Characteristics of the Vasa Gene in Silurus asotus and Its Expression Response to Letrozole Treatment
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals Treatment and Samples Collection
2.2. RNA Extraction and cDNA Synthesis
2.3. Isolation of Savasa cDNA Sequence
2.4. Sequence Analysis
2.5. qRT-PCR Analyses
2.6. In Situ Hybridization
2.7. LT Treatment
2.8. Statistical Analysis
3. Results
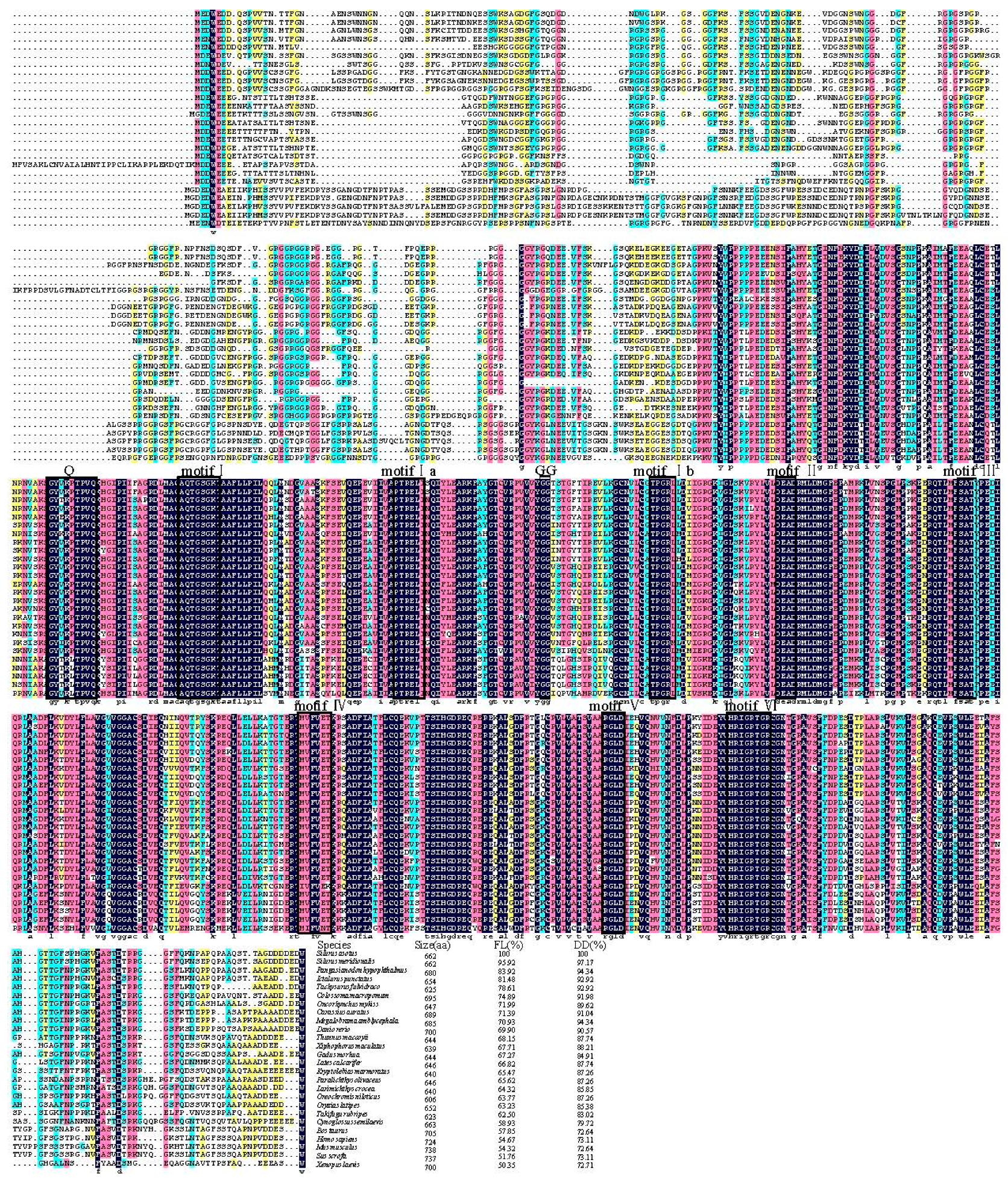
3.1. Identification and Molecular Characterization of Savasa
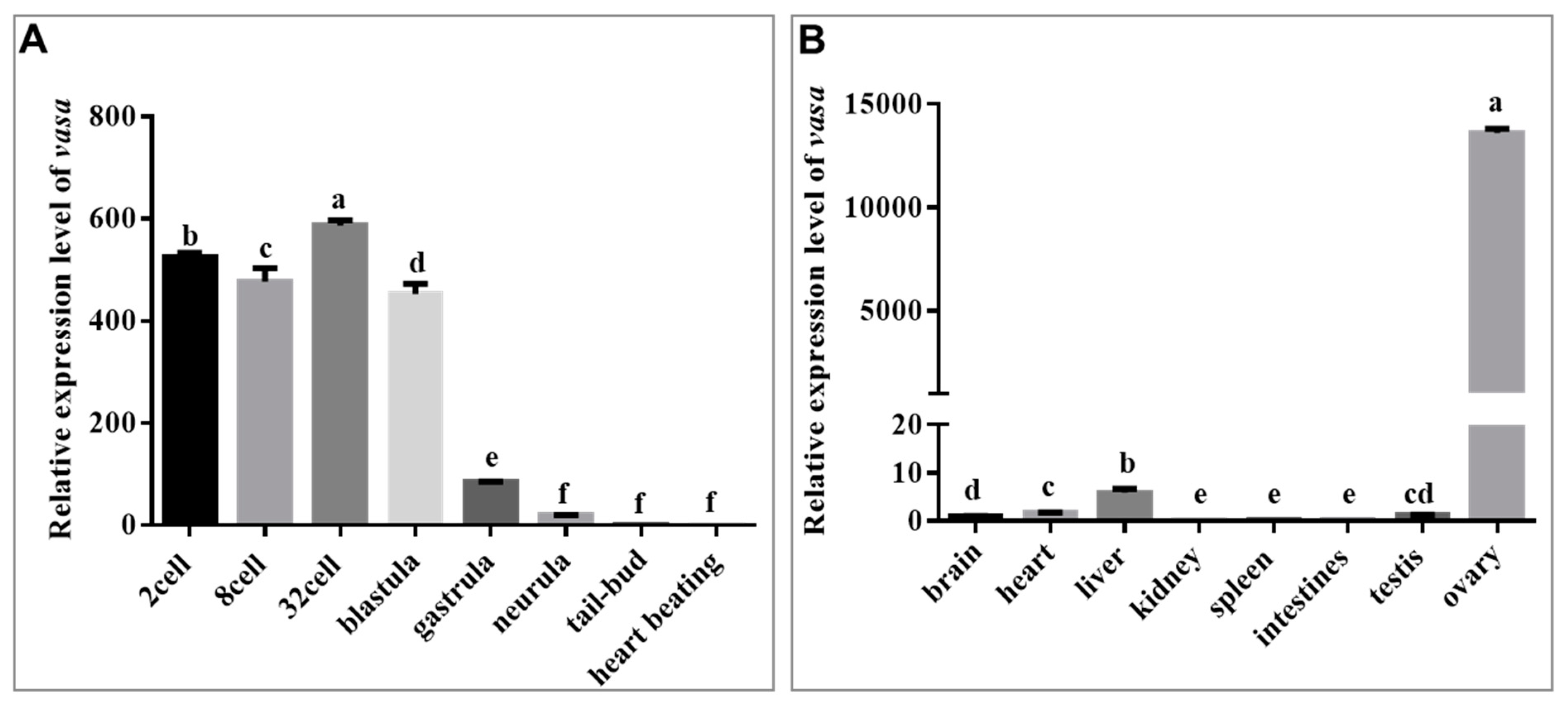
3.2. Maternal and Gonad-Predominant Expression of Savasa
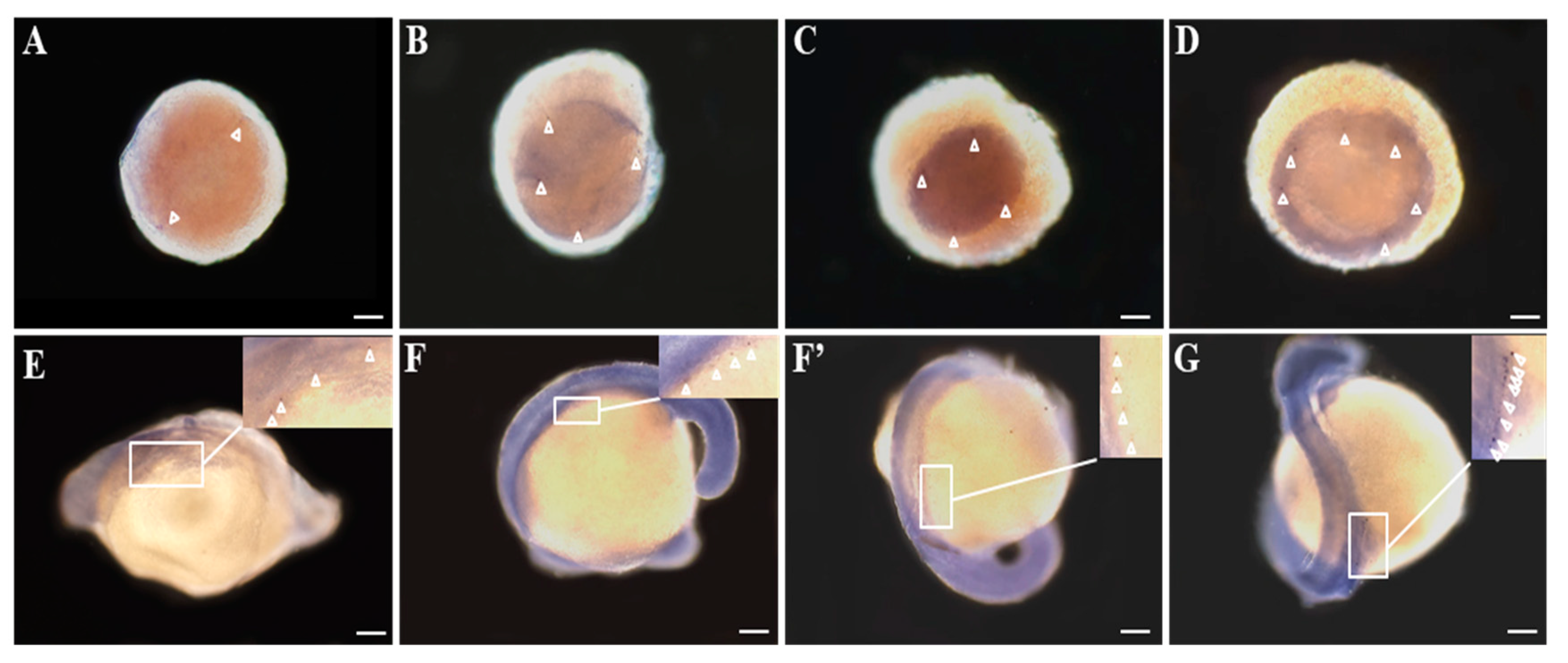
3.3. Distribution of Savasa mRNA in the Developmental Embryos
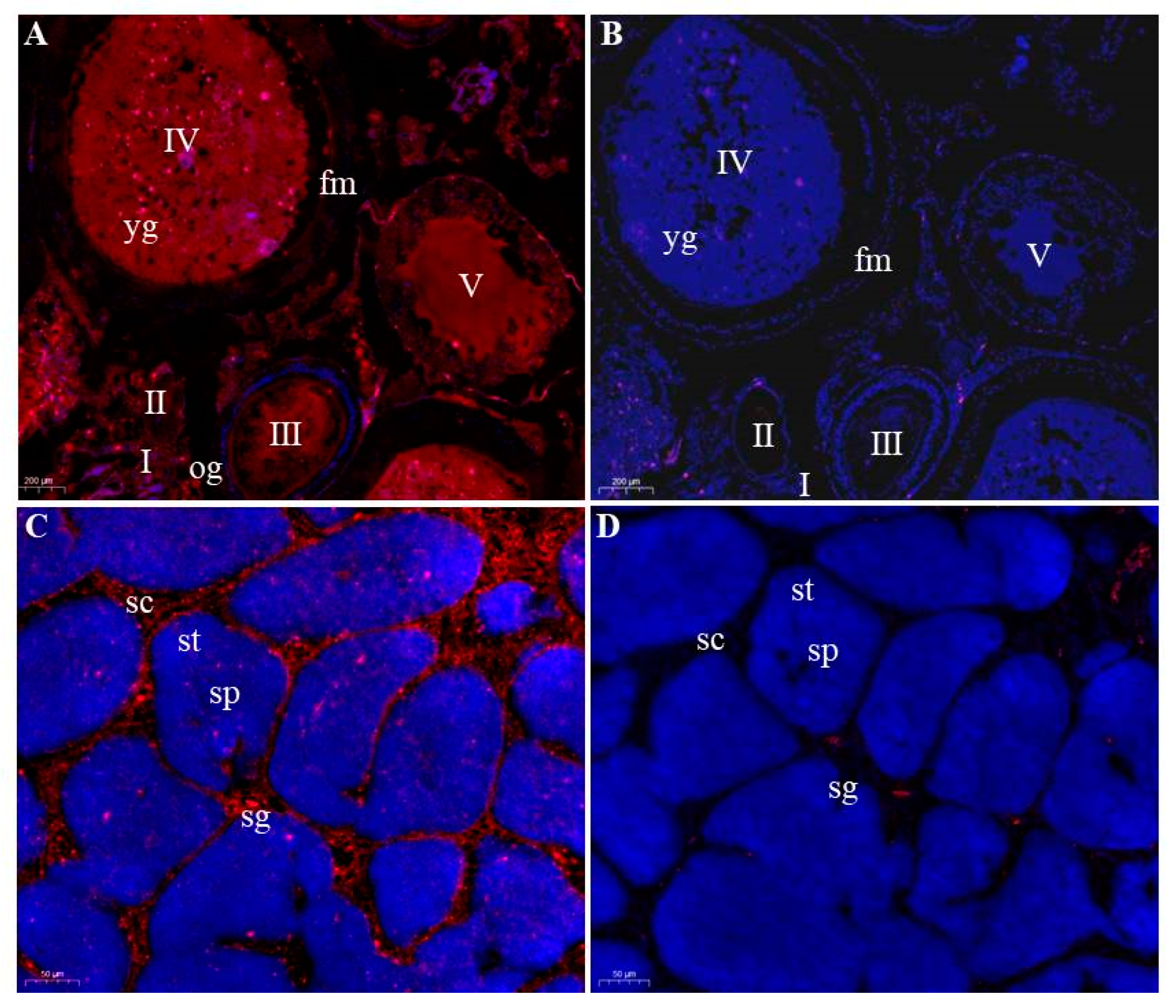
3.4. Germ Cells-Specific Expression of Savasa in Adult Gonads
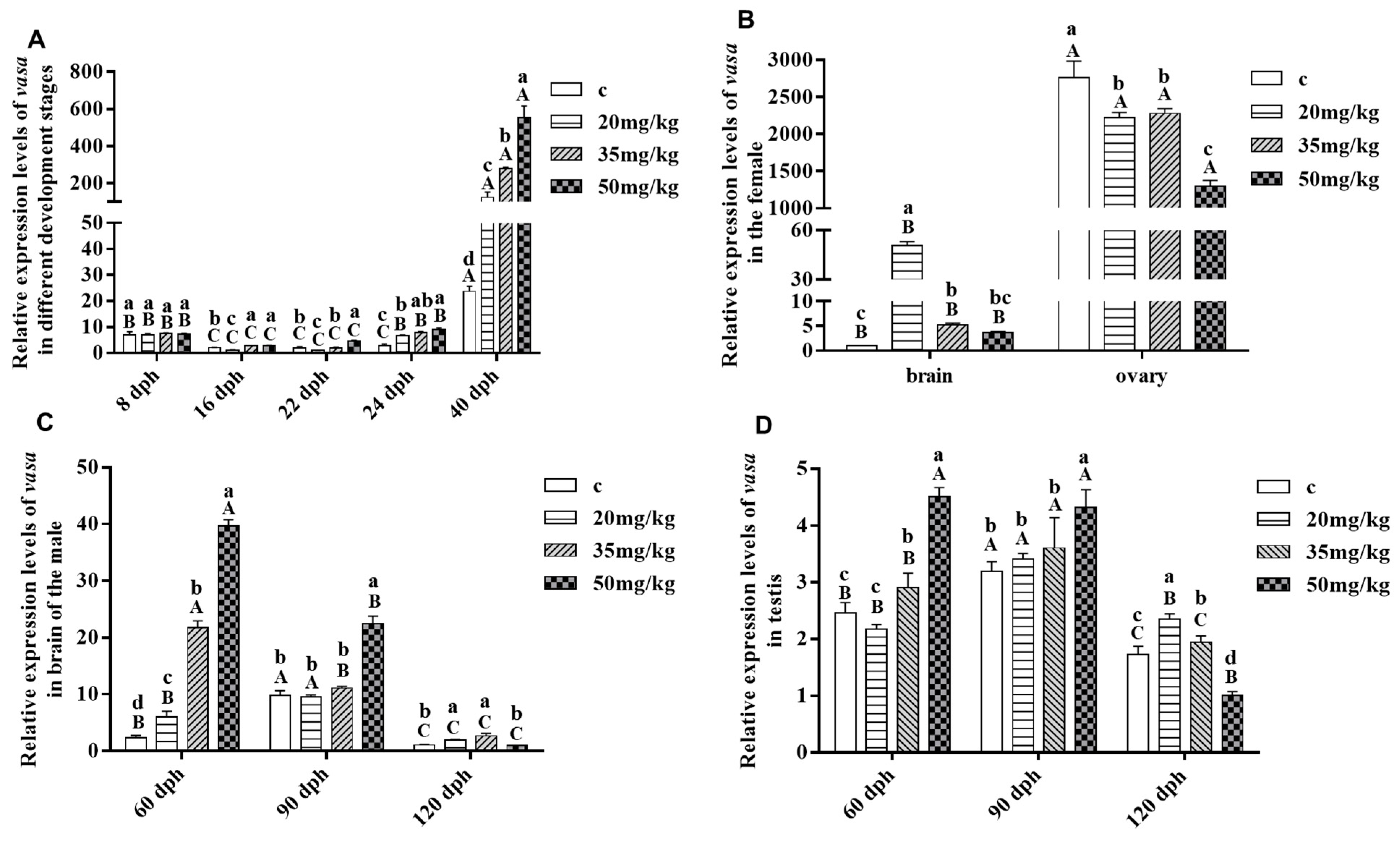
3.5. Response of Savasa Expression to Letrozole Treatment
4. Discussion
4.1. Identification of Savasa
4.2. The Spatio-Temporal Expression Pattern of Savasa
4.3. Response of Savasa to Letrozole Treatment and the Possible Primary Mechanism
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hay, B.; Jan, L.Y.; Jan, Y.N. A protein component of Drosophila polar granules is encoded by vasa and has extensive sequence similarity to ATP-dependent helicases. Cell 1988, 55, 577–587. [Google Scholar] [CrossRef] [PubMed]
- Kuniya, A.; Toshiaki, N. A DEAD-family protein gene, Ddx4, encoding a murine homolog of Drosophila vasa maps to the distal end of mouse Chromosome 13. Mamm. Genome 1997, 8, 622–623. [Google Scholar] [CrossRef] [PubMed]
- Ikenishi, K.; Tanaka, T.S. Spatio-temporal expression of Xenopus vasa homolog, XVLG1, in oocytes and embryos: The presence of XVLG1 RNA in somatic cells as well as germline cells. Dev. Growth Differ. 2000, 42, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Nakao, H.; Hatakeyama, M.; Lee, J.M.; Shimoda, M.; Kanda, T. Expression pattern of Bombyx vasa-like (BmVLG) protein and its implications in germ cell development. Dev. Genes. Evol. 2006, 216, 94–99. [Google Scholar] [CrossRef] [PubMed]
- Castrillon, D.H.; Quade, B.J.; Wang, T.Y.; Quigley, C.; Crum, C.P. The human VASA gene is specifically expressed in the germ cell lineage. Proc. Natl. Acad. Sci. USA 2000, 97, 9585–9590. [Google Scholar] [CrossRef] [PubMed]
