Unlocking the Genetic Identity of Endangered Paphiopedilum Orchids: A DNA Barcoding Approach
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. DNA Isolation
2.3. Polymerase Chain Reaction (PCR)
2.4. Agarose Gel Electrophoresis
2.5. Sequencing and Data Analyses
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guo, Y.-Y.; Luo, Y.-B.; Liu, Z.-J.; Wang, X.-Q. Reticulate Evolution and Sea-Level Fluctuations Together Drove Species Diversification of Slipper Orchids (Paphiopedilum) in South-East Asia. Mol. Ecol. 2015, 24, 2838–2855. [Google Scholar] [CrossRef] [PubMed]
- Teoh, E.-S. Orchids of Asia, 3rd ed.; Times Editions-Marshall Cavendish: Singapore, 2005; ISBN 978-981-261-015-7. [Google Scholar]
- Naik, S.K.; Usha Bharathi, T.; Barman, D.; De Rampal, L.C.; Medhi, R.P. Basics of Orchid Nutrition; ICAR-NRC for Orchids: Sikkim, India, 2010. [Google Scholar]
- Gurung, D.B.; Gyeltshen, N.; Tobgay, K.; Dalström, S.; Wangdi, J.; Ghalley, B.B.; Chaida, L.; Phuntsho, P.; Gyeltshen, N.; Dawa, K.; et al. Distribution and Habitats of Paphiopedilum Pfitzer (Orchidaceae) Known to Occur in Bhutan. J. Threat. Taxa 2019, 11, 14101–14111. [Google Scholar] [CrossRef]
- Lertnitikul, N.; Jittham, P.; Khankhampoch, L.; Pattamadilok, C.; Sukrong, S.; Suttisri, R. Cytotoxic Stilbenes from the Roots of Paphiopedilum godefroyae. J. Asian Nat. Prod. Res. 2016, 18, 1143–1150. [Google Scholar] [CrossRef] [PubMed]
- Naphatsawan, P.; Nonthalert, L.; Rutt, S. Stilbenes from Paphiopedilum Exul Roots. Thai J. Pharm. Sci. 2016, 40, 116–119. [Google Scholar]
- Nwe, S.Y.; Tungphatthong, C.; Laorpaksa, A.; Sritularak, B.; Thanakijcharoenpath, W.; Tanasupawat, S.; Sukrong, S. Bioassay Guided Isolation of Topoisomerase Ι Poison from Paphiopedilum Callosum (Rchb.f.) Stein. Rec. Nat. Prod. 2019, 14, 89–97. [Google Scholar] [CrossRef]
- Guo, Y.-Y.; Huang, L.-Q.; Liu, Z.-J.; Wang, X.-Q. Promise and Challenge of DNA Barcoding in Venus Slipper (Paphiopedilum). PLoS ONE 2016, 11, e0146880. [Google Scholar] [CrossRef] [PubMed]
- Hollingsworth, P.M.; Graham, S.W.; Little, D.P. Choosing and Using a Plant DNA Barcode. PLoS ONE 2011, 6, e19254. [Google Scholar] [CrossRef] [PubMed]
