Y Chromosome Story—Ancient Genetic Data as a Supplementary Tool for the Analysis of Modern Croatian Genetic Pool
Abstract
:1. Introduction
2. Modern Y-Chromosome Diversity of Southeastern Europe
2.1. Early Studies of Y-Chromosome Markers
2.2. Comprehensive Reviews and Recent Genetic Contributions
2.3. Settlement Patterns and Genetic Transitions in SEE
2.4. Dominant Y-Chromosome Haplogroups in Modern SEE Population
2.4.1. Haplogroup I2a
2.4.2. Haplogroup R1
2.4.3. Haplogroup E
2.4.4. Haplogroup G and J
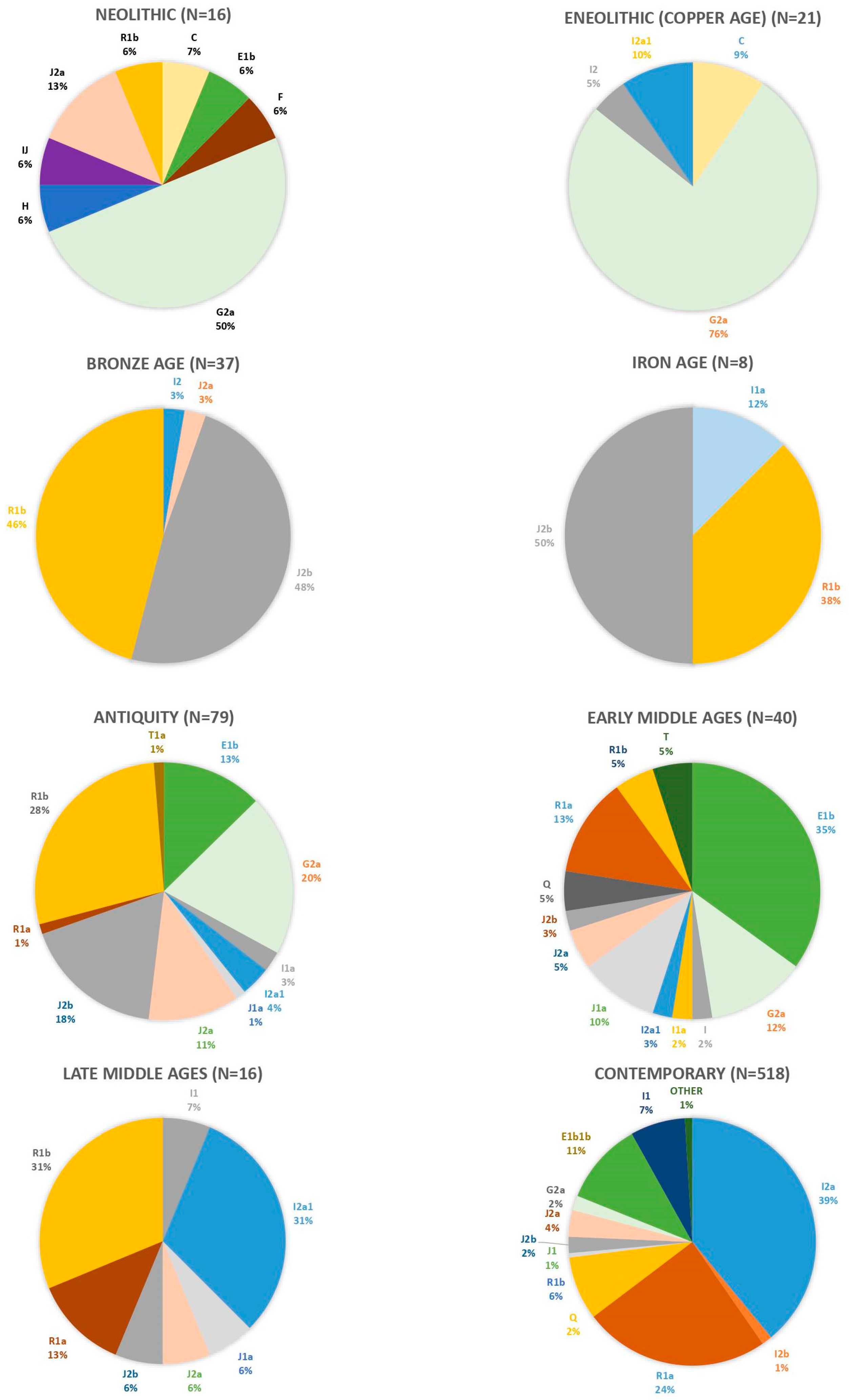
3. Ancient Y-Chromosome Diversity of Croatia and the Neighboring Populations
4. Future Developments in Y-Chromosome Mapping of Southeastern Europe
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zuckerkandl, E. Molecular Evolution as a Pathway to Man. Z. Morphol. Anthropol. 1978, 69, 117–142. [Google Scholar] [CrossRef] [PubMed]
- Jobling, M.A.; Tyler-Smith, C. The Human Y Chromosome: An Evolutionary Marker Comes of Age. Nat. Rev. Genet. 2003, 4, 598–612. [Google Scholar] [CrossRef]
- Underhill, P.A.; Kivisild, T. Use of y Chromosome and Mitochondrial DNA Population Structure in Tracing Human Migrations. Annu. Rev. Genet. 2007, 41, 539–564. [Google Scholar] [CrossRef]
- Karavanić, I.; Janković, I. The Middle and Early Upper Paleolithic in Croatia. Opvscvla Archaeol. 2006, 30, 21–54. [Google Scholar]
- Vujević, D. Adriatic Connections: Exploring Relationships from the Middle Palaeolithic to the Mesolithic. In Croatia at the Crossroads: A Consideration of Archaeological and Historical Connectivity; Davison, D., Gaffney, V., Miracle, P.T., Sofaer, J., Eds.; Archaeopress: Oxford, UK, 2016; pp. 19–32. [Google Scholar]
- Forenbaher, S.; Kaiser, T.; Miracle, P.T. Dating the East Adriatic Neolithic. Eur. J. Archaeol. 2013, 16, 589–609. [Google Scholar] [CrossRef]
- Forenbaher, S. The Spread of Farming in the Eastern Adriatic. Antiquity 2005, 79, 514–528. [Google Scholar] [CrossRef]
- Mathieson, I.; Alpaslan-Roodenberg, S.; Posth, C.; Szécsényi-Nagy, A.; Rohland, N.; Mallick, S.; Olalde, I.; Broomandkhoshbacht, N.; Candilio, F.; Cheronet, O.; et al. The Genomic History of Southeastern Europe. Nature 2018, 555, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Reed, K. Agricultural Change in Copper Age Croatia (ca. 4500–2500 Cal B.C)? Archaeol. Anthropol. Sci. 2017, 9, 1745–1765. [Google Scholar] [CrossRef]
- Balen, J.; Miloglav, I.; Rajković, D. Back to the Past. Copper Age in Northern Croatia; Arheološki Muzej u Zagrebu, Filozofski Fakultet Sveučilišta u Zagrebu, Arheološki Muzej Osijek: Zagreb, Croatia; Osijek, Croatia, 2018. [Google Scholar]
- Novak, M.; Olalde, I.; Ringbauer, H.; Rohland, N.; Ahern, J.; Balen, J.; Janković, I.; Potrebica, H.; Pinhasi, R.; Reich, D. Genome-Wide Analysis of Nearly All the Victims of a 6200 Year Old Massacre. PLoS ONE 2021, 16, e0247332. [Google Scholar] [CrossRef] [PubMed]