- Sundaram Buitrago, P.A.; Rao, K.; Yajima, M. Vasa, a regulator of localized mRNA translation on the spindle. Bioessays 2023, 45, e2300004. [Google Scholar] [CrossRef] [PubMed]
- Olsen, L.C.; Aasland, R.; Fjose, A. A vasa-like gene in zebrafish identifies putative primordial germ cells. Mech. Dev. 1997, 66, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Yoon, C.; Kawakami, K.; Hopkins, N. Zebrafish vasa homologue RNA is localized to the cleavage planes of 2-and 4-cell-stage embryos and is expressed in the primordial germ cells. Development 1997, 124, 3157–3165. [Google Scholar] [CrossRef] [PubMed]
- Shinomiya, A.; Tanaka, M.; Kobayashi, T.; Nagahama, Y.; Hamaguchi, S. The vasa-like gene, olvas, identifies the migration path of primordial germ cells during embryonic body formation stage in the medaka, Oryzias latipes. Dev. Growth Differ. 2000, 42, 317–326. [Google Scholar] [CrossRef]
- Shen, J.L.; Yokota, S.; Yokoi, H.; Suzuki, T. Diethylnitrosamine-induced expression of germline-specific genes and pluripotency factors, including vasa and oct4, in medaka somatic cells. Biochem. Biophys. Res. Commun. 2016, 478, 858–863. [Google Scholar] [CrossRef]
- Kobayashi, T.; Kajiura-Kobayashi, H.; Nagahama, Y. Two isoforms of vasa homologs in a teleost fish: Their differential expression during germ cell differentiation. Mech. Dev. 2002, 111, 167–171. [Google Scholar] [CrossRef]
- Xu, H.Y.; Gui, J.F.; Hong, Y.H. Differential expression of vasa RNA and protein during spermatogenesis and oogenesis in the gibel carp (Carassius auratus gibelio), a bisexually and gynogenetically reproducing vertebrate. Dev. Dyn. 2005, 233, 872–882. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.J.; Wu, F.R.; Liu, Z.H.; Huang, B.F.; Zhang, Y.G.; Wang, D.S. Molecular cloning and expression of two isoforms of Vasa gene in Southern catfish Silurus meridionalis. Curr. Zool. 2008, 54, 1051–1060. [Google Scholar]
- Lin, R.J. Gonad Transcriptome of Discus Fish (Symphysodon haraldi) and Expression of vasa Gene Dissertation. Master’s Thesis, Shanghai Ocean University, Shanghai, China, 2017. [Google Scholar]
- Xue, T. Identification and Application of vasa Gene in Mylopharyngodon Piceus. Ph.D. Thesis, Huazhong Agricultural University, Wuhan, China, 2017. [Google Scholar]