- CBOL Plant Working Group; Hollingsworth, P.M.; Forrest, L.L.; Spouge, J.L.; Hajibabaei, M.; Ratnasingham, S.; Van Der Bank, M.; Chase, M.W.; Cowan, R.S.; Erickson, D.L.; et al. A DNA Barcode for Land Plants. Proc. Natl. Acad. Sci. USA 2009, 106, 12794–12797. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.-F.; Ma, H.; Zhang, X.-Y.; Ci, X.-Q.; Li, L.; Hu, J.-L.; Zhang, C.-Y.; Xiao, J.-H.; Li, H.-W.; Conran, J.G.; et al. Do Taxon-Specific DNA Barcodes Improve Species Discrimination Relative to Universal Barcodes in Lauraceae? Bot. J. Linn. Soc. 2022, 199, 741–753. [Google Scholar] [CrossRef]
- Chase, M.W.; Fay, M.F. Barcoding of Plants and Fungi. Science 2009, 325, 682–683. [Google Scholar] [CrossRef] [PubMed]
- Walker, T. Plant Diversity and Evolution. Genotypic and Phenotypic Variation in Higher Plants; Henry, R.J., Ed.; CABI Publishing: Wallingford, UK, 2005; p. 332. ISBN 0-85199-904-2. [Google Scholar] [CrossRef]
- Ismail, M.; Ahmad, A.; Nadeem, M.; Javed, M.A.; Khan, S.H.; Khawaish, I.; Sthanadar, A.A.; Qari, S.H.; Alghanem, S.M.; Khan, K.A.; et al. Development of DNA Barcodes for Selected Acacia Species by Using rbcL and matK DNA Markers. Saudi J. Biol. Sci. 2020, 27, 3735–3742. [Google Scholar] [CrossRef] [PubMed]
- Selvaraj, D.; Sarma, R.K.; Sathishkumar, R. Phylogenetic Analysis of Chloroplast matK Gene from Zingiberaceae for Plant DNA Barcoding. Bioinformation 2008, 3, 24–27. [Google Scholar] [CrossRef] [PubMed]
- Kress, W.J.; Erickson, D.L. A Two-Locus Global DNA Barcode for Land Plants: The Coding rbcL Gene Complements the Non-Coding trnH-psbA Spacer Region. PLoS ONE 2007, 2, e508. [Google Scholar] [CrossRef] [PubMed]
- Vu, T.H.T.; Le, T.L.; Nguyen, T.K.; Tran, D.D.; Tran, H.D. Review on molecular markers for identification of Orchids. Vietnam. J. Sci. Technol. Eng. 2017, 59, 62–75. [Google Scholar] [CrossRef] [PubMed]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; deWaard, J.R. Biological Identifications through DNA Barcodes. Proc. R. Soc. Lond. B 2003, 270, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Zeng, S.; Wu, K.; Teixeira Da Silva, J.A.; Zhang, J.; Chen, Z.; Xia, N.; Duan, J. Asymbiotic Seed Germination, Seedling Development and Reintroduction of Paphiopedilum Wardii Sumerh., an Endangered Terrestrial Orchid. Sci. Hortic. 2012, 138, 198–209. [Google Scholar] [CrossRef]
- CITES—Convention on International Trade in Endangered Species of Wild Fauna and Flora. Available online: https://cites.org/eng (accessed on 27 May 2023).
- Doyle, J. DNA Protocols for Plants. In Molecular Techniques in Taxonomy; Hewitt, G.M., Johnston, A.W.B., Young, J.P.W., Eds.; Springer Berlin Heidelberg: Berlin/Heidelberg, Germany, 1991; pp. 283–293. ISBN 978-3-642-83964-1. [Google Scholar]
