Robertsonian Translocation between Human Chromosomes 21 and 22, Inherited across Three Generations, without Any Phenotypic Effect
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of Chromosomes and Nuclei and Chromosomal Banding
2.2. DNA Probes, In Situ Hybridization, and Detection
2.3. Radial Nuclear Location Analysis
3. Results
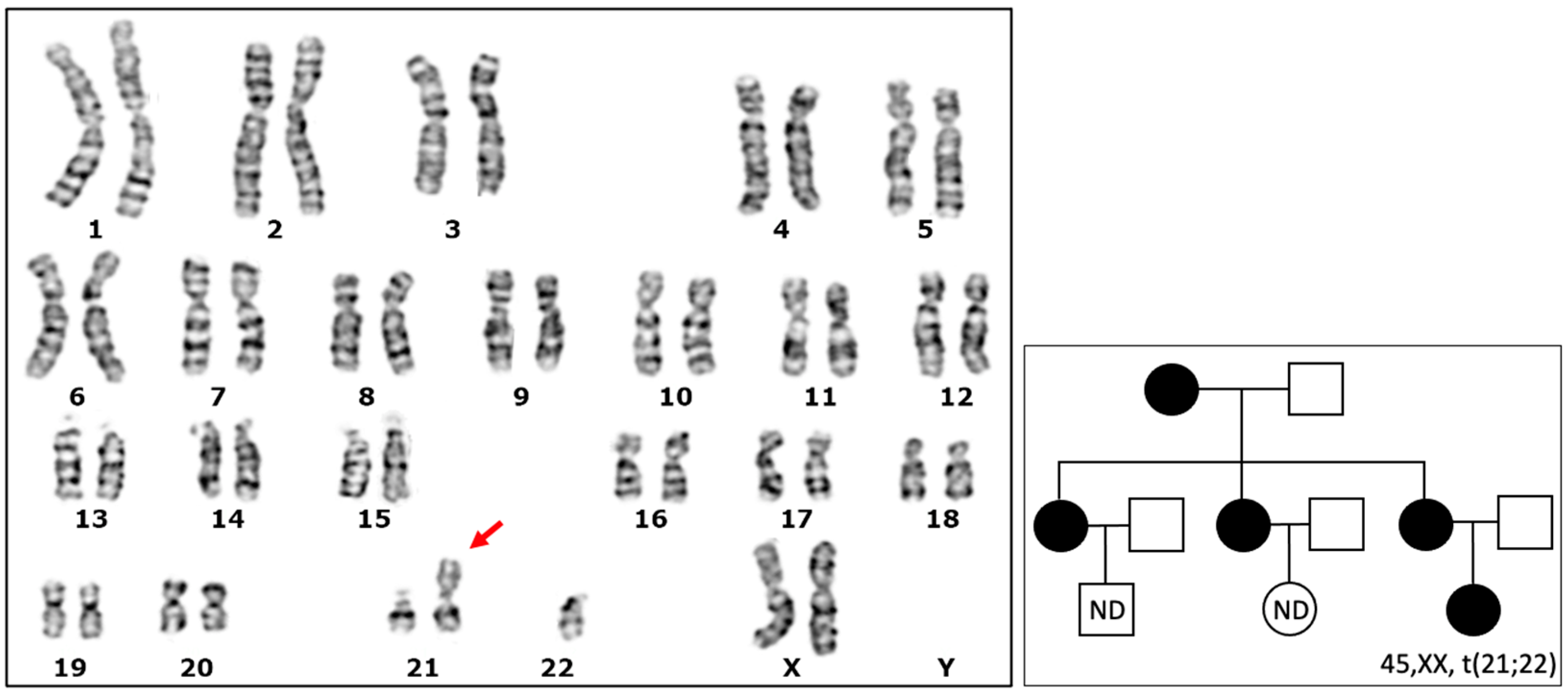
3.1. Detection of a Karyotype with a Robertsonian Translocation
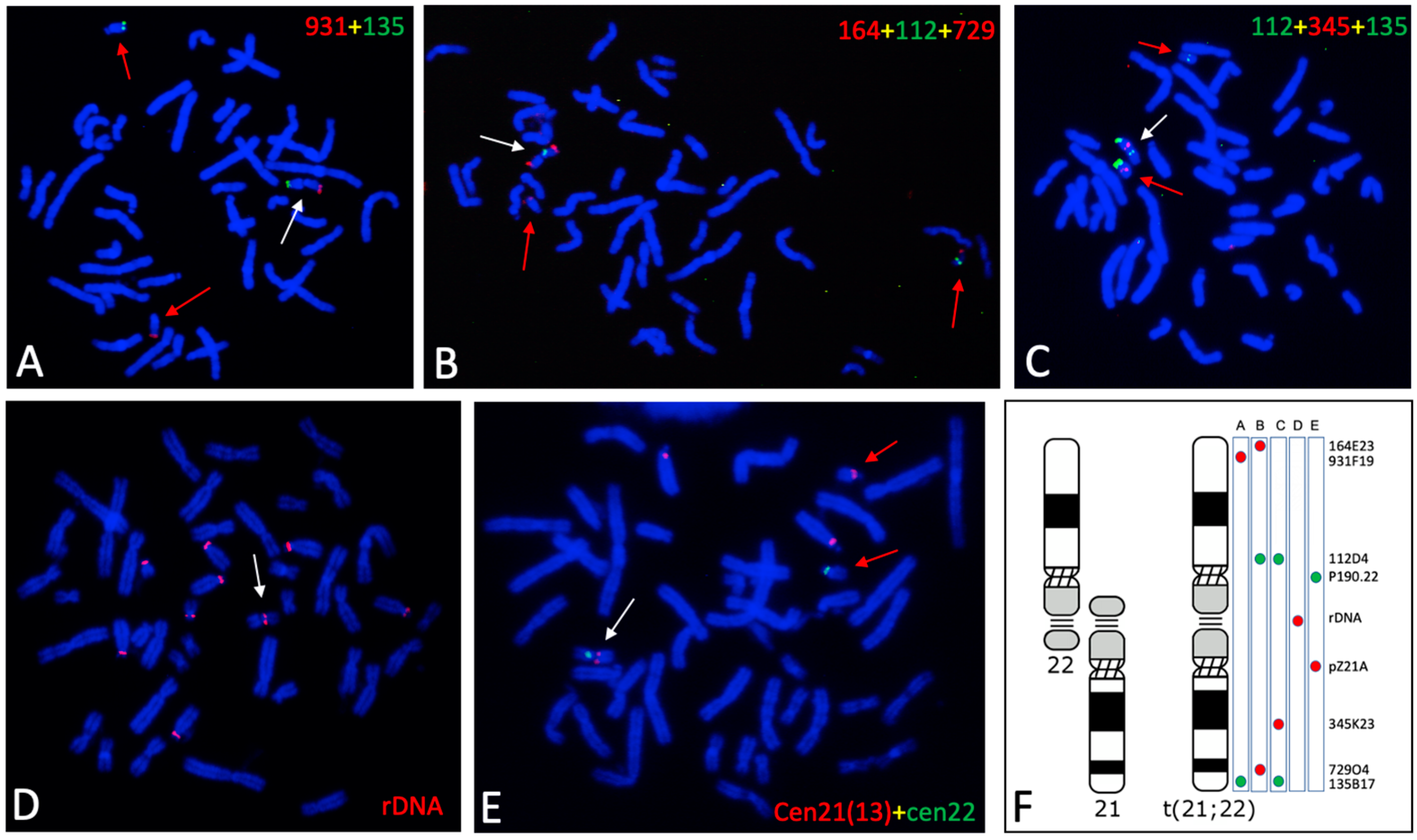
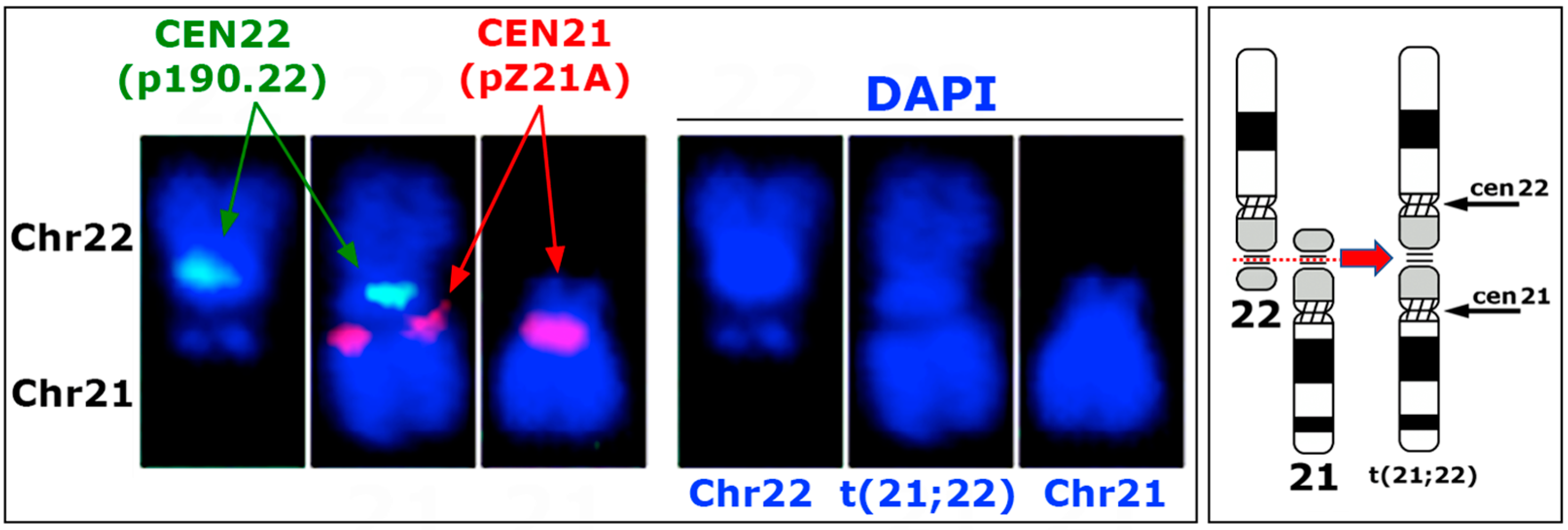
3.2. Genomic Features of the t(21:22) Chromosome
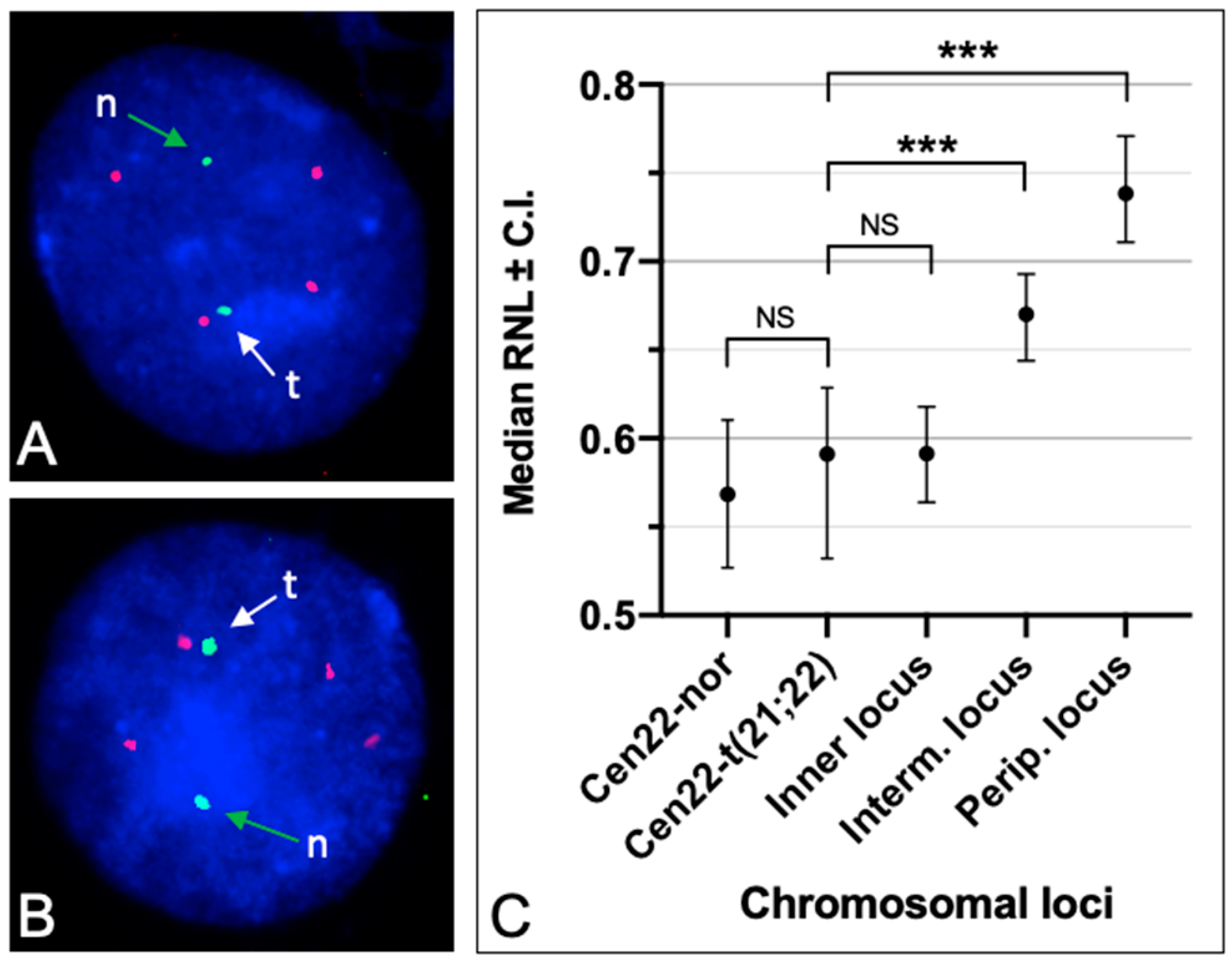
3.3. Radial Nuclear Positioning of the t(21;22) Chromosome
4. Discussion
4.1. Chromosomes and Genes in the Interphase Nucleus
4.2. Chromosomal Rearrangements and Centromere Fate
4.3. Centromere Formation and Inactivation
4.4. The Position Effect of Genes in the Nucleus
4.5. Robertsonian Translocation and Chromosome Transmission
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Frönicke, L.; Scherthan, H. Zoo-fluorescence in situ hybridization analysis of human and Indian muntjac karyotypes (Muntiacus muntjak vaginalis) reveals satellite DNA clusters at the margins of conserved syntenic segments. Chromosome Res. 1997, 5, 254–261. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; O’Brien, P.C.M.; Wienberg, J.; Neitzel, H.; Lin, C.C.; Ferguson-Smith, M.A. Chromosomal evolution of the Chinese muntjac (Muntiacus reevesi). Chromosoma 1997, 106, 37–43. [Google Scholar] [CrossRef]
- Mudd, A.B.; Bredeson, J.V.; Baum, R.; Hockemeyer, D.; Rokhsar, D.S. Analysis of muntjac deer genome and chromatin architecture reveals rapid karyotype evolution. Commun. Biol. 2020, 3, 480. [Google Scholar] [CrossRef] [PubMed]
- Iannuzzi, L.; Meo, G.P.; Perucatti, A. An improved characterization of goat chromosomes by means of G- and R-band comparison. Hereditas 1994, 120, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Iannuzzi, L.; Di Meo, G.P. Chromosomal evolution in bovids: A comparison of cattle, sheep and goat G- and R-banded chromosomes and cytogenetic divergences among cattle, goat and river buffalo sex chromosomes. Chromosome Res. 1995, 3, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Deakin, J.E.; Kruger-Andrzejewska, M. Marsupials as models for understanding the role of chromosome rearrangements in evolution and disease. Chromosoma 2016, 125, 633–644. [Google Scholar] [CrossRef] [PubMed]
- Burt, D.W. Origin and evolution of avian microchromosomes. Cytogenet. Cell Genet. 2002, 96, 97–112. [Google Scholar] [CrossRef] [PubMed]