- Xiol, J.; Spinelli, P.; Laussmann, M.A.; Homolka, D.; Yang, Z.L.; Cora, E.; Couté, Y.; Conn, S.; Kadlec, J.; Sachidanandam, R. RNA clamping by vasa assembles a pirna amplifier complex on transposon transcripts. Cell 2014, 157, 1698–1711. [Google Scholar] [CrossRef] [PubMed]
- Durdevic, Z.; Ephrussi, A. Germ Cell Lineage Homeostasis in Drosophila Requires the Vasa RNA Helicase. Genetics 2019, 213, 911–922. [Google Scholar] [CrossRef]
- Knaut, H.; Pelegri, F.; Bohmann, K.; Schwarz, H.; Nüsslein-Volhard, C. Zebrafish vasa RNA but not its protein is a component of the germ plasm and segregates asymmetrically before germline specification. J. Cell Biol. 2000, 149, 875–888. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Diehl-Jones, W.; Lasko, P. Localization of vasa protein to the Drosophila pole plasm is independent of its RNA-binding and helicase activities. Development 1994, 120, 1201–1211. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Fang, D.A.; Sun, J.L.; Wang, Y.; Wang, J.; Liu, L.H. Characterization of the vasa gene in the Chinese mitten crab Eriocheir sinensis: A germ line molecular marker. J. Insect Physiol. 2012, 58, 960–965. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.Y.; Ding, H.; Wu, S.X.; Wang, M.Y.; Wei, C.; Wang, B.; Bao, Z.M.; Hu, J.J. Vasa Is a Potential Germ Cell Marker in Leopard Coral Grouper (Plectropomus leopardus). Genes 2022, 13, 1077. [Google Scholar] [CrossRef]
- Tomancak, P.; Guichet, A.; Zavorszky, P.; Ephrussi, A. Oocyte polarity depends on regulation of gurken by Vasa. Development 1998, 125, 1723–1732. [Google Scholar] [CrossRef]
- Lasko, P. The DEAD-box helicase Vasa: Evidence for a multiplicity of functions in RNA processes and developmental biology. Biochim. Biophys. Acta 2013, 1829, 810–816. [Google Scholar] [CrossRef] [PubMed]
- Hartung, O.; Forbes, M.M.; Marlow, F.L. Zebrafish vasa is required for germ-cell differentiation and maintenance. Mol. Reprod. Dev. 2014, 81, 946–961. [Google Scholar] [CrossRef] [PubMed]
- Braat, A.K.; van de Water, S.; Korving, J.; Zivkovic, D. A zebrafish vasa morphant abolishes Vasa protein but does not affect the establishment of the germline. Genesis 2001, 30, 183–185. [Google Scholar] [CrossRef] [PubMed]