- Haak, W.; Lazaridis, I.; Patterson, N.; Rohland, N.; Mallick, S.; Llamas, B.; Brandt, G.; Nordenfelt, S.; Harney, E.; Stewardson, K.; et al. Massive Migration from the Steppe Was a Source for Indo-European Languages in Europe. Nature 2015, 522, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Freilich, S.; Ringbauer, H.; Los, D.; Novak, M.; Pavičić, D.T.; Schiffels, S.; Pinhasi, R. Reconstructing Genetic Histories and Social Organisation in Neolithic and Bronze Age Croatia. Sci. Rep. 2021, 11, 16729. [Google Scholar] [CrossRef] [PubMed]
- Lazaridis, I.; Alpaslan-Roodenberg, S.; Acar, A.; Açıkkol, A.; Agelarakis, A.; Aghikyan, L.; Akyüz, U.; Andreeva, D.; Andrijašević, G.; Antonović, D.; et al. The Genetic History of the Southern Arc: A Bridge between West Asia and Europe. Science 2022, 377, eabm4247. [Google Scholar] [CrossRef]
- Patterson, N.; Isakov, M.; Booth, T.; Büster, L.; Fischer, C.-E.; Olalde, I.; Ringbauer, H.; Akbari, A.; Cheronet, O.; Bleasdale, M.; et al. Large-Scale Migration into Britain during the Middle to Late Bronze Age. Nature 2022, 601, 588–594. [Google Scholar] [CrossRef] [PubMed]
- Andrades Valtueña, A.; Mittnik, A.; Key, F.M.; Haak, W.; Allmäe, R.; Belinskij, A.; Daubaras, M.; Feldman, M.; Jankauskas, R.; Janković, I.; et al. The Stone Age Plague and Its Persistence in Eurasia. Curr. Biol. 2017, 27, 3683–3691.e8. [Google Scholar] [CrossRef] [PubMed]
- Geraghty, R.M. The Impact of Globalization in the Roman Empire, 200 BC-AD 100. J. Econ. Hist. 2007, 67, 1036–1061. [Google Scholar] [CrossRef]
- Hingley, R. Globalizing Roman Culture: Unity, Diversity and Empire; Routledge: London, UK, 2005. [Google Scholar]
- Olalde, I.; Carrión, P.; Mikić, I.; Rohland, N.; Mallick, S.; Lazaridis, I.; Mah, M.; Korać, M.; Golubović, S.; Petković, S.; et al. A Genetic History of the Balkans from Roman Frontier to Slavic Migrations. Cell 2023, 186, 5472–5485.e9. [Google Scholar] [CrossRef] [PubMed]
- Antonio, M.L.; Weiß, C.L.; Gao, Z.; Sawyer, S.; Oberreiter, V.; Moots, H.M.; Spence, J.P.; Cheronet, O.; Zagorc, B.; Praxmarer, E.; et al. Stable Population Structure in Europe since the Iron Age, despite High Mobility. Elife 2024, 13, e79714. [Google Scholar] [CrossRef]
- Harper, K. The Fate of Rome: Climate, Disease, and the End of an Empire; Princeton University Press: Princeton, NJ, USA, 2017. [Google Scholar]
- Amorim, C.E.G.; Vai, S.; Posth, C.; Modi, A.; Koncz, I.; Hakenbeck, S.; La Rocca, M.C.; Mende, B.; Bobo, D.; Pohl, W.; et al. Understanding 6th-Century Barbarian Social Organization and Migration through Paleogenomics. Nat. Commun. 2018, 9, 3547. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, D.; Sirak, K.; Cheronet, O.; Howcroft, R.; Čavka, M.; Los, D.; Burmaz, J.; Pinhasi, R.; Novak, M. Cranial Deformation and Genetic Diversity in Three Adolescent Male Individuals from the Great Migration Period from Osijek, Eastern Croatia. PLoS ONE 2019, 14, e0216366. [Google Scholar] [CrossRef] [PubMed]