- Kiazim, L.G.; O’connor, R.E.; Larkin, D.M.; Romanov, M.N.; Narushin, V.G.; Brazhnik, E.A.; Griffin, D.K. Comparative Mapping of the Macrochromosomes of Eight Avian Species Provides Further Insight into Their Phylogenetic Relationships and Avian Karyotype Evolution. Cells 2021, 10, 362. [Google Scholar] [CrossRef] [PubMed]
- De Boer, L.E.M.; Sinoo, R.P. A karyological study of Accipitridae (Aves: Falconiformes), with karyotypic description of 16 species new to cytology. Genetica 1984, 65, 89–107. [Google Scholar] [CrossRef]
- Bed’Hom, B.; Coullin, P.; Guillier-Gencik, Z.; Moulin, S.; Bernheim, A.; Volobouev, V. Characterization of the atypical karyotype of the black-winged kite Elanus caeruleus (Falconiformes: Accipitridae) by means of classical and molecular cytogenetic techniques. Chromosome Res. 2003, 11, 335–343. [Google Scholar] [CrossRef]
- Federico, C.; Cantarella, C.D.; Scavo, C.; Saccone, S.; Bed’Hom, B.; Bernardi, G. Avian Genomes: Different karyotypes but a similar distribution of the GC-richest chromosome regions at interphase. Chromosome Res. 2005, 13, 785–793. [Google Scholar] [CrossRef] [PubMed]
- Deakin, J.E.; Ezaz, T. Understanding the Evolution of Reptile Chromosomes through Applications of Combined Cytogenetics and Genomics Approaches. Cytogenet. Genome Res. 2019, 157, 7–20. [Google Scholar] [CrossRef] [PubMed]
- Session, S.K. Evolutionary cytogenetics in salamanders. Chromosome Res. 2008, 16, 183–201. [Google Scholar] [CrossRef]
- Bertollo, L.A.C. Chromosome evolution in the Neotropical Erythrinidae fish family: An overview. In Fish Cytogenetics; Pizano, E., Ozouf-Costaz, C., Foresti, F., Kapoor, B.G., Eds.; Science Publishers: Enfield, UK, 2007; pp. 195–211. [Google Scholar]
- Cioffi, M.; Liehr, T.; Trifonov, V.; Molina, W.; Bertollo, L. Independent sex chromosome evolution in lower vertebrates: A molecular cytogenetic overview in the Erythrinidae fish family. Cytogenet. Genome Res. 2013, 141, 86–194. [Google Scholar] [CrossRef]
- de Oliveira, E.A.; Bertollo, L.A.C.; Yano, C.F.; Liehr, T.; Cioffi, M.d.B. Comparative cytogenetics in the genus Hoplias (Characiformes, Erythrinidae) highlights contrasting karyotype evolution among congeneric species. Mol. Cytogenet. 2015, 8, 56. [Google Scholar] [CrossRef]
- Seuánez, H.N. The Phylogeny of Human Chromosomes; Springer: Berlin/Heidelberg, Germany, 1979. [Google Scholar]
- Müller, S.; Stanyon, R.; O’Brien, P.C.M.; Ferguson-Smith, M.A.; Plesker, R.; Wienberg, J. Defining the ancestral karyotype of all primates by multidirectional chromosome painting between tree shrews, lemurs and humans. Chromosoma 1999, 108, 393–400. [Google Scholar] [CrossRef]