- Li, M.Y.; Hong, N.; Xu, H.Y.; Yi, M.S.; Li, C.M.; Gui, J.F.; Hong, Y.H. Medaka vasa is required for migration but not survival of primordial germ cells. Mech. Dev. 2009, 126, 366–381. [Google Scholar] [CrossRef] [PubMed]
- Miwa, T.; Yoshizaki, G.; Naka, H.; Nakatani, M.; Sakai, K.; Kobayashi, M.; Takeuchi, T. Ovarian steroid synthesis during oocyte maturation and ovulation in Japanese catfish (Silurus asotus). Aquaculture 2001, 198, 179–191. [Google Scholar] [CrossRef]
- Qiao, Z.G.; LI, X.J. Biology and New Breeding Techniques of Silurus asotus, 1st ed.; China Agricultural Science and Technology Press: Beijing, China, 2007; pp. 1–35. [Google Scholar]
- Yang, Y.H.; He, Y.L.; Zhou, J.S.; Li, L.; Wang, L.Z.; Yang, J.N.; Li, H.B.; Chen, B.; Wang, Y.L. Quality evaluation of the meat of Silurus lanzhouensis, Silurus asotus and Cyrinus carpio. Acta Hydrobiol. Sin. 2013, 37, 54–61. [Google Scholar]
- Shen, F.F.; Long, Y.; Li, F.Y.; Ge, G.D.; Song, G.L.; Li, Q.; Qiao, Z.G.; Cui, Z.B. De novo transcriptome assembly and sex-biased gene expression in the gonads of Amur catfish (Silurus asotus). Genomics 2020, 112, 2603–2614. [Google Scholar] [CrossRef] [PubMed]
- Piferrer, F. Endocrine sex control strategies for the feminization of teleost fish. Aquaculture 2001, 197, 229–281. [Google Scholar] [CrossRef]
- Wang, H.P.; Shen, Z.G. Sex Control in Aquaculture: Concept to Practice. In Sex Control in Aquaculture, 1st ed.; John Wiley & Sons: Hoboken, NJ, USA, 2018; pp. 1–34. [Google Scholar]
- Yu, Y.; Chen, M.; Shen, Z.G. Molecular biological, physiological, cytological, and epigenetic mechanisms of environmental sex differentiation in teleosts: A systematic review. Ecotoxicol. Environ. Saf. 2023, 267, 115654. [Google Scholar] [CrossRef]
- Li, G.L.; Liu, X.C.; Lin, H.R. Aromatase inhibitor letrozole induces sex inversion in the protogynous red spotted grouper (Epinephelus akaara). Acta Physiol. Sin. 2005, 4, 473–479. [Google Scholar]
- Yang, F. Effect of 17α-Methyltestosterone and Letrozole on the Sex Differentiation in Two Croakers. Master’s Thesis, Zhejiang Ocean University, Zhoushan, China, 2017. [Google Scholar]