- Srivastava, D.; Manjunath, K. DNA Barcoding of Endemic and Endangered Orchids of India: A Molecular Method of Species Identification. Phcog Mag. 2020, 16, 290. [Google Scholar] [CrossRef]
- Feng, S.; Jiang, Y.; Wang, S.; Jiang, M.; Chen, Z.; Ying, Q.; Wang, H. Molecular Identification of Dendrobium Species (Orchidaceae) Based on the DNA Barcode ITS2 Region and Its Application for Phylogenetic Study. Int. J. Mol. Sci. 2015, 16, 21975–21988. [Google Scholar] [CrossRef] [PubMed]
- Hollingsworth, M.L.; Andra Clark, A.; Forrest, L.L.; Richardson, J.; Pennington, R.T.; Long, D.G.; Cowan, R.; Chase, M.W.; Gaudeul, M.; Hollingsworth, P.M. Selecting Barcoding Loci for Plants: Evaluation of Seven Candidate Loci with Species-level Sampling in Three Divergent Groups of Land Plants. Mol. Ecol. Resour. 2009, 9, 439–457. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. England. OUP 1999, 95–98. [Google Scholar]
- Tamura; Stecher; Kumar MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [CrossRef] [PubMed]
- Kuzmina, M.L.; Johnson, K.L.; Barron, H.R.; Hebert, P.D. Identification of the Vascular Plants of Churchill, Manitoba, Using a DNA Barcode Library. BMC Ecol. 2012, 12, 25. [Google Scholar] [CrossRef] [PubMed]
- Rajaram, M.C.; Yong, C.S.Y.; Gansau, J.A.; Go, R. DNA Barcoding of Endangered Paphiopedilum Species (Orchidaceae) of Peninsular Malaysia. Phytotaxa 2019, 387, 94–104. [Google Scholar] [CrossRef]
- Johnson, R.N. The Use of DNA Identification in Prosecuting Wildlife-Traffickers in Australia: Do the Penalties Fit the Crimes? Forensic Sci. Med. Pathol. 2010, 6, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.; Lee, Y.; Moon, S.; Kim, N.; Kim, S.; Yang, M.; Choi, D.; Han, M. The Identification of Elephant Ivory Evidences of Illegal Trade with Mitochondrial Cytochrome b Gene and Hypervariable D-Loop Region. J. Forensic Leg. Med. 2013, 20, 174–178. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, P.F.M.; Oliveira-Marques, A.R.; Matsumoto, T.E.; Miyaki, C.Y. DNA Barcoding Identifies Illegal Parrot Trade: Figure 1. JHERED 2015, 106, 560–564. [Google Scholar] [CrossRef] [PubMed]
- Aubriot, X.; Lowry, P.P.; Cruaud, C.; Couloux, A.; Haevermans, T. DNA Barcoding in a Biodiversity Hot Spot: Potential Value for the Identification of M AlagasyE Uphorbia L. Listed in CITES Appendices I and II. Mol. Ecol. Resour. 2013, 13, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Yesson, C.; Bárcenas, R.T.; Hernández, H.M.; De La Luz Ruiz-Maqueda, M.; Prado, A.; Rodríguez, V.M.; Hawkins, J.A. DNA Barcodes for Mexican Cactaceae, Plants under Pressure from Wild Collecting: DNA Barcodes for Mexican Cactaceae. Mol. Ecol. Resour. 2011, 11, 775–783. [Google Scholar] [CrossRef] [PubMed]