- Neusser, M.; Schubel, V.; Koch, A.; Cremer, T.; Müller, S. Evolutionary conserved, cell type and species-specific higher order chromatin arrangements in interphase nuclei of primates. Chromosoma 2007, 116, 307–320. [Google Scholar] [CrossRef]
- Stanyon, R.; Rocchi, M.; Capozzi, O.; Roberto, R.; Misceo, D.; Ventura, M.; Cardone, M.F.; Bigoni, F.; Archidiacono, N. Primate chromosome evolution: Ancestral karyotypes, marker order and neocentromeres. Chromosome Res. 2008, 16, 17–39. [Google Scholar] [CrossRef] [PubMed]
- Ijdo, J.W.; Baldini, A.; Ward, D.C.; Reeders, S.T.; A Wells, R. Origin of human chromosome 2: An ancestral telomere-telomere fusion. Proc. Natl. Acad. Sci. USA 1991, 88, 9051–9055. [Google Scholar] [CrossRef]
- Baldini, A.; Ried, T.; Shridhar, V.; Ogura, K.; D’Aiuto, L.; Rocchi, M.; Ward, D.C. An alphoid DNA sequence conserved in all human and great ape chromosomes: Evidence for ancient centromeric sequences at human chromosomal regions 2q21 and 9q13. Hum. Genet. 1993, 90, 577–583. [Google Scholar] [CrossRef]
- Müller, S.; Finelli, P.; Neusser, M.; Wienberg, J. The evolutionary history of human chromosome 7. Genomics 2004, 84, 458–467. [Google Scholar] [PubMed]
- Petersen, M.B.; Adelsberger, P.A.; Schinzel, A.A.; Binkert, F.; Hinkel, G.K.; Antonarakis, S.E. Down syndrome due to de novo Robertsonian translocation t(14q;21q): DNA polymorphism analysis suggests that the origin of the extra 21q is maternal. Am. J. Hum. Genet. 1991, 49, 529–536. [Google Scholar] [PubMed]
- Kolgeci, S.; Azemi, M.; Ahmeti, H.; Dervishi, Z.; Sopjani, M.; Kolgeci, J. Recurrent abortions and down syndrome resulting from Robertsonian translocation 21q; 21q. Med. Arch. 2012, 66, 350–352. [Google Scholar] [CrossRef] [PubMed]
- Molyneux, E.M.; Rochford, R.; Griffin, B.; Newton, R.; Jackson, G.; Menon, G.; Harrison, C.J.; Israels, T.; Bailey, S. Burkitt’s lymphoma. Lancet 2012, 379, 1234–1244. [Google Scholar] [CrossRef]
- Said, J.; Lones, M.; Yea, S. Burkitt lymphoma and MYC: What else is new? Adv. Anat. Pathol. 2014, 21, 160–165. [Google Scholar] [CrossRef]
- LLin, X.; Qureshi, M.Z.; Attar, R.; Khalid, S.; Tahir, F.; Yaqub, A.; Aslam, A.; Yaylim, I.; Back, L.K.D.C.; A Farooqi, A.; et al. Targeting of BCR-ABL: Lessons learned from BCR-ABL inhibition. Cell Mol. Biol. 2016, 62, 129–137. [Google Scholar]
