.2 | 8.2 | 8.10 * | 8.9 |
| #12 | 9.12 | 9.1 | 9.8 | 4.11 | |
| #13 | 9.2.6 | 3.14 | 9.2.2 | 10.2 | |
| #14 | 11.13 | 11.1 | 11.9 | 4.5 | |
| #15 | 13.4 | 12.7* | 12.3 | 13.1 * | |
| #16 | 14.3 | 11.14 | 11.2 | ||
| #17 | 15.6 | 15.2 | 16.2 | ||
| #18 | 16.7 | 16.1 | 16.3 | 16.2.2 * | |
| #19 | 17.5 | 2.4* | |||
| #20 | 18.5 | 1.16 | 18.4 | ||
| #21 | 19.10 | 20.1 | 19.9* | ||
| #22 | 21.15 | 21.1 | |||
| #23 | 21.3.1 | 5.8 * | 22.1 | 20.4 | |
| #24 | 23.11 * | 20.2.2 * | |||
| #25 | 24.10 * | 24.1 | 21.5 | ||
| #26 | 25.16 | 25.1 | |||
| #27 | 25.2.1 | 4.17 | 21.12 | ||
| #28 | 26.17 * | 26.1 | |||
| #29 | 27.9 | 27.1 | 20.7 | ||
Appendix C
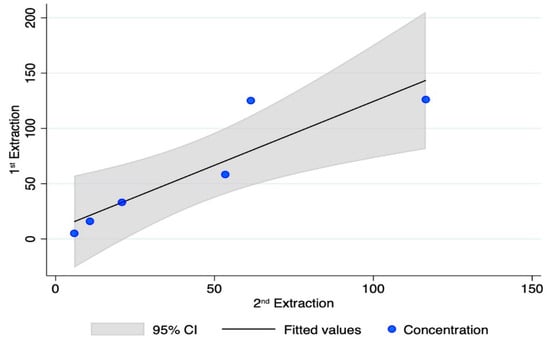
Figure A1.
DNA recovery as assessed by the Quantifiler kit during two different extraction sessions; x-axis: ng DNA/g bone in the 2nd extraction; y-axis: ng DNA/g bone in the 1st extraction.
Figure A1.
DNA recovery as assessed by the Quantifiler kit during two different extraction sessions; x-axis: ng DNA/g bone in the 2nd extraction; y-axis: ng DNA/g bone in the 1st extraction.
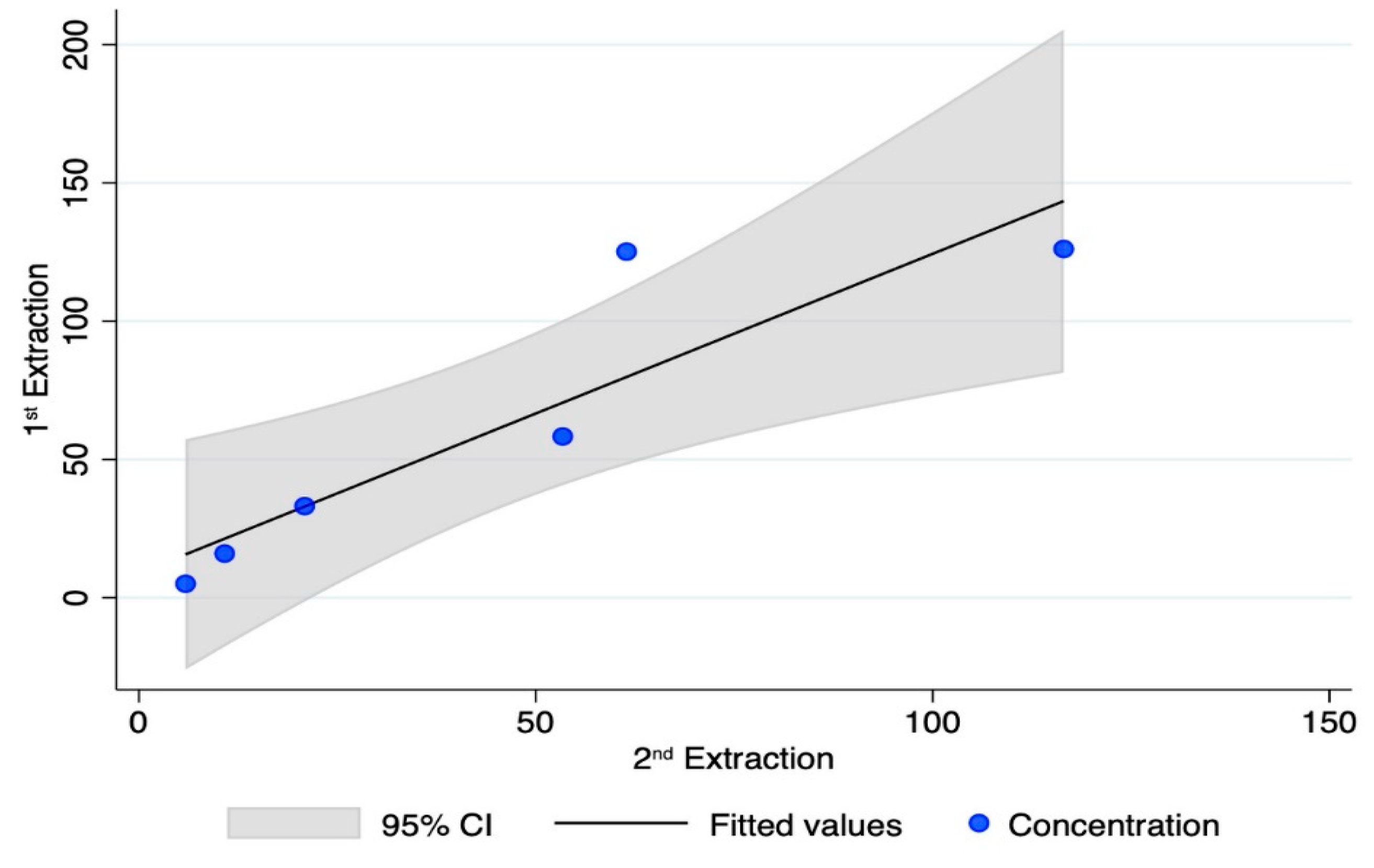
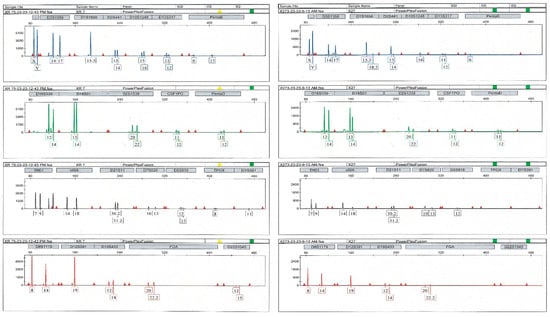
Figure A2.
Genetic profiles from two different bone elements of the same skeleton: left, petrous bone (full profile); right, metacarpal (partial profile). Due to DNA degradation, the ski-slope effect is evident in both traces.
Figure A2.
Genetic profiles from two different bone elements of the same skeleton: left, petrous bone (full profile); right, metacarpal (partial profile). Due to DNA degradation, the ski-slope effect is evident in both traces.
References
- Butler, J.M. Recent advances in forensic biology and forensic DNA typing: INTERPOL review 2019–2022. Forensic Sci. Int. Synerg. 2023, 6, 100311. [Google Scholar] [CrossRef] [PubMed]
- Hagelberg, E.; Hofreiter, M.; Keyser, C. Introduction. Ancient DNA: The first three decades. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 370, 20130371. [Google Scholar] [CrossRef]
- Hofreiter, M.; Sneberger, J.; Pospisek, M.; Vanek, D. Progress in forensic bone DNA analysis: Lessons learned from ancient DNA. Forensic Sci. Int. Genet. 2021, 54, 102538. [Google Scholar] [CrossRef] [PubMed]
- Holland, M.M.; Cave, C.A.; Holland, C.A.; Bille, T.W. Development of a quality, high throughput DNA analysis procedure for skeletal samples to assist with the identification of victims from the World Trade Center attacks. Croat. Med. J. 2003, 44, 264–272. [Google Scholar] [PubMed]
- Andelinović, S.; Sutlović, D.; Erceg Ivkosić, I.; Skaro, V.; Ivkosić, A.; Paić, F.; Rezić, B.; Definis-Gojanović, M.; Primorac, D. Twelve-year experience in identification of skeletal remains from mass graves. Croat. Med. J. 2005, 46, 530–539. [Google Scholar]