- Jeanson, M.L.; Labat, J.-N.; Little, D.P. DNA Barcoding: A New Tool for Palm Taxonomists? Ann. Bot. 2011, 108, 1445–1451. [Google Scholar] [CrossRef] [PubMed]
- Muellner, A.N.; Schaefer, H.; Lahaye, R. Evaluation of Candidate DNA Barcoding Loci for Economically Important Timber Species of the Mahogany Family (Meliaceae). Mol. Ecol. Resour. 2011, 11, 450–460. [Google Scholar] [CrossRef] [PubMed]
- Burgess, K.S.; Fazekas, A.J.; Kesanakurti, P.R.; Graham, S.W.; Husband, B.C.; Newmaster, S.G.; Percy, D.M.; Hajibabaei, M.; Barrett, S.C.H. Discriminating Plant Species in a Local Temperate Flora Using the rbcL + matK DNA Barcode: Barcoding Plants in a Local Flora. Methods Ecol. Evol. 2011, 2, 333–340. [Google Scholar] [CrossRef]
- Vasconcelos, S.; Nunes, G.L.; Dias, M.C.; Lorena, J.; Oliveira, R.R.M.; Lima, T.G.L.; Pires, E.S.; Valadares, R.B.S.; Alves, R.; Watanabe, M.T.C.; et al. Unraveling the Plant Diversity of the Amazonian Canga through DNA Barcoding. Ecol. Evol. 2021, 11, 13348–13362. [Google Scholar] [CrossRef] [PubMed]
- Dunning, L.T.; Savolainen, V. Broad-Scale Amplification of matK for DNA Barcoding Plants, a Technical Note: Amplification of matK for DNA Barcoding Plants. Bot. J. Linn. Soc. 2010, 164, 1–9. [Google Scholar] [CrossRef]
- Wu, S.; Yin, L.; Deng, Z.; Chen, Q.; Fu, Y.; Xue, H. Using DNA Barcoding to Identify the Genus Lolium. Not. Bot. Horti Agrobo 2015, 43, 536–541. [Google Scholar] [CrossRef]
- Yan, L.-J.; Liu, J.; Möller, M.; Zhang, L.; Zhang, X.-M.; Li, D.-Z.; Gao, L.-M. DNA Barcoding of Rhododendron (Ericaceae), the Largest Chinese Plant Genus in Biodiversity Hotspots of the Himalaya-Hengduan Mountains. Mol. Ecol. Resour. 2015, 15, 932–944. [Google Scholar] [CrossRef]
- Fazekas, A.J.; Kuzmina, M.L.; Newmaster, S.G.; Hollingsworth, P.M. DNA Barcoding Methods for Land Plants. In DNA Barcodes; Kress, W.J., Erickson, D.L., Eds.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2012; Volume 858, pp. 223–252. ISBN 978-1-61779-590-9. [Google Scholar]
- Yu, J.; Xue, J.-H.; Zhou, S.-L. New Universal matK Primers for DNA Barcoding Angiosperms. J. Syst. Evol. 2011, 49, 176–181. [Google Scholar] [CrossRef]