- Tosi, S.; Harbott, J.; Teigler-schlegel, A.; Haas, O.A.; Pirc-Danoewinata, H.; Harrison, C.J.; Biondi, A.; Cazzaniga, G.; Kempski, H.; Scherer, S.W.; et al. t(7;12)(q36;p13), a new recurrent translocation involving ETV6 in infant leukemia. Genes Chromosomes Cancer 2000, 29, 325–332. [Google Scholar] [CrossRef]
- Ballabio, E.; Cantarella, C.D.; Federico, C.; Di Mare, P.; Hall, G.; Harbott, J.; Hughes, J.; Saccone, S.; Tosi, S. Ectopic expression of the HLXB9 gene is associated with an altered nuclear position in t(7;12) leukaemias. Leukemia 2009, 23, 1179–1182. [Google Scholar] [CrossRef]
- Gulino, G.M.; Bruno, F.; Sturiale, V.; Brancato, D.; Ragusa, D.; Tosi, S.; Saccone, S.; Federico, C. From FISH to Hi-C: The chromatin architecture of the chromosomal region 7q36.3, frequently rearranged in leukemic cells, is evolutionary conserved. Int. J. Mol. Sci. 2021, 22, 2338. [Google Scholar] [CrossRef]
- Ragusa, D.; Cicirò, Y.; Federico, C.; Saccone, S.; Bruno, F.; Saeedi, R.; Sisu, C.; Pina, C.; Sala, A.; Tosi, S. Engineered model of t(7;12)(q36;p13) AML recapitulates patient-specific features and gene expression profiles. Oncogenesis 2022, 11, 50. [Google Scholar] [CrossRef]
- Garimberti, E.; Federico, C.; Ragusa, D.; Bruno, F.; Saccone, S.; Bridger, J.M.; Tosi, S. Alterations of the genome organization in lymphoma cell nuclei due to the presence of t(14;18) translocation. Int. J. Mol. Sci. 2024, 25, 2377. [Google Scholar] [CrossRef] [PubMed]
- Federico, C.; Owoka, T.; Ragusa, D.; Sturiale, V.; Caponnetto, D.; Leotta, C.G.; Bruno, F.; Foster, H.A.; Rigamonti, S.; Giudici, G.; et al. Deletions of Chromosome 7q Affect Nuclear Organization and HLXB9Gene Expression in Hematological Disorders. Cancers 2019, 11, 585. [Google Scholar] [CrossRef] [PubMed]
- Federico, C.; Pappalardo, A.M.; Ferrito, V.; Tosi, S.; Saccone, S. Genomic properties of chromosomal bands are linked to evolutionary rearrangements and new centromere formation in primates. Chromosome Res. 2017, 25, 261–276. [Google Scholar] [CrossRef] [PubMed]
- Kowalczyk, M.; Srebniak, M.; Tomaszewska, A. Chromosome abnormalities without phenotypic consequences. J. Appl. Genet. 2007, 48, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Archidiacono, N.; Antonacci, R.; Marzella, R.; Finelli, P.; Lonoce, A.; Rocchi, M. Comparative mapping of human alphoid sequences in great apes, using fluorescence in situ hybridization. Genomics 1995, 25, 477–484. [Google Scholar] [PubMed]
- Rocchi, M.; Archidiacono, N.; Antonacci, R.; Finelli, P.; D’Aiuto, L.; Carbone, R.; Lindsay, E.; Baldini, A. Cloning and comparative mapping of recently evolved human chromosome 22-specific α satellite DNA. Somat. Cell Mol. Genet. 1994, 20, 443–448. [Google Scholar] [CrossRef]
