Establishment and Application of a Novel Genetic Detection Panel for SNPs in Mongolian Gerbils
Highlights
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Samples
2.2. DNA Extraction
2.3. Sequencing and Quality Control
2.4. SNP Locus Selection
2.5. Primer Design and Sequencing
2.6. Population Genetic Analysis
2.7. Compilation of Principal Component Analyses and Population Structure
3. Results
3.1. Establishment and Optimization of the SNP Detection System
3.2. Genetic Analysis of the Outbred Mongolian Gerbil Populations
3.3. Structure Analysis of the Three Mongolian Gerbil Populations
3.4. Principal Component Analysis in the Three Mongolian Gerbil Populations
3.5. Genetic Analysis of the Outbred Mongolian Gerbil Populations
3.6. Genetic Analysis of the Inbred Mongolian Gerbil Lines
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abe, Y.; Toyama, K.; Shinohara, A.; Nagura-Kato, G.A.; Ikai, Y.; Koshimoto, C.; Spin, J.M.; Hato, N. Message to researchers: The characteristic absence of a posterior communicating artery is easily lost in the gerbil. Anat. Sci. Int. 2023, 98, 426–433. [Google Scholar] [CrossRef]
- Du, X.; Wang, D.; Li, Y.; Huo, X.; Li, C.; Lu, J.; Wang, Y.; Guo, M.; Chen, Z. Newly breeding an inbred strain of ischemia-prone Mongolian gerbils and its reproduction and genetic characteristics. Exp. Anim. 2018, 67, 83–90. [Google Scholar] [CrossRef]
- Kondo, T.; Yoshida, S.; Nagai, H.; Takeshita, A.; Mino, M.; Morioka, H.; Nakajima, T.; Kusakabe, K.T.; Okada, T. Transient forebrain ischemia induces impairment in cognitive performance prior to extensive neuronal cell death in Mongolian gerbil (Meriones unguiculatus). J. Vet. Sci. 2018, 19, 505–511. [Google Scholar] [CrossRef]
- Selvaraj, B.; Le, T.T.; Kim, D.W.; Jung, B.H.; Yoo, K.Y.; Ahn, H.R.; Thuong, P.T.; Tran, T.T.T.; Pae, A.N.; Jung, S.H.; et al. Neuroprotective Effects of Ethanol Extract of Polyscias fruticosa (EEPF) against Glutamate-Mediated Neuronal Toxicity in HT22 Cells. Int. J. Mol. Sci. 2023, 24, 3969. [Google Scholar] [CrossRef]
- Shen, Y.; Lu, H.; Xu, R.; Tian, H.; Xia, X.; Zhou, F.H.; Wang, L.; Dong, J.; Sun, L. The Expression of GLAST and GLT1 in a Transient Cerebral Ischemia Mongolian Gerbil Model. Neuropsychiatr. Dis. Treat. 2020, 16, 789–800. [Google Scholar] [CrossRef]
- Ye, Q.; Hai, K.; Liu, W.; Wang, Y.; Zhou, X.; Ye, Z.; Liu, X. Investigation of the protective effect of heparin pre-treatment on cerebral ischaemia in gerbils. Pharm. Biol. 2019, 57, 519–528. [Google Scholar] [CrossRef]
- Noto, J.M.; Rose, K.L.; Hachey, A.J.; Delgado, A.G.; Romero-Gallo, J.; Wroblewski, L.E.; Schneider, B.G.; Shah, S.C.; Cover, T.L.; Wilson, K.T.; et al. Carcinogenic Helicobacter pylori Strains Selectively Dysregulate the In Vivo Gastric Proteome, Which May Be Associated with Stomach Cancer Progression. Mol. Cell Proteom. 2019, 18, 352–371. [Google Scholar] [CrossRef]