- Maróti, Z.; Neparáczki, E.; Schütz, O.; Maár, K.; Varga, G.I.B.; Kovács, B.; Kalmár, T.; Nyerki, E.; Nagy, I.; Latinovics, D.; et al. The Genetic Origin of Huns, Avars, and Conquering Hungarians. Curr. Biol. 2022, 32, 2858–2870.e7. [Google Scholar] [CrossRef] [PubMed]
- Rosser, Z.H.; Zerjal, T.; Hurles, M.E.; Adojaan, M.; Alavantic, D.; Amorim, A.; Amos, W.; Armenteros, M.; Arroyo, E.; Barbujani, G.; et al. Y-Chromosomal Diversity in Europe Is Clinal and Influenced Primarily by Geography, Rather than by Language. Am. J. Hum. Genet. 2000, 67, 1526–1543. [Google Scholar] [CrossRef] [PubMed]
- Semino, O.; Passarino, G.; Oefner, P.J.; Lin, A.A.; Arbuzova, S.; Beckman, L.E.; De Benedictis, G.; Francalacci, P.; Kouvatsi, A.; Limborska, S.; et al. The Genetic Legacy of Paleolithic Homo Sapiens Sapiens in Extant Europeans: A Y Chromosome Perspective. Science 2000, 290, 1155–1159. [Google Scholar] [CrossRef] [PubMed]
- Semino, O.; Magri, C.; Benuzzi, G.; Lin, A.A.; Al-Zahery, N.; Battaglia, V.; Maccioni, L.; Triantaphyllidis, C.; Shen, P.; Oefner, P.J.; et al. Origin, Diffusion, and Differentiation of Y-Chromosome Haplogroups E and J: Inferences on the Neolithization of Europe and Later Migratory Events in the Mediterranean Area. Am. J. Hum. Genet. 2004, 74, 1023–1034. [Google Scholar] [CrossRef] [PubMed]
- Barać, L.; Pericić, M.; Klarić, I.M.; Rootsi, S.; Janićijević, B.; Kivisild, T.; Parik, J.; Rudan, I.; Villems, R.; Rudan, P. Y Chromosomal Heritage of Croatian Population and Its Island Isolates. Eur. J. Hum. Genet. 2003, 11, 535–542. [Google Scholar] [CrossRef] [PubMed]
- Marjanovic, D.; Fornarino, S.; Montagna, S.; Primorac, D.; Hadziselimovic, R.; Vidovic, S.; Pojskic, N.; Battaglia, V.; Achilli, A.; Drobnic, K.; et al. The Peopling of Modern Bosnia-Herzegovina: Y-Chromosome Haplogroups in the Three Main Ethnic Groups. Ann. Hum. Genet. 2005, 69, 757–763. [Google Scholar] [CrossRef] [PubMed]
- Pericić, M.; Barać Lauc, L.; Martinović Klarić, I.; Janićijević, B.; Rudan, P. Review of Croatian Genetic Heritage as Revealed by Mitochondrial DNA and Y Chromosomal Lineages. Croat. Med. J. 2005, 46, 502–513. [Google Scholar] [PubMed]
- Bosch, E.; Calafell, F.; González-Neira, A.; Flaiz, C.; Mateu, E.; Scheil, H.-G.; Huckenbeck, W.; Efremovska, L.; Mikerezi, I.; Xirotiris, N.; et al. Paternal and Maternal Lineages in the Balkans Show a Homogeneous Landscape over Linguistic Barriers, except for the Isolated Aromuns. Ann. Hum. Genet. 2006, 70, 459–487. [Google Scholar] [CrossRef] [PubMed]