- Bucksch, M.; Ziegler, M.; Kosayakova, N.; Mulatinho, M.V.; Llerena, J.C.; Morlot, S.; Fischer, W.; Polityko, A.D.; Kulpanovich, A.I.; Petersen, M.B.; et al. A new multicolor fluorescence in situ hybridization probe set directed against heterochromatin: HCM-FISH. J. Histochem. Cytochem. 2012, 60, 530–536. [Google Scholar] [CrossRef] [PubMed]
- Federico, C.; Cantarella, C.D.; Di Mare, P.; Tosi, S.; Saccone, S. The radial arrangement of the human chromosome 7 in the lymphocyte cell nucleus is associated with chromosomal band gene density. Chromosoma 2008, 117, 399–410. [Google Scholar] [CrossRef]
- Saccone, S.; Federico, C.; Bernardi, G. Localization of the gene-richest and the gene-poorest isochores in the interphase nuclei of mammals and birds. Gene 2002, 300, 169–178. [Google Scholar] [CrossRef]
- Sadoni, N.; Langer, S.; Fauth, C.; Bernardi, G.; Cremer, T.; Turner, B.M.; Zink, D. Nuclear organization of mammalian genomes: Polar chromosome territories build up functionally distinct higher order compartments. J. Cell Biol. 1999, 146, 1211–1226. [Google Scholar] [CrossRef]
- Szczerbal, I.; Foster, H.A.; Bridger, J.M. The spatial repositioning of adipogenesis genes is correlated with their expression status in a porcine mesenchymal stem cell adipogenesis model system. Chromosoma 2009, 118, 647–663. [Google Scholar] [CrossRef] [PubMed]
- Foster, H.A.; Bridger, J.M. The genome and the nucleus: A marriage made by evolution. Genome organization and nuclear architecture. Chromosoma 2005, 114, 212–229. [Google Scholar] [CrossRef] [PubMed]
- Mora, L.; Sánchez, I.; Garcia, M.; Ponsà, M. Chromosome territory positioning of conserved homologous chromosomes in different primate species. Chromosoma 2006, 115, 367–375. [Google Scholar] [CrossRef] [PubMed]
- Bridger, J.; Boyle, S.; Kill, I.; Bickmore, W. Re-modelling of nuclear architecture in quiescent and senescent human fibroblasts. Curr. Biol. 2000, 10, 149–152. [Google Scholar] [CrossRef]
- Simonis, M.; Klous, P.; Splinter, E.; Moshkin, Y.; Willemsen, R.; de Wit, E.; van Steensel, B.; de Laat, W. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture on-chip (4C). Nat. Genet. 2006, 38, 1348–1354. [Google Scholar] [CrossRef] [PubMed]
- Lieberman-Aiden, E.; Van Berkum, N.L.; Williams, L.; Imakaev, M.; Ragoczy, T.; Telling, A.; Amit, I.; Lajoie, B.R.; Sabo, P.J.; Dorschner, M.O.; et al. Comprehensive mapping of long range interactions reveals folding principles of the human genome. Science 2009, 326, 289–293. [Google Scholar] [CrossRef]