- Lin, C.Y.; Huang, T.Y.; Shih, H.C.; Yuan, C.H.; Chen, L.J.; Tsai, H.S.; Pan, C.H.; Chiang, H.M.; Liu, H.L.; Su, W.C.; et al. The strategies to DVI challenges in Typhoon Morakot. Int. J. Leg. Med. 2011, 125, 637–641. [Google Scholar] [CrossRef] [PubMed]
- Sozer, A.; Baird, M.; Beckwith, M.; Harmon, B.; Lee, D.; Riley, G.; Schmitt, S. Guidelines for Mass Fatality DNA Identification Operations; AABB, Ed.; AABB: Bethesda, MD, USA, 2010; p. 52. [Google Scholar]
- Prinz, M.; Carracedo, A.; Mayr, W.R.; Morling, N.; Parsons, T.J.; Sajantila, A.; Scheithauer, R.; Schmitter, H.; Schneider, P.M. DNA Commission of the International Society for Forensic Genetics (ISFG): Recommendations regarding the role of forensic genetics for disaster victim identification (DVI). Forensic Sci. Int. Genet. 2007, 1, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Emmons, A.L.; Davoren, J.; DeBruyn, J.M.; Mundorff, A.Z. Inter and intra-individual variation in skeletal DNA preservation in buried remains. Forensic Sci. Int. Genet. 2020, 44, 102193. [Google Scholar] [CrossRef] [PubMed]
- Parsons, T.J.; Huel, R.M.L.; Bajunovic, Z.; Rizvic, A. Large scale DNA identification: The ICMP experience. Forensic Sci. Int. Genet. 2019, 38, 236–244. [Google Scholar] [CrossRef]
- Antinick, T.C.; Foran, D.R. Intra- and Inter-Element Variability in Mitochondrial and Nuclear DNA from Fresh and Environmentally Exposed Skeletal Remains. J. Forensic Sci. 2019, 64, 88–97. [Google Scholar] [CrossRef]
- Hedges, R.E.M. Bone diagenesis: An overview of processes. Archaeometry 2002, 44, 319–328. [Google Scholar] [CrossRef]
- Benedik Bevc, T.; Bozic, L.; Podovsovnik, E.; Zupanc, T.; Zupanic Pajnic, I. Intra-bone nuclear DNA variability and STR typing success in Second World War 12th thoracic vertebrae. Forensic Sci. Int. Genet. 2021, 55, 102587. [Google Scholar] [CrossRef] [PubMed]
- Finaughty, C.; Heathfield, L.J.; Kemp, V.; Marquez-Grant, N. Forensic DNA extraction methods for human hard tissue: A systematic literature review and meta-analysis of technologies and sample type. Forensic Sci. Int. Genet. 2023, 63, 102818. [Google Scholar] [CrossRef] [PubMed]
- Gaudio, D.; Fernandes, D.M.; Schmidt, R.; Cheronet, O.; Mazzarelli, D.; Mattia, M.; O’Keeffe, T.; Feeney, R.N.M.; Cattaneo, C.; Pinhasi, R. Genome-Wide DNA from Degraded Petrous Bones and the Assessment of Sex and Probable Geographic Origins of Forensic Cases. Sci. Rep. 2019, 9, 8226. [Google Scholar] [CrossRef]
- Gonzalez, A.; Cannet, C.; Zvenigorosky, V.; Geraut, A.; Koch, G.; Delabarde, T.; Ludes, B.; Raul, J.S.; Keyser, C. The petrous bone: Ideal substrate in legal medicine? Forensic Sci. Int. Genet. 2020, 47, 102305. [Google Scholar] [CrossRef] [PubMed]