- Shen, F.F. Inducing Sex Reversal by Using Drug, and Finging the Sex-Specific Genes in the Silurus asotus Base on RNA-seq. Master’s Thesis, Henan Normal University, Xinxiang, China, 2018. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Xue, T.; Chen, T.S.; Fang, J.; Pan, Q.H.; Deng, Y.; Li, L.Y.; Chen, K.; Wang, Y.Z. Maternal inheritance of Nanog ortholog in blunt-snout bream. J. Exp. Zool. Part. B 2017, 328, 749–759. [Google Scholar] [CrossRef] [PubMed]
- Blaser, H.; Eisenbeiss, S.; Neumann, M.; Reichman-Fried, M.; Thisse, B.; Thisse, C.; Raz, E. Transition from non-motile behaviour to directed migration during early PGC development in zebrafish. J. Cell Sci. 2005, 118, 4027–4038. [Google Scholar] [CrossRef] [PubMed]
- Cordin, O.; Banroques, J.; Tanner, N.K.; Linder, P. The DEAD-box protein family of RNA helicases. Gene 2006, 367, 17–37. [Google Scholar] [CrossRef] [PubMed]
- Sengoku, T.; Nureki, O.; Nakamura, A.; Kobayashi, S.; Yokoyama, S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell 2006, 125, 287–300. [Google Scholar] [CrossRef] [PubMed]
- Li, C.J.; Liu, L.; Chen, X.H.; Zhang, T.; Gan, F.; Cheng, B.L. Identification of a vasa homologue gene in grass carp and its expression pattern in tissues and during embryogenesis. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2010, 157, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, Y.; Komiya, T.; Kawabata, H.; Sato, M.; Fujimoto, H.; Furusawa, M.; Noce, T. Isolation of a DEAD-family protein gene that encodes a murine homolog of Drosophila vasa and its specific expression in germ cell lineage. Proc. Natl. Acad. Sci. USA 1994, 91, 12258–12262. [Google Scholar] [CrossRef] [PubMed]
- Yoshizaki, G.; Sakatani, S.; Tominaga, H.; Takeuchi, T. Cloning and characterization of a vasa-like gene in rainbow trout and its expression in the germ cell lineage. Mol. Reprod. Dev. 2000, 55, 364–371. [Google Scholar]
- Raghuveer, K.; Senthilkumaran, B. Cloning and differential expression pattern of vasa in the developing and recrudescing gonads of catfish, Clarias gariepinus. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2010, 157, 79–85. [Google Scholar] [CrossRef]
- Wu, X.M.; Wang, Z.K.; Jiang, J.J.; Gao, J.N.; Wang, J.; Zhou, X.S.; Zhang, Q.Q. Cloning, expression promoter analysis of vasa gene in Japanese flounder (Paralichthys olivaceus). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2014, 167, 41–50. [Google Scholar] [CrossRef]