- Rao, S.S.P.; Huntley, M.H.; Durand, N.C.; Stamenova, E.K.; Bochkov, I.D.; Robinson, J.T.; Sanborn, A.L.; Machol, I.; Omer, A.D.; Lander, E.S.; et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 2014, 159, 1665–1680. [Google Scholar] [CrossRef] [PubMed]
- Bickmore, W.A.; van der Maarel, S.M. Perturbations of chromatin structure in human genetic disease: Recent advances. Hum. Mol. Genet. 2003, 12, 207–213. [Google Scholar] [CrossRef]
- Federico, C.; Bruno, F.; Ragusa, D.; Clements, C.S.; Brancato, D.; Henry, M.P.; Bridger, J.M.; Tosi, S.; Saccone, S. Chromosomal rearrangements and altered nuclear organization: Recent mechanistic models in cancer. Cancers 2021, 13, 5869. [Google Scholar] [CrossRef]
- Therman, E.; Susman, B.; Denniston, C. The nonrandom participation of human acrocentric chromosomes in Robertsonian translocations. Ann. Hum. Genet. 1989, 53, 49–65. [Google Scholar] [CrossRef]
- Heng, H.H. Genome Chaos: Rethinking Genetics, Evolution, and Molecular Medicine; Academic Press Elsevier: Cambridge, MA, USA, 2019; ISBN 978-012-8136-35-5. [Google Scholar]
- Ye, C.J.; Stilgenbauer, L.; Moy, A.; Liu, G.; Heng, H.H. What Is Karyotype Coding and Why Is Genomic Topology Important for Cancer and Evolution? Front. Genet. 2019, 10, 1082. [Google Scholar] [CrossRef] [PubMed]
- Bernardi, G. Structural and Evolutionary Genomics: Natural Selection in Genome Evolution; Elsevier: Amsterdam, The Netherlands, 2005; ISBN 9780080461878. [Google Scholar]
- Jabbari, K.; Bernardi, G. An Isochore Framework Underlies Chromatin Architecture. PLoS ONE 2017, 12, e0168023. [Google Scholar] [CrossRef] [PubMed]
- Heng, H.H.; Liu, G.; Stevens, J.B.; Abdallah, B.Y.; Horne, S.D.; Ye, K.J.; Bremer, S.W.; Chowdhury, S.K.; Ye, C.J. Karyotype heterogeneity and unclassified chromosomal abnormalities. Cytogenet Genome Res. 2013, 139, 144–157. [Google Scholar] [CrossRef] [PubMed]
- Federico, C.; Scavo, C.; Cantarella, C.D.; Motta, S.; Saccone, S.; Bernardi, G. Gene-rich and gene-poor chromosomal regions have different locations in the interphase nuclei of cold-blooded vertebrates. Chromosoma 2006, 115, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Derenzini, M.; Pasquinelli, G.; O’Donohue, M.-F.; Ploton, D.; Thiry, M. Structural and Functional Organization of Ribosomal Genes within the Mammalian Cell Nucleolus. J. Histochem. Cytochem. 2006, 54, 131–145. [Google Scholar] [CrossRef] [PubMed]
- Stults, D.; Killen, M.W.; Williamson, E.P.; Hourigan, J.S.; Vargas, H.D.; Arnold, S.M.; Moscow, J.A.; Pierce, A.J. Human rRNA gene clusters are recombinational hotspots in cancer. Cancer Res. 2009, 69, 9096–9104. [Google Scholar] [PubMed]