- King, R.J.; Ozcan, S.S.; Carter, T.; Kalfoğlu, E.; Atasoy, S.; Triantaphyllidis, C.; Kouvatsi, A.; Lin, A.A.; Chow, C.-E.T.; Zhivotovsky, L.A.; et al. Differential Y-Chromosome Anatolian Influences on the Greek and Cretan Neolithic. Ann. Hum. Genet. 2008, 72, 205–214. [Google Scholar] [CrossRef]
- Battaglia, V.; Fornarino, S.; Al-Zahery, N.; Olivieri, A.; Pala, M.; Myres, N.M.; King, R.J.; Rootsi, S.; Marjanovic, D.; Primorac, D.; et al. Y-Chromosomal Evidence of the Cultural Diffusion of Agriculture in Southeast Europe. Eur. J. Hum. Genet. 2009, 17, 820–830. [Google Scholar] [CrossRef] [PubMed]
- Mirabal, S.; Varljen, T.; Gayden, T.; Regueiro, M.; Vujovic, S.; Popovic, D.; Djuric, M.; Stojkovic, O.; Herrera, R.J. Human Y-Chromosome Short Tandem Repeats: A Tale of Acculturation and Migrations as Mechanisms for the Diffusion of Agriculture in the Balkan Peninsula. Am. J. Phys. Anthropol. 2010, 142, 380–390. [Google Scholar] [CrossRef]
- Boattini, A.; Martinez-Cruz, B.; Sarno, S.; Harmant, C.; Useli, A.; Sanz, P.; Yang-Yao, D.; Manry, J.; Ciani, G.; Luiselli, D.; et al. Uniparental Markers in Italy Reveal a Sex-Biased Genetic Structure and Different Historical Strata. PLoS ONE 2013, 8, e65441. [Google Scholar] [CrossRef]
- Primorac, D.; Marjanović, D.; Rudan, P.; Villems, R.; Underhill, P.A. Croatian Genetic Heritage: Y-Chromosome Story. Croat. Med. J. 2011, 52, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Regueiro, M.; Rivera, L.; Damnjanovic, T.; Lukovic, L.; Milasin, J.; Herrera, R.J. High Levels of Paleolithic Y-Chromosome Lineages Characterize Serbia. Gene 2012, 498, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Šarac, J.; Šarić, T.; Havaš Auguštin, D.; Novokmet, N.; Vekarić, N.; Mustać, M.; Grahovac, B.; Kapović, M.; Nevajda, B.; Glasnović, A.; et al. Genetic Heritage of Croatians in the Southeastern European Gene Pool-Y Chromosome Analysis of the Croatian Continental and Island Population. Am. J. Hum. Biol. 2016, 28, 837–845. [Google Scholar] [CrossRef]
- Primorac, D.; Škaro, V.; Projić, P.; Missoni, S.; Horjan Zanki, I.; Merkaš, S.; Šarac, J.; Novokmet, N.; Ledić, A.; Makar, A.; et al. Croatian Genetic Heritage: An Updated Y-Chromosome Story. Croat. Med. J. 2022, 63, 273–286. [Google Scholar] [CrossRef] [PubMed]
- Karmin, M.; Saag, L.; Vicente, M.; Wilson Sayres, M.A.; Järve, M.; Talas, U.G.; Rootsi, S.; Ilumäe, A.-M.; Mägi, R.; Mitt, M.; et al. A Recent Bottleneck of Y Chromosome Diversity Coincides with a Global Change in Culture. Genome Res. 2015, 25, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Scorrano, G.; Finocchio, A.; De Angelis, F.; Martínez-Labarga, C.; Šarac, J.; Contini, I.; Scano, G.; Novokmet, N.; Frezza, D.; Rickards, O. The Genetic Landscape of Serbian Populations through Mitochondrial DNA Sequencing and Non-Recombining Region of the Y Chromosome Microsatellites. Coll. Antropol. 2017, 41, 275–296. [Google Scholar]