- Kulstein, G.; Hadrys, T.; Wiegand, P. As solid as a rock-comparison of CE- and MPS-based analyses of the petrosal bone as a source of DNA for forensic identification of challenging cranial bones. Int. J. Leg. Med. 2018, 132, 13–24. [Google Scholar] [CrossRef]
- Misner, L.M.; Halvorson, A.C.; Dreier, J.L.; Ubelaker, D.H.; Foran, D.R. The correlation between skeletal weathering and DNA quality and quantity. J. Forensic Sci. 2009, 54, 822–828. [Google Scholar] [CrossRef] [PubMed]
- Pilli, E.; Vai, S.; Caruso, M.G.; D’Errico, G.; Berti, A.; Caramelli, D. Neither femur nor tooth: Petrous bone for identifying archaeological bone samples via forensic approach. Forensic Sci. Int. 2018, 283, 144–149. [Google Scholar] [CrossRef] [PubMed]
- Pinhasi, R.; Fernandes, D.; Sirak, K.; Novak, M.; Connell, S.; Alpaslan-Roodenberg, S.; Gerritsen, F.; Moiseyev, V.; Gromov, A.; Raczky, P.; et al. Optimal Ancient DNA Yields from the Inner Ear Part of the Human Petrous Bone. PLoS ONE 2015, 10, e0129102. [Google Scholar] [CrossRef]
- Zupanic Pajnic, I.; Inkret, J.; Zupanc, T.; Podovsovnik, E. Comparison of nuclear DNA yield and STR typing success in Second World War petrous bones and metacarpals III. Forensic Sci. Int. Genet. 2021, 55, 102578. [Google Scholar] [CrossRef]
- Fernandes, D.M.; Sirak, K.A.; Cheronet, O.; Novak, M.; Bruck, F.; Zelger, E.; Llanos-Lizcano, A.; Wagner, A.; Zettl, A.; Mandl, K.; et al. Density separation of petrous bone powders for optimized ancient DNA yields. Genome Res. 2023, 33, 622–631. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, J.; Brumfeld, V.; Addadi, Y.; Rubin, S.; Weiner, S.; Boaretto, E. The petrous bone contains high concentrations of osteocytes: One possible reason why ancient DNA is better preserved in this bone. PLoS ONE 2022, 17, e0269348. [Google Scholar] [CrossRef] [PubMed]
- Sirak, K.; Fernandes, D.; Cheronet, O.; Harney, E.; Mah, M.; Mallick, S.; Rohland, N.; Adamski, N.; Broomandkhoshbacht, N.; Callan, K.; et al. Human auditory ossicles as an alternative optimal source of ancient DNA. Genome Res. 2020, 30, 427–436. [Google Scholar] [CrossRef] [PubMed]
- Correa, H.; Cortellini, V.; Franceschetti, L.; Verzeletti, A. Large fragment demineralization: An alternative pretreatment for forensic DNA typing of bones. Int. J. Leg. Med. 2021, 135, 1417–1424. [Google Scholar] [CrossRef] [PubMed]
- Calacal, G.C.; Gallardo, B.G.; Apaga, D.L.T.; De Ungria, M.C.A. Improved autosomal STR typing of degraded femur samples extracted using a custom demineralization buffer and DNA IQ. Forensic Sci. Int. Synerg. 2021, 3, 100131. [Google Scholar] [CrossRef]
- Duijs, F.E.; Sijen, T. A rapid and efficient method for DNA extraction from bone powder. Forensic Sci. Int. Rep. 2020, 2, 100099. [Google Scholar] [CrossRef]