- Grummt, I.; Pikaard, C.S. Epigenetic silencing of RNA polymerase I transcription. Nat. Rev. Mol. Cell Biol. 2003, 4, 641–649. [Google Scholar] [CrossRef] [PubMed]
- Sanij, E.; Poortinga, G.; Sharkey, K.; Hung, S.; Holloway, T.P.; Quin, J.; Robb, E.; Wong, L.H.; Thomas, W.G.; Stefanovsky, V.; et al. UBF levels determine the number of active ribosomal RNA genes in mammals. J. Cell Biol. 2008, 183, 1259–1274. [Google Scholar] [CrossRef]
- Song, J.; Li, X.; Sun, L.; Xu, S.; Liu, N.; Yao, Y.; Liu, Z.; Wang, W.; Rong, H.; Wang, B. A family with Robertsonian translocation: A potential mechanism of speciation in humans. Mol. Cytogenet. 2016, 9, 48. [Google Scholar] [CrossRef]
- Choo, K.H. Centromere DNA dynamics: Latent centromeres and neocentromere formation. Am. J. Hum. Genet. 1997, 61, 1225–1233. [Google Scholar] [CrossRef]
- Ventura, M.; Weigl, S.; Carbone, L.; Cardone, M.F.; Misceo, D.; Teti, M.; D’Addabbo, P.; Wandall, A.; Björck, E.; de Jong, P.J.; et al. Recurrent sites for new centromere seeding. Genome Res. 2004, 14, 1696–1703. [Google Scholar] [CrossRef] [PubMed]
- Amor, D.J.; Choo, K.A. Neocentromeres: Role in human disease, evolution, and centromere study. Am. J. Hum. Genet. 2002, 71, 695–714. [Google Scholar] [CrossRef]
- Palazzo, A.; Piccolo, I.; Minervini, C.F.; Purgato, S.; Capozzi, O.; D’addabbo, P.; Cumbo, C.; Albano, F.; Rocchi, M.; Catacchio, C.R. Genome characterization and CRISPR-Cas9 editing of a human neocentromere. Chromosoma 2022, 131, 239–251. [Google Scholar] [CrossRef]
- Amor, D.J.; Bentley, K.; Ryan, J.; Perry, J.; Wong, L.; Slater, H.; Choo, K.H.A. Human centromere repositioning “in progress”. Proc. Natl. Acad. Sci. USA 2004, 101, 6542–6547. [Google Scholar] [CrossRef]
- Tolomeo, D.; Capozzi, O.; Stanyon, R.R.; Archidiacono, N.; D’addabbo, P.; Catacchio, C.R.; Purgato, S.; Perini, G.; Schempp, W.; Huddleston, J.; et al. Epigenetic origin of evolutionary novel centromeres. Sci. Rep. 2017, 7, 41980. [Google Scholar] [CrossRef]
- Ewers, E.; Yoda, K.; Hamid, A.B.; Weise, A.; Manvelyan, M.; Liehr, T. Centromere activity in dicentric small supernumerary marker chromosomes. Chromosome Res. 2010, 18, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Spittel, H.; Kubek, F.; Kreskowski, K.; Ziegler, M.; Klein, E.; Hamid, A.B.; Kosyakova, N.; Radhakrishnan, G.; Junge, A.; Kozlowski, P.; et al. Mitotic Stability of Small Supernumerary Marker Chromosomes: A Study Based on 93 Immortalized Cell Lines. Cytogenet. Genome Res. 2014, 142, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Lalchev, S.; Tzancheva, M.; Markova, R. A case of trisomy 22 with a probable Robertsonian translocation 21/22. Hum. Genet. 1978, 45, 219–223. [Google Scholar] [CrossRef] [PubMed]