- Tan, S.-L.; Luo, Y.-H.; Hollingsworth, P.M.; Burgess, K.S.; Xu, K.; Li, D.-Z.; Gao, L.-M. DNA Barcoding Herbaceous and Woody Plant Species at a Subalpine Forest Dynamics Plot in Southwest China. Ecol. Evol. 2018, 8, 7195–7205. [Google Scholar] [CrossRef] [PubMed]
- Sass, C.; Little, D.P.; Stevenson, D.W.; Specht, C.D. DNA Barcoding in the Cycadales: Testing the Potential of Proposed Barcoding Markers for Species Identification of Cycads. PLoS ONE 2007, 2, e1154. [Google Scholar] [CrossRef] [PubMed]
- Pang, X.; Luo, H.; Sun, C. Assessing the Potential of Candidate DNA Barcodes for Identifying Non-Flowering Seed Plants: DNA Barcoding in Non-Flowering Seed Plants. Plant Biol. 2012, 14, 839–844. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-T.; Gupta, S.K.; Wang, A.Z.-M.; Lo, S.-F.; Kuo, C.-L.; Ko, Y.-J.; Chen, C.-L.; Hsieh, C.-C.; Tsay, H.-S. Internal Transcribed Spacer Sequence Based Identification and Phylogenic Relationship of Herba Dendrobii. J. Food Drug Anal. 2020, 20. [Google Scholar] [CrossRef]
- Tsai, C.-C.; Liao, P.-C.; Ko, Y.-Z.; Chen, C.-H.; Chiang, Y.-C. Phylogeny and Historical Biogeography of Paphiopedilum Pfitzer (Orchidaceae) Based on Nuclear and Plastid DNA. Front. Plant Sci. 2020, 11, 126. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Yao, H.; Han, J.; Liu, C.; Song, J.; Shi, L.; Zhu, Y.; Ma, X.; Gao, T.; Pang, X.; et al. Validation of the ITS2 Region as a Novel DNA Barcode for Identifying Medicinal Plant Species. PLoS ONE 2010, 5, e8613. [Google Scholar] [CrossRef] [PubMed]
- China Plant BOL Group; Li, D.-Z.; Gao, L.-M.; Li, H.-T.; Wang, H.; Ge, X.-J.; Liu, J.-Q.; Chen, Z.-D.; Zhou, S.-L.; Chen, S.-L.; et al. Comparative Analysis of a Large Dataset Indicates That Internal Transcribed Spacer (ITS) Should Be Incorporated into the Core Barcode for Seed Plants. Proc. Natl. Acad. Sci. USA 2011, 108, 19641–19646. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Tong, Y.; Xing, F. DNA Barcoding Evaluation and Its Taxonomic Implications in the Recently Evolved Genus Oberonia Lindl. (Orchidaceae) in China. Front. Plant Sci. 2016, 7I, 1791. [Google Scholar] [CrossRef] [PubMed]
- Vu, H.-T.; Vu, Q.-L.; Nguyen, T.-D.; Tran, N.; Nguyen, T.-C.; Luu, P.-N.; Tran, D.-D.; Nguyen, T.-K.; Le, L. Genetic Diversity and Identification of Vietnamese Paphiopedilum Species Using DNA Sequences. Biology 2019, 9, 9. [Google Scholar] [CrossRef] [PubMed]
- Hochbach, A.; Linder, H.P.; Röser, M. Nuclear Genes, matK and the Phylogeny of the Poales. Taxon 2018, 67, 521–536. [Google Scholar] [CrossRef]