- Amalia, R.; Panenggak, N.S.R.; Doohan, D.; Rezkitha, Y.A.A.; Waskito, L.A.; Syam, A.F.; Lubis, M.; Yamaoka, Y.; Miftahussurur, M. A comprehensive evaluation of an animal model for Helicobacter pylori-associated stomach cancer: Fact and controversy. Helicobacter 2023, 28, e12943. [Google Scholar] [CrossRef]
- Lee, T.K.; Hong, J.; Lee, J.W.; Kim, S.S.; Sim, H.; Lee, J.C.; Kim, D.W.; Lim, S.S.; Kang, I.J.; Won, M.H. Ischemia-Induced Cognitive Impairment Is Improved via Remyelination and Restoration of Synaptic Density in the Hippocampus after Treatment with COG-Up(®) in a Gerbil Model of Ischemic Stroke. Vet. Sci. 2021, 8, 321. [Google Scholar] [CrossRef]
- Zhou, L.; Jia, R.; Zeng, W.; Cai, Q.; Qu, Y. Study on the Difference of Protective Efficacy and Mechanism of Radix Aconiti Coreani and Rhizoma Typhonii in Gerbils with Ischemic Stroke. ACS Chem. Neurosci. 2023, 14, 3686–3693. [Google Scholar] [CrossRef]
- Jeong, D.Y.; Jeong, S.Y.; Zhang, T.; Wu, X.; Qiu, J.Y.; Park, S. Chungkookjang, a soy food, fermented with Bacillus amyloliquefaciens protects gerbils against ishcmeic stroke injury, and post-stroke hyperglycemia. Food Res. Int. 2020, 128, 108769. [Google Scholar] [CrossRef]
- Kim, B.; Lee, T.K.; Park, C.W.; Kim, D.W.; Ahn, J.H.; Sim, H.; Lee, J.C.; Yang, G.E.; Kim, J.D.; Shin, M.C.; et al. Pycnogenol(®) Supplementation Attenuates Memory Deficits and Protects Hippocampal CA1 Pyramidal Neurons via Antioxidative Role in a Gerbil Model of Transient Forebrain Ischemia. Nutrients 2020, 12, 2477. [Google Scholar] [CrossRef]
- Li, X.; Lu, J.; Wang, Y.; Huo, X.; Li, Z.; Zhang, S.; Li, C.; Guo, M.; Du, X.; Chen, Z. Establishment and Characterization of a Newly Established Diabetic Gerbil Line. PLoS ONE 2016, 11, e0159420. [Google Scholar] [CrossRef]
- Gong, J.; Du, X.; Li, Z.; Li, X.; Guo, M.; Lu, J.; Wang, Y.; Chen, Z.; Li, C. Differential expression of genes identified by suppression subtractive hybridization in liver and adipose tissue of gerbils with diabetes. PLoS ONE 2018, 13, e0191212. [Google Scholar] [CrossRef]
- Zhang, X.; He, Y.; Zhang, W.; Wang, Y.; Liu, X.; Cui, A.; Gong, Y.; Lu, J.; Liu, X.; Huo, X.; et al. Development of Microsatellite Marker System to Determine the Genetic Diversity of Experimental Chicken, Duck, Goose, and Pigeon Populations. Biomed. Res. Int. 2021, 2021, 8851888. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, P.; Song, Z.; Du, X.; Huo, X.; Lu, J.; Liu, X.; Lv, J.; Li, C.; Guo, M.; et al. Generation of Gene-Knockout Mongolian Gerbils via CRISPR/Cas9 System. Front. Bioeng. Biotechnol. 2020, 8, 780. [Google Scholar] [CrossRef]
- Li, J. Full-Length Transcriptome Analysis of Mongolian Gerbils. 2023. Available online: https://link.cnki.net/doi/10.27229/d.cnki.gnmnu.2023.001321 (accessed on 15 June 2024).