- Cannizzaro, L.; Aronson, M.; Emanuel, B. In situ hybridization and translocation breakpoint mapping. II. Two unusual t(21;22) translocations. Cytogenet. Cell Genet. 1985, 39, 173–178. [Google Scholar] [CrossRef]
- Syme, R.; Martin, R. Meiotic segregation of a 21;22 Robertsonian translocation. Hum. Reprod. 1992, 7, 825–829. [Google Scholar] [CrossRef]
- Coyne, J.A.; Orr, H.A. The evolutionary genetics of speciation. Phil. Trans. R. Soc. Lond. B 1998, 353, 287–305. [Google Scholar] [CrossRef] [PubMed]

| BAC | Position (a) | Start (b) | End (b) | Size (c) |
|---|---|---|---|---|
| RP11-345K23 | 21q11.2 | 16,368,352 | 16,556,943 | 188,592 |
| RP11-729O4 | 21q22.2 | 40,844,581 | 41,029,286 | 184,706 |
| RP11-135B17 | 21q22.3 | 47,931,911 | 48,108,188 | 176,278 |
| RP11-112D4 | 22q11.21 | 18,202,100 | 18,385,443 | 183,343 |
| RP11-931F19 | 22q13.33 | 50,425,014 | 50,627,374 | 202,361 |
| RP11-164E23 | 22q13.33 | 50,841,120 | 50,995,394 | 154,275 |
| RP11-79O21 | 7p21.3 | 12,053,525 | 12,194,425 | 140,901 |
| RP11-17H7 | 7q15.1 | 28,171,887 | 28,341,999 | 170,112 |
| RP11-213E22 | 7q22.1 | 100,705,096 | 100,899,914 | 194,819 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Federico, C.; Brancato, D.; Bruno, F.; Galvano, D.; Caruso, M.; Saccone, S. Robertsonian Translocation between Human Chromosomes 21 and 22, Inherited across Three Generations, without Any Phenotypic Effect. Genes 2024, 15, 722. https://doi.org/10.3390/genes15060722
Federico C, Brancato D, Bruno F, Galvano D, Caruso M, Saccone S. Robertsonian Translocation between Human Chromosomes 21 and 22, Inherited across Three Generations, without Any Phenotypic Effect. Genes. 2024; 15(6):722. https://doi.org/10.3390/genes15060722
Chicago/Turabian StyleFederico, Concetta, Desiree Brancato, Francesca Bruno, Daiana Galvano, Mariella Caruso, and Salvatore Saccone. 2024. "Robertsonian Translocation between Human Chromosomes 21 and 22, Inherited across Three Generations, without Any Phenotypic Effect" Genes 15, no. 6: 722. https://doi.org/10.3390/genes15060722
APA StyleFederico, C., Brancato, D., Bruno, F., Galvano, D., Caruso, M., & Saccone, S. (2024). Robertsonian Translocation between Human Chromosomes 21 and 22, Inherited across Three Generations, without Any Phenotypic Effect. Genes, 15(6), 722. https://doi.org/10.3390/genes15060722