- Melo, L.H.; Melo, R.M.C.; Luz, R.K.; Bazzoli, N.; Rizzo, E. Expression of Vasa, Nanos2 and Sox9 during initial testicular development in Nile tilapia (Oreochromis niloticus) submitted to sex reversal. Reprod. Fertil. Dev. 2019, 31, 1637–1646. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, Y.; Yoshizaki, G.; Kobayashi, T.; Takeuchi, T. Mass isolation of primordial germ cells from transgenic rainbow trout carrying the green fluorescent protein gene driven by the vasa gene promoter. Biol. Reprod. 2002, 67, 1087–1092. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.Q.; Chen, S.L.; Liu, Y.; Shao, C.W.; Lin, F.; Wang, N.; Hu, Q.M. Molecular characterization, sexually dimorphic expression, and functional analysis of 3′-untranslated region of vasa gene in half-smooth tongue sole (Cynoglossus semilaevis). Theriogenology 2014, 82, 213–224. [Google Scholar] [CrossRef]
- Santos, A.C.; Lehmann, R. Germ cell specification and migration in Drosophila and beyond. Curr. Biol. 2004, 14, R578–R589. [Google Scholar] [CrossRef] [PubMed]
- Ikenishi, K. Germ plasm in Caenorhabditis elegans, Drosophila and Xenopus. Dev. Growth Differ. 1998, 40, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Braat, A.K.; van de Water, S.; Goos, H.; Bogerd, J.; Zivkovic, D. Vasa protein expression and localization in the zebrafish. Mech. Dev. 2000, 95, 271–274. [Google Scholar] [CrossRef] [PubMed]
- Blázquez, M.; González, A.; Mylonas, C.C.; Piferrer, F. Cloning and sequence analysis of a vasa homolog in the European sea bass (Dicentrarchus labrax): Tissue distribution and mRNA expression levels during early development and sex differentiation. Gen. Comp. Endocrinol. 2011, 170, 322–333. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.; Xu, S.H.; Ma, D.Y.; Xiao, Z.Z.; Zhao, C.Y.; Xiao, Y.S.; Chi, L.; Liu, Q.H.; Li, J. Germ line specific expression of a vasa homologue gene in turbot (Scophthalmus maximus): Evidence for vasa localization at cleavage furrows in euteleostei. Mol. Reprod. Dev. 2012, 79, 803–813. [Google Scholar] [CrossRef] [PubMed]