- Liu, X.; Yu, X.; Xu, Y.; Du, X.; Huo, X.; Li, C.; Lv, J.; Guo, M.; Lu, J.; Chen, Z. Development of an effective microsatellite marker system to determine the genetic structure of Meriones meridianus populations. Exp. Anim. 2020, 69, 224–232. [Google Scholar] [CrossRef]
- Deng, K.; Liu, W.; Wang, D.H. Relatedness and spatial distance modulate intergroup interactions: Experimental evidence from a social rodent. Curr. Zool. 2019, 65, 527–534. [Google Scholar] [CrossRef]
- Du, X.Y.; Li, W.; Sa, X.Y.; Li, C.L.; Lu, J.; Wang, Y.Z.; Chen, Z.W. Selection of an effective microsatellite marker system for genetic control and analysis of gerbil populations in China. Genet. Mol. Res. 2015, 14, 11030–11042. [Google Scholar] [CrossRef]
- Wang, G.; Liu, W.; Wang, Y.; Wan, X.; Zhong, W. Restricted dispersal determines fine-scale spatial genetic structure of Mongolian gerbils. Curr. Zool. 2017, 63, 687–691. [Google Scholar] [CrossRef]
- Mekada, K.; Yoshiki, A. Substrains matter in phenotyping of C57BL/6 mice. Exp. Anim. 2021, 70, 145–160. [Google Scholar] [CrossRef]
- Liu, Y.; Chudgar, N.; Mastrogiacomo, B.; He, D.; Lankadasari, M.B.; Bapat, S.; Jones, G.D.; Sanchez-Vega, F.; Tan, K.S.; Schultz, N.; et al. A germline SNP in BRMS1 predisposes patients with lung adenocarcinoma to metastasis and can be ameliorated by targeting c-fos. Sci. Transl. Med. 2022, 14, eabo1050. [Google Scholar] [CrossRef]
- Brynildsen, J.K.; Yang, K.; Lemchi, C.; Dani, J.A.; De Biasi, M.; Blendy, J.A. A common SNP in Chrna5 enhances morphine reward in female mice. Neuropharmacology 2022, 218, 109218. [Google Scholar] [CrossRef]
- Chen, C.L.; Rodiger, J.; Chung, V.; Viswanatha, R.; Mohr, S.E.; Hu, Y.; Perrimon, N. SNP-CRISPR: A Web Tool for SNP-Specific Genome Editing. G3 2020, 10, 489–494. [Google Scholar] [CrossRef]
- Wang, X.; Lu, C.; Chen, Y.; Wang, Q.; Bao, X.; Zhang, Z.; Huang, X. Resveratrol promotes bone mass in ovariectomized rats and the SIRT1 rs7896005 SNP is associated with bone mass in women during perimenopause and early postmenopause. Climacteric 2023, 26, 25–33. [Google Scholar] [CrossRef]
- Fowler, S.; Wang, T.; Munro, D.; Kumar, A.; Chitre, A.S.; Hollingsworth, T.J.; Garcia Martinez, A.; St Pierre, C.L.; Bimschleger, H.; Gao, J.; et al. Genome-wide association study finds multiple loci associated with intraocular pressure in HS rats. Front. Genet. 2022, 13, 1029058. [Google Scholar] [CrossRef]
- Devyatkin, V.A.; Redina, O.E.; Kolosova, N.G.; Muraleva, N.A. Single-Nucleotide Polymorphisms Associated with the Senescence-Accelerated Phenotype of OXYS Rats: A Focus on Alzheimer’s Disease-Like and Age-Related-Macular-Degeneration-Like Pathologies. J. Alzheimers Dis. 2020, 73, 1167–1183. [Google Scholar] [CrossRef]
- Kitagawa, A.; Kizub, I.; Jacob, C.; Michael, K.; D’Alessandro, A.; Reisz, J.A.; Grzybowski, M.; Geurts, A.M.; Rocic, P.; Gupte, R.; et al. CRISPR-Mediated Single Nucleotide Polymorphism Modeling in Rats Reveals Insight Into Reduced Cardiovascular Risk Associated With Mediterranean G6PD Variant. Hypertension 2020, 76, 523–532. [Google Scholar] [CrossRef]
- GB 14923-2022; Laboratory Animal—Genetic Quality Control. CHINESE GB Standards: Beijing, China, 2022. Available online: http://c.gb688.cn/bzgk/gb/showGb?type=download&hcno=D5AD6845D0955F91D1F53D3FE8DE2FDB (accessed on 15 June 2024).
- Wang, K.; Li, H.; Xu, Y.; Shao, Q.; Yi, J.; Wang, R.; Cai, W.; Hang, X.; Zhang, C.; Cai, H.; et al. MFEprimer-3.0: Quality control for PCR primers. Nucleic Acids Res. 2019, 47, W610–W613. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Xue, J.; Tan, M.; Chen