- Hilu, K.W.; Borsch, T.; Müller, K.; Soltis, D.E.; Soltis, P.S.; Savolainen, V.; Chase, M.W.; Powell, M.P.; Alice, L.A.; Evans, R.; et al. Angiosperm Phylogeny Based on Mat K Sequence Information. Am. J. Bot. 2003, 90, 1758–1776. [Google Scholar] [CrossRef] [PubMed]
- Poovitha, S.; Stalin, N.; Balaji, R.; Parani, M. Multi-Locus DNA Barcoding Identifies matK as a Suitable Marker for Species Identification in Hibiscus L. Genome 2016, 59, 1150–1156. [Google Scholar] [CrossRef] [PubMed]
- Parveen, I.; Singh, H.K.; Raghuvanshi, S.; Pradhan, U.C.; Babbar, S.B. DNA Barcoding of Endangered Indian Paphiopedilum Species. Mol. Ecol. Resour. 2012, 12, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Cabelin, V.L.; Alejandro, G.D. Efficiency of matK, rbcL, trnH-psbA, and trnL-F (cpDNA) to Molecularly Authenticate Philippine Ethnomedicinal Apocynaceae through DNA Barcoding. Phcog Mag. 2016, 12, 384. [Google Scholar] [CrossRef]
- Algarni, A.A. Molecular Identification and Phylogenetic Analysis of Aloe Shadensis from Saudi Arabia Based on matK, rbcL and ITS DNA Barcode Sequence. Saudi J. Biol. Sci. 2022, 29, 1125–1133. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.-Q.; Zhu, Y.-J.; Guo, J.-L.; Peng, B.; Bai, M.-M.; Hang, Y.-Y. DNA Barcoding the Dioscorea in China, a Vital Group in the Evolution of Monocotyledon: Use of matK Gene for Species Discrimination. PLoS ONE 2012, 7, e32057. [Google Scholar] [CrossRef]
- Alves, T.L.S.; Chauveau, O.; Eggers, L.; De Souza-Chies, T.T. Species Discrimination in Sisyrinchium (Iridaceae): Assessment of DNA Barcodes in a Taxonomically Challenging Genus. Mol. Ecol. Resour. 2014, 14, 324–335. [Google Scholar] [CrossRef] [PubMed]
- Raskoti, B.B.; Ale, R. DNA Barcoding of Medicinal Orchids in Asia. Sci. Rep. 2021, 11, 23651. [Google Scholar] [CrossRef] [PubMed]

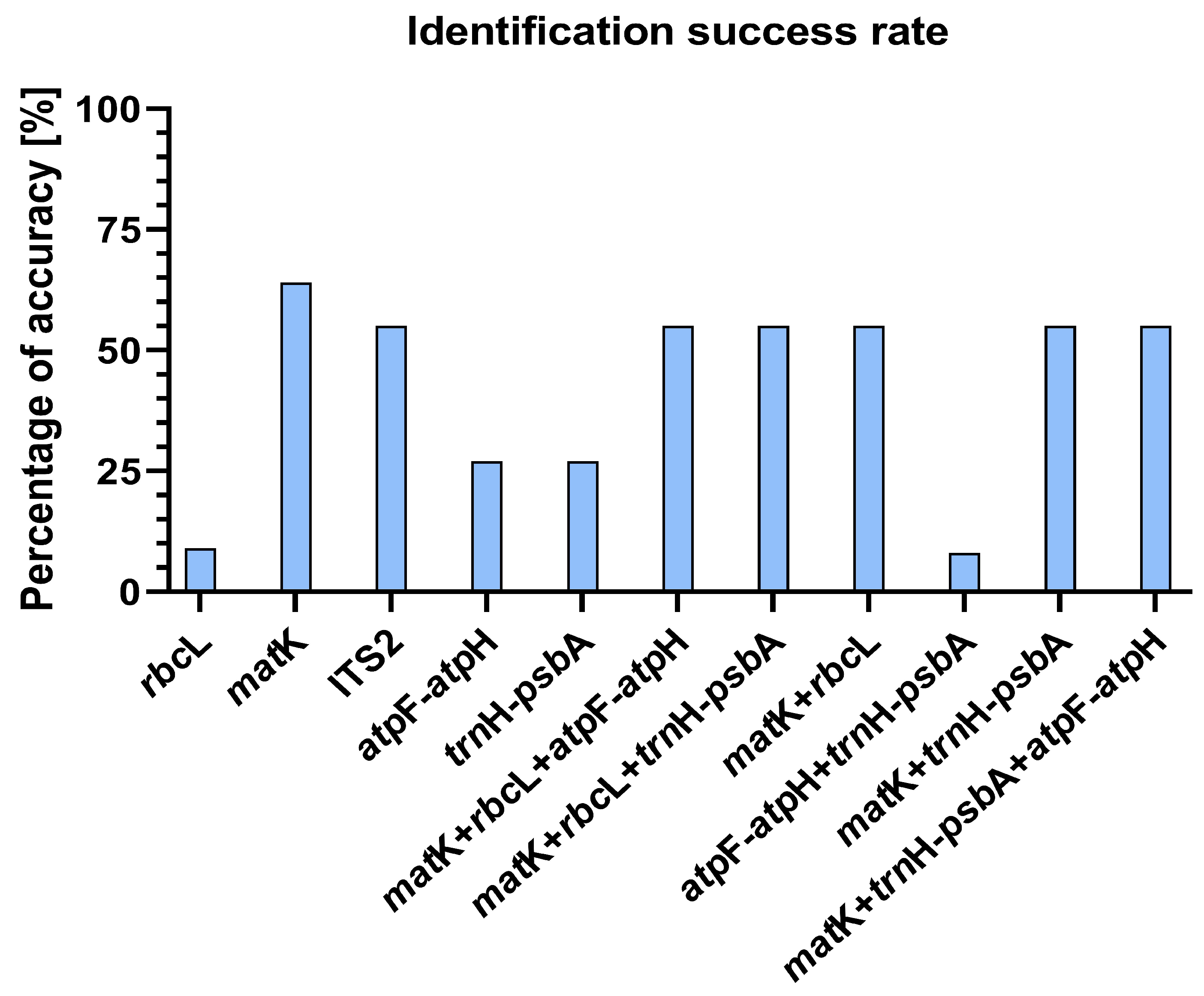
| rbcL | ITS2 | atpF-atpH | trnH-psbA | matK | |
|---|---|---|---|---|---|
| PCR amplification success (%) | 100 | 100 | 100 | 100 | 63.6 |
| Sequencing success (%) | 100 | 100 | 100 | 100 | 100 |
| Sequence length (bp) | 267–528 | 474–490 | 262–494 | 550–921 | 834–873 |
| Aligned sequence length (bp) | 536 | 504 | 500 | 987 | 883 |
| Number of conservative sites | 497/536 | 433/504 | 480/500 | 786/987 | 825/883 |
| Number of variable sites | 28/536 | 53/504 | 18/500 | 130/987 | 53/883 |
| Guanine and cytosine content in nucleotide sequences (%) | 40.8–43.9 | 52.0–53.3 | 34.4–36.9 | 31.8–34.0 | 31.6–33.1 |