- Pacchiarini, T.; Cross, I.; Leite, R.B.; Gavaia, P.; Ortiz-Delgado, J.B.; Pousão-Ferreira, P.; Rebordinos, L.; Sarasquete, C.; Cabrita, E. Solea senegalensis vasa transcripts: Molecular characterisation, tissue distribution and developmental expression profiles. Reprod. Fertil. Dev. 2013, 25, 646–660. [Google Scholar] [CrossRef]
- Kobayashi, T.; Kajiura-Kobayashi, H.; Nagahama, Y. Differential expression of vasa homologue gene in the germ cells during oogenesis and spermatogenesis in a teleost fish, tilapia, Oreochromis niloticus. Mech. Dev. 2000, 99, 139–142. [Google Scholar] [CrossRef]
- Xu, H.Y.; Lim, M.; Dwarakanath, M.; Hong, Y.H. Vasa identifies germ cells and critical stages of oogenesis in the Asian seabass. Int. J. Biol. Sci. 2014, 10, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.G.; Ye, D.; Song, P.; Lv, D.Y.; Gui, J.F. Expression and Distributing of vasa Gene During Gametogenesis of Goldfish (Carassius auratus). Zool. Res. 2005, 26, 179–183. [Google Scholar]
- Qin, Z.Q. Induction of Neo-Male and Screening Analysis of Sex-Related Genes in Yellow Drum. Master’s Thesis, Zhejiang Ocean University, Zhoushan, China, 2020. [Google Scholar]
- Shen, Z.G.; Fan, Q.X.; Yang, W.; Zhang, Y.L.; Wang, H.P. Effects of 17α-methyltestosterone and aromatase inhibitor letrozole on sex reversal, gonadal structure, and growth in yellow catfish Pelteobagrus fulvidraco. Biol. Bull. 2015, 228, 108–117. [Google Scholar] [CrossRef] [PubMed]
- Wu, G. Expression of Sex Determining Gene and Sex Development Gene in Protandrous Black Porgy (Acanthpagrus schlegeli): The Mechanism of Sex Change. Master’s Thesis, NTOU: Institute of Marine Biology, Jilong, China, 2003. [Google Scholar]
- Yajima, M.; Wessel, G.M. The multiple hats of Vasa: Its functions in the germline and in cell cycle progression. Mol. Reprod. Dev. 2011, 78, 861–867. [Google Scholar] [CrossRef] [PubMed]
- Zhan, B.J. The Effects of Fadrozole and Finasteride on the Expression Levels of cyp19a, foxl2, vasa Genes in Loach. Master’s Thesis, Fujian Normal University, Fuzhou, China, 2013. [Google Scholar]
- Zhang, D.; Duarte-Guterman, P.; Langlois, V.S.; Trudeau, V.L. Temporal expression and steroidal regulation of piRNA pathway genes (mael, piwi, vasa) during Silurana (Xenopus) tropicalis embryogenesis and early larval development. Comp. Biochem. Physiol. Part. C Toxicol. Pharmacol. 2010, 152, 202–206. [Google Scholar] [CrossRef]