- Haarkotter, C.; Galvez, X.; Vinueza-Espinosa, D.C.; Medina-Lozano, M.I.; Saiz, M.; Lorente, J.A.; Alvarez, J.C. A comparison of five DNA extraction methods from degraded human skeletal remains. Forensic Sci. Int. 2023, 348, 111730. [Google Scholar] [CrossRef] [PubMed]
- Xavier, C.; Eduardoff, M.; Bertoglio, B.; Amory, C.; Berger, C.; Casas-Vargas, A.; Pallua, J.; Parson, W. Evaluation of DNA Extraction Methods Developed for Forensic and Ancient DNA Applications Using Bone Samples of Different Age. Genes 2021, 12, 146. [Google Scholar] [CrossRef]
- Zupanic Pajnic, I.; Leskovar, T.; Zupanc, T.; Podovsovnik, E. A fast and highly efficient automated DNA extraction method from small quantities of bone powder from aged bone samples. Forensic Sci. Int. Genet. 2023, 65, 102882. [Google Scholar] [CrossRef]
- Fattorini, P.; Marrubini, G.; Ricci, U.; Gerin, F.; Grignani, P.; Cigliero, S.S.; Xamin, A.; Edalucci, E.; La Marca, G.; Previdere, C. Estimating the integrity of aged DNA samples by CE. Electrophoresis 2009, 30, 3986–3995. [Google Scholar] [CrossRef]
- Pajnic, I.Z. Extraction of DNA from Human Skeletal Material. Methods Mol. Biol. 2016, 1420, 89–108. [Google Scholar] [CrossRef]
- Zupanic Pajnic, I.; Fattorini, P. Strategy for STR typing of bones from the Second World War combining CE and NGS technology: A pilot study. Forensic Sci. Int. Genet. 2021, 50, 102401. [Google Scholar] [CrossRef]
- Zupanic Pajnic, I.; Zupanc, T.; Leskovar, T.; Cresnar, M.; Fattorini, P. Eye and Hair Color Prediction of Ancient and Second World War Skeletal Remains Using a Forensic PCR-MPS Approach. Genes 2022, 13, 1432. [Google Scholar] [CrossRef]
- Alaeddini, R.; Walsh, S.J.; Abbas, A. Forensic implications of genetic analyses from degraded DNA—A review. Forensic Sci. Int. Genet. 2010, 4, 148–157. [Google Scholar] [CrossRef]
- Llamas, B.; Valverde, G.; Fehren-Schmitz, L.; Weyrich, L.S.; Cooper, A.; Haak, W. From the field to the laboratory: Controlling DNA contamination in human ancient DNA research in the high-throughput sequencing era. STAR Sci. Technol. Archaeol. Res. 2017, 3, 1–14. [Google Scholar] [CrossRef]
- Paabo, S.; Poinar, H.; Serre, D.; Jaenicke-Despres, V.; Hebler, J.; Rohland, N.; Kuch, M.; Krause, J.; Vigilant, L.; Hofreiter, M. Genetic analyses from ancient DNA. Annu. Rev. Genet. 2004, 38, 645–679. [Google Scholar] [CrossRef]
- Taberlet, P.; Griffin, S.; Goossens, B.; Questiau, S.; Manceau, V.; Escaravage, N.; Waits, L.P.; Bouvet, J. Reliable genotyping of samples with very low DNA quantities using PCR. Nucleic Acids Res. 1996, 24, 3189–3194. [Google Scholar] [CrossRef]
- Lindahl, T. Instability and decay of the primary structure of DNA. Nature 1993, 362, 709–715. [Google Scholar] [CrossRef]
- Lee, S.B.; McCord, B.; Buel, E. Advances in forensic DNA quantification: A review. Electrophoresis 2014, 35, 3044–3052. [Google Scholar] [CrossRef]