| matK | rbcL | |||||||
|---|---|---|---|---|---|---|---|---|
| A | T | C | G | A | T | C | G | |
| A | - | 8.08% | 3.72% | 15.19% | - | 7.74% | 5.54% | 9.60% |
| T | 6.38% | - | 3.95% | 3.30% | 7.11% | - | 10.66% | 4.57% |
| C | 6.38% | 8.57% | - | 3.30% | 7.11% | 14.91% | - | 4.57% |
| G | 29.33% | 8.08% | 3.72% | - | 14.91% | 7.74% | 5.54% | - |
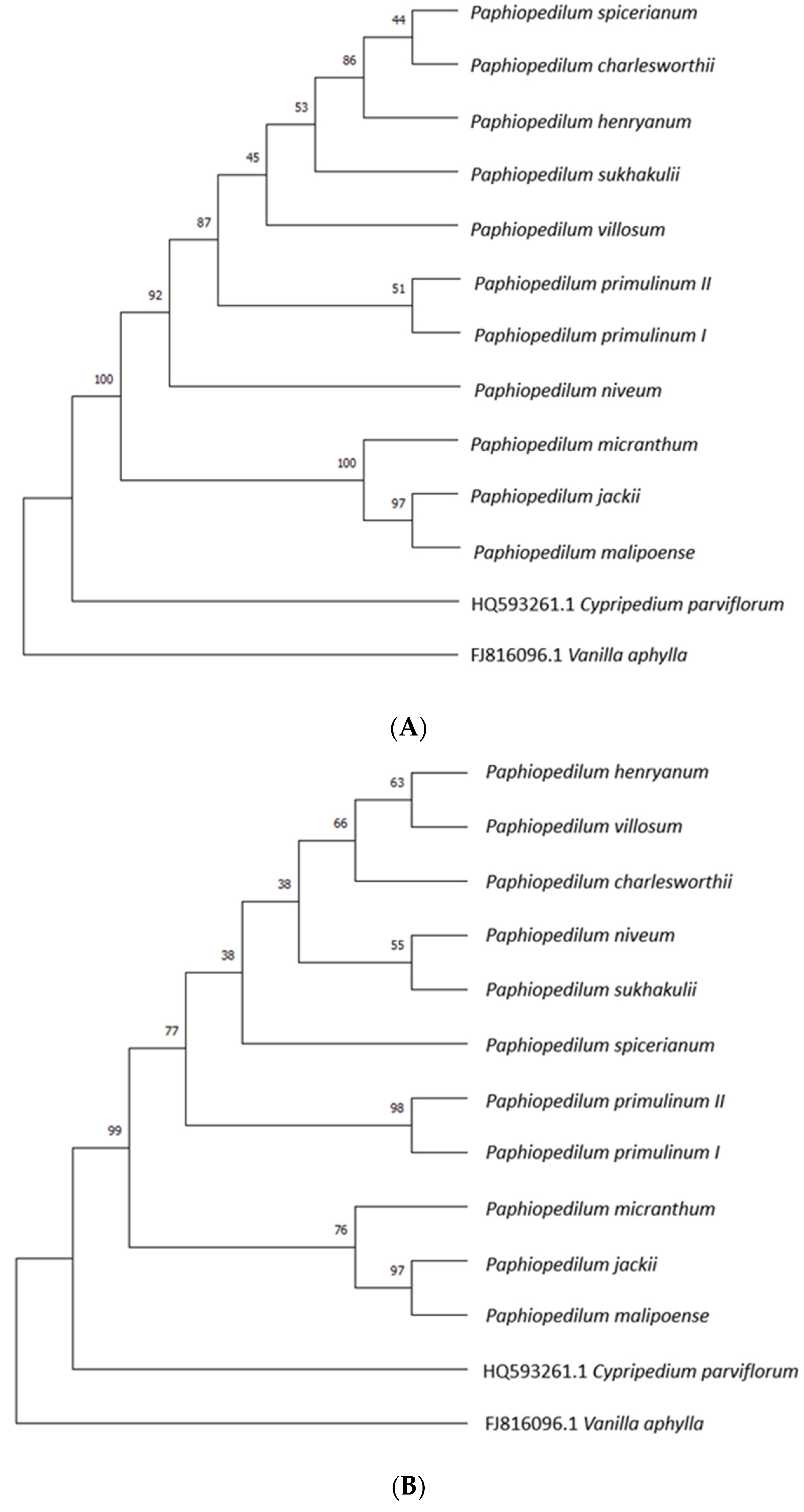
| Species | P. henryanum | P. villosum | P. spicerianum | P. primulinum II | P. jackii | P. charlesworthii | P. malipoense | P. micranthum | P. niveum | P. primulinum I | P. sukhakulii |
|---|---|---|---|---|---|---|---|---|---|---|---|
| P. henryanum | 97.9% | 97.9% | 95.6% | 93.8% | 98.5% | 94.2% | 95.4% | 91.0% | 95.8% | 95.6% | |
| P. villosum | 99.0% | 97.1% | 94.8% | 93.0% | 97.7% | 93.4% | 94.6% | 90.4% | 95.0% | 96.4% | |
| P. spicerianum | 98.6% | 98.6% | 95.8% | 94.2% | 98.1% | 94.6% | 95.8% | 90.8% | 96.0% | 95.8% | |
| P. primulinum II | 96.0% | 95.8% | 95.0% | 92.5% | 95.6% | 92.5% | 94.0% | 88.6% | 99.7% | 93.4% | |
| P. jackii | 94.2% | 93.5% | 92.9% | 96.3% | 94.0% | 98.9% | 96.2% | 87.2% | 92.3% | 91.7% | |
| P. charlesworthii | 96.5% | 95.6% | 95.2% | 98.8% | 96.7% | 94.8% | 95.6% | 90.8% | 95.8% | 95.8% | |
| P. malipoense | 93.7% | 93.0% | 92.5% | 95.7% | 99.4% | 96.2% | 96.2% | 87.6% | 92.7% | 92.3% | |
| P. micranthum | 95.5% | 95.5% | 95.3% | 92.4% | 94.5% | 92.2% | 94.2% | 88.6% | 93.8% | 93.6% | |
| P. niveum | 96.5% | 96.6% | 95.8% | 93.9% | 92.2% | 93.2% | 91.7% | 95.6% | 88.8% | 89.4% | |
| P. primulinum I | 97.3% | 97.3% | 96.4% | 98.3% | 95.8% | 97.7% | 95.3% | 93.8% | 95.1% | 93.6% | |
| P. sukhakulii | 95.3% | 94.8% | 94.0% | 98.1% | 95.2% | 97.9% | 94.7% | 91.2% | 92.5% | 97.0% |
| Molecular Identification | rbcL | |||
|---|---|---|---|---|
| Accession Number | Similarity | Best Match Sequence | E Value | |
| P. sukhakulii | OR786325 | 98.27% | NC_069897.1 | 0.0 |
| P. primulinum I | OR786326 | 99.62% | KX755536.1 | 0.0 |
| P. niveum | OR786327 | 99.62% | MG522891.1 | 0.0 |
| P. micranthum | OR786328 | 99.39% | NC_045278.1 | 0.0 |
| P. malipoense | OR786329 | 97.46% | KX264992.1 | 0.0 |
| P. charlesworthii | OR786330 | 98.63% | OL875129.1 | 0.0 |
| P. jackii | OR786331 | 99.42% | NC_069882.1 | 0.0 |
| P. primulinum II | OR786332 | 99.41% | KX755536.1 | 0.0 |
| P. spicerianum | OR786333 | 98.86% | OM066324.1 | 0.0 |
| P. villosum | OR786334 | 98.15% | NC_069906.1 | 3.00 × 10−127 |
| P. henryanum | OR786335 | 97.65% | OM066293.1 | 0.0 |