- Marjanović, D. Preci u Nama—Genetičko Blago Bosne i Hercegovine. Available online: https://shop.skolskaknjiga.hr/preci-u-nama-geneticko-blago-bosne-i-hercegovine.html (accessed on 19 January 2024).
- Kovacevic, L.; Tambets, K.; Ilumäe, A.-M.; Kushniarevich, A.; Yunusbayev, B.; Solnik, A.; Bego, T.; Primorac, D.; Skaro, V.; Leskovac, A.; et al. Standing at the Gateway to Europe--the Genetic Structure of Western Balkan Populations Based on Autosomal and Haploid Markers. PLoS ONE 2014, 9, e105090. [Google Scholar] [CrossRef] [PubMed]
- Rootsi, S.; Magri, C.; Kivisild, T.; Benuzzi, G.; Help, H.; Bermisheva, M.; Kutuev, I.; Barać, L.; Pericić, M.; Balanovsky, O.; et al. Phylogeography of Y-Chromosome Haplogroup I Reveals Distinct Domains of Prehistoric Gene Flow in Europe. Am. J. Hum. Genet. 2004, 75, 128–137. [Google Scholar] [CrossRef] [PubMed]
- Kivisild, T. The Study of Human Y Chromosome Variation through Ancient DNA. Hum. Genet. 2017, 136, 529–546. [Google Scholar] [CrossRef]
- Myres, N.M.; Rootsi, S.; Lin, A.A.; Järve, M.; King, R.J.; Kutuev, I.; Cabrera, V.M.; Khusnutdinova, E.K.; Pshenichnov, A.; Yunusbayev, B.; et al. A Major Y-Chromosome Haplogroup R1b Holocene Era Founder Effect in Central and Western Europe. Eur. J. Hum. Genet. 2011, 19, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Underhill, P.A.; Poznik, G.D.; Rootsi, S.; Järve, M.; Lin, A.A.; Wang, J.; Passarelli, B.; Kanbar, J.; Myres, N.M.; King, R.J.; et al. The Phylogenetic and Geographic Structure of Y-Chromosome Haplogroup R1a. Eur. J. Hum. Genet. 2015, 23, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Underhill, P.A.; Myres, N.M.; Rootsi, S.; Metspalu, M.; Zhivotovsky, L.A.; King, R.J.; Lin, A.A.; Chow, C.-E.T.; Semino, O.; Battaglia, V.; et al. Separating the Post-Glacial Coancestry of European and Asian Y Chromosomes within Haplogroup R1a. Eur. J. Hum. Genet. 2010, 18, 479–484. [Google Scholar] [CrossRef] [PubMed]
- Haber, M.; Jones, A.L.; Connell, B.A.; Asan; Arciero, E.; Yang, H.; Thomas, M.G.; Xue, Y.; Tyler-Smith, C. A Rare Deep-Rooting D0 African Y-Chromosomal Haplogroup and Its Implications for the Expansion of Modern Humans Out of Africa. Genetics 2019, 212, 1421–1428. [Google Scholar] [CrossRef] [PubMed]
- Cruciani, F.; La Fratta, R.; Trombetta, B.; Santolamazza, P.; Sellitto, D.; Colomb, E.B.; Dugoujon, J.-M.; Crivellaro, F.; Benincasa, T.; Pascone, R.; et al. Tracing Past Human Male Movements in Northern/Eastern Africa and Western Eurasia: New Clues from Y-Chromosomal Haplogroups E-M78 and J-M12. Mol. Biol. Evol. 2007, 24, 1300–1311. [Google Scholar] [CrossRef] [PubMed]