- Chen, Y.D.; Hua, Z.Z.; Wang, Y.; Zhang, Z.P. Progress in studies of vasa gene. Chin. J. Zool. 2010, 45, 173–180. [Google Scholar]
- Liang, Y.Q.; Jing, Z.X.; Pan, C.G.; Lin, Z.; Zhen, Z.; Hou, L.P.; Dong, Z.D. The progestin norethindrone alters growth, reproductive histology and gene expression in zebrafish (Danio rerio). Chemosphere 2020, 242, 125285. [Google Scholar] [CrossRef]


| Name | Sequence (5′-3′) | Usage |
|---|---|---|
| Sa-actin-F | AAGATCATTGCCCCACCTGA | qRT-PCR |
| Sa-actin-R | CCTGCTTGCTGATCCACATC | qRT-PCR |
| Sa-q-vasa-F | CCGAGAAGTGTTGAAAGGTT | qRT-PCR |
| Sa-q-vasa-R | CCAGCACCAAATAACGAACT | qRT-PCR |
| Sa-vasa-F | CTATCTTCAACTATGGAGGACTGG | ORF amplification |
| Sa-vasa-R | CAGTTTCCTCCAGCCATGAAG | ORF amplification |
| Sa-v-GSP | ACATCAATCAAGTCACTCGGTA | 3′RACE PCR |
| Sa-v-NGSP | CCTACCGAAATACATTGACGAG | 3′RACE PCR |
| Sa-vasa-situ F | AAAGAATTCGAACTCTTGGAACAACGGGAA | probe synthesis |
| Sa-vasa-situ R | AAATCTAGAGTAAATCCTGTGCTTGTACCTC | probe synthesis |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, M.; Wang, F.; Li, M.; Wang, Y.; Gao, X.; Zhang, H.; Liu, Z.; Zhou, Z.; Zhao, D.; Zhang, M.; et al. Characteristics of the Vasa Gene in Silurus asotus and Its Expression Response to Letrozole Treatment. Genes 2024, 15, 756. https://doi.org/10.3390/genes15060756
Yu M, Wang F, Li M, Wang Y, Gao X, Zhang H, Liu Z, Zhou Z, Zhao D, Zhang M, et al. Characteristics of the Vasa Gene in Silurus asotus and Its Expression Response to Letrozole Treatment. Genes. 2024; 15(6):756. https://doi.org/10.3390/genes15060756
Chicago/Turabian StyleYu, Miao, Fangyuan Wang, Muzi Li, Yuan Wang, Xiangzhe Gao, Hanhan Zhang, Zhenzhu Liu, Zhicheng Zhou, Daoquan Zhao, Meng Zhang, and et al. 2024. "Characteristics of the Vasa Gene in Silurus asotus and Its Expression Response to Letrozole Treatment" Genes 15, no. 6: 756. https://doi.org/10.3390/genes15060756
APA StyleYu, M., Wang, F., Li, M., Wang, Y., Gao, X., Zhang, H., Liu, Z., Zhou, Z., Zhao, D., Zhang, M., Wang, L., Jiang, H., & Qiao, Z. (2024). Characteristics of the Vasa Gene in Silurus asotus and Its Expression Response to Letrozole Treatment. Genes, 15(6), 756. https://doi.org/10.3390/genes15060756