- Thomas, J.T.; Cavagnino, C.; Kjelland, K.; Anderson, E.; Sturk-Andreaggi, K.; Daniels-Higginbotham, J.; Amory, C.; Spatola, B.; Moran, K.; Parson, W.; et al. Evaluating the Usefulness of Human DNA Quantification to Predict DNA Profiling Success of Historical Bone Samples. Genes 2023, 14, 994. [Google Scholar] [CrossRef]
- Haarkotter, C.; Vinueza-Espinosa, D.C.; Galvez, X.; Saiz, M.; Medina-Lozano, M.I.; Lorente, J.A.; Alvarez, J.C. A comparison between petrous bone and tooth, femur and tibia DNA analysis from degraded skeletal remains. Electrophoresis 2023, 44, 1559–1568. [Google Scholar] [CrossRef]
- Baeta, M.; Nunez, C.; Cardoso, S.; Palencia-Madrid, L.; Herrasti, L.; Etxeberria, F.; de Pancorbo, M.M. Digging up the recent Spanish memory: Genetic identification of human remains from mass graves of the Spanish Civil War and posterior dictatorship. Forensic Sci. Int. Genet. 2015, 19, 272–279. [Google Scholar] [CrossRef]
- Campos, P.F.; Craig, O.E.; Turner-Walker, G.; Peacock, E.; Willerslev, E.; Gilbert, M.T. DNA in ancient bone—Where is it located and how should we extract it? Ann. Anat. 2012, 194, 7–16. [Google Scholar] [CrossRef]
- Kendall, C.; Eriksen, A.M.H.; Kontopoulos, I.; Collins, M.J.; Turner-Walker, G. Diagenesis of archaeological bone and tooth. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2018, 491, 21–37. [Google Scholar] [CrossRef]
- Kontopoulos, I.; Penkman, K.; Mullin, V.E.; Winkelbach, L.; Unterlander, M.; Scheu, A.; Kreutzer, S.; Hansen, H.B.; Margaryan, A.; Teasdale, M.D.; et al. Screening archaeological bone for palaeogenetic and palaeoproteomic studies. PLoS ONE 2020, 15, e0235146. [Google Scholar] [CrossRef]
- Mulligan, C. Anthropological Applications of Ancient DNA: Problems and Prospects. Am. Antiq. 2006, 71, 365. [Google Scholar] [CrossRef]
- Kontopoulos, I.; Penkman, K.; McAllister, G.; Lynnerup, N.; Damgaard, P.; Hansen, H.; Allentoft, M.; Collins, M. Petrous bone diagenesis: A multi-analytical approach. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2019, 518, 143–154. [Google Scholar] [CrossRef]
- de Boer, H.H.; Blau, S.; Delabarde, T.; Hackman, L. The role of forensic anthropology in disaster victim identification (DVI): Recent developments and future prospects. Forensic Sci. Res. 2019, 4, 303–315. [Google Scholar] [CrossRef]
- Tripp, J.A.; Squire, M.E.; Hedges, R.E.M.; Stevens, R.E. Use of micro-computed tomography imaging and porosity measurements as indicators of collagen preservation in archaeological bone. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2018, 511, 462–471. [Google Scholar] [CrossRef]
- Zupanič Pajnič, I.; Leskovar, T.; Jerman, I. ATR-FTIR spectroscopy and chemometric complexity: Unfolding the intra-skeleton variability. J. Chemom. 2022, 36, e3448. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).