| Molecular Identification | matK | |||
| Accession Number | Similarity | Best Match Sequence | E Value | |
| P. sukhakulii | OR772083 | 99.66% | NC_069897.1 | 0.0 |
| P. primulinum I | OR772084 | 99.53% | NC_069888.1 | 0.0 |
| P. niveum | OR772085 | 98.82% | NC_026776.1 | 0.0 |
| P. micranthum | OR772086 | 99.02% | KX886268.1 | 0.0 |
| P. malipoense | OR772087 | 99.65% | NC_069881.1 | 0.0 |
| P. charlesworthii | OR772088 | 99.89% | OL875129.1 | 0.0 |
| P. jackii | OR772089 | 99.88% | NC_069882.1 | 0.0 |
| P. primulinum II | OR772090 | 99.66% | NC_069888.1 | 0.0 |
| P. spicerianum | OR772091 | 99.28% | OM066324.1 | 0.0 |
| P. villosum | OR772092 | 99.64% | NC_069906.1 | 0.0 |
| P. henryanum | OR772093 | 99.88% | KY966920.1 | 0.0 |
| Molecular Identification | ITS2 | |||
| Accession Number | Similarity | Best Match Sequence | E Value | |
| P. sukhakulii | OR671174 | 99.43% | JQ929349.1 | 1.00 × 10−178 |
| P. primulinum I | OR671175 | 99.45% | AY643439.1 | 0.0 |
| P. niveum | OR671176 | 99.45% | AY643436.1 | 0.0 |
| P. micranthum | OR671177 | 99.50% | KX931039.1 | 0.0 |
| P. malipoense | OR671178 | 99.20% | HQ123427.1 | 0.0 |
| P.charlesworthii | OR671179 | 99.44% | JQ929310.1 | 0.0 |
| P. jackii | OR671180 | 99.44% | MH550872.1 | 8.00 × 10−180 |
| P. primulinum II | OR671181 | 99.45% | AY643439.1 | 0.0 |
| P. spicerianum | OR671182 | 99.45% | HQ998468.1 | 0.0 |
| P. villosum | OR671183 | 99.71% | HQ998477.1 | 7.00 × 10−176 |
| P. henryanum | OR671184 | 99.46% | AY643445.1 | 0.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karbarz, M.; Szlachcikowska, D.; Zapał, A.; Leśko, A. Unlocking the Genetic Identity of Endangered Paphiopedilum Orchids: A DNA Barcoding Approach. Genes 2024, 15, 689. https://doi.org/10.3390/genes15060689
Karbarz M, Szlachcikowska D, Zapał A, Leśko A. Unlocking the Genetic Identity of Endangered Paphiopedilum Orchids: A DNA Barcoding Approach. Genes. 2024; 15(6):689. https://doi.org/10.3390/genes15060689
Chicago/Turabian StyleKarbarz, Małgorzata, Dominika Szlachcikowska, Angelika Zapał, and Agnieszka Leśko. 2024. "Unlocking the Genetic Identity of Endangered Paphiopedilum Orchids: A DNA Barcoding Approach" Genes 15, no. 6: 689. https://doi.org/10.3390/genes15060689
APA StyleKarbarz, M., Szlachcikowska, D., Zapał, A., & Leśko, A. (2024). Unlocking the Genetic Identity of Endangered Paphiopedilum Orchids: A DNA Barcoding Approach. Genes, 15(6), 689. https://doi.org/10.3390/genes15060689