- Rootsi, S.; Myres, N.M.; Lin, A.A.; Järve, M.; King, R.J.; Kutuev, I.; Cabrera, V.M.; Khusnutdinova, E.K.; Varendi, K.; Sahakyan, H.; et al. Distinguishing the Co-Ancestries of Haplogroup G Y-Chromosomes in the Populations of Europe and the Caucasus. Eur. J. Hum. Genet. 2012, 20, 1275–1282. [Google Scholar] [CrossRef] [PubMed]
- Lacan, M.; Keyser, C.; Ricaut, F.-X.; Brucato, N.; Duranthon, F.; Guilaine, J.; Crubézy, E.; Ludes, B. Ancient DNA Reveals Male Diffusion through the Neolithic Mediterranean Route. Proc. Natl. Acad. Sci. USA 2011, 108, 9788–9791. [Google Scholar] [CrossRef]
- Lazaridis, I.; Patterson, N.; Mittnik, A.; Renaud, G.; Mallick, S.; Kirsanow, K.; Sudmant, P.H.; Schraiber, J.G.; Castellano, S.; Lipson, M.; et al. Ancient Human Genomes Suggest Three Ancestral Populations for Present-Day Europeans. Nature 2014, 513, 409–413. [Google Scholar] [CrossRef] [PubMed]
- Skoglund, P.; Sjödin, P.; Skoglund, T.; Lascoux, M.; Jakobsson, M. Investigating Population History Using Temporal Genetic Differentiation. Mol. Biol. Evol. 2014, 31, 2516–2527. [Google Scholar] [CrossRef] [PubMed]
- Mathieson, I.; Lazaridis, I.; Rohland, N.; Mallick, S.; Patterson, N.; Roodenberg, S.A.; Harney, E.; Stewardson, K.; Fernandes, D.; Novak, M.; et al. Genome-Wide Patterns of Selection in 230 Ancient Eurasians. Nature 2015, 528, 499–503. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; Posth, C.; Hajdinjak, M.; Petr, M.; Mallick, S.; Fernandes, D.; Furtwängler, A.; Haak, W.; Meyer, M.; Mittnik, A.; et al. The Genetic History of Ice Age Europe. Nature 2016, 534, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Szécsényi-Nagy, A.; Brandt, G.; Haak, W.; Keerl, V.; Jakucs, J.; Möller-Rieker, S.; Köhler, K.; Mende, B.G.; Oross, K.; Marton, T.; et al. Tracing the Genetic Origin of Europe’s First Farmers Reveals Insights into Their Social Organization. Proc. Biol. Sci. 2015, 282, 20150339. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Primorac, D.; Šarac, J.; Havaš Auguštin, D.; Novokmet, N.; Bego, T.; Pinhasi, R.; Šlaus, M.; Novak, M.; Marjanović, D. Y Chromosome Story—Ancient Genetic Data as a Supplementary Tool for the Analysis of Modern Croatian Genetic Pool. Genes 2024, 15, 748. https://doi.org/10.3390/genes15060748
Primorac D, Šarac J, Havaš Auguštin D, Novokmet N, Bego T, Pinhasi R, Šlaus M, Novak M, Marjanović D. Y Chromosome Story—Ancient Genetic Data as a Supplementary Tool for the Analysis of Modern Croatian Genetic Pool. Genes. 2024; 15(6):748. https://doi.org/10.3390/genes15060748
Chicago/Turabian StylePrimorac, Dragan, Jelena Šarac, Dubravka Havaš Auguštin, Natalija Novokmet, Tamer Bego, Ron Pinhasi, Mario Šlaus, Mario Novak, and Damir Marjanović. 2024. "Y Chromosome Story—Ancient Genetic Data as a Supplementary Tool for the Analysis of Modern Croatian Genetic Pool" Genes 15, no. 6: 748. https://doi.org/10.3390/genes15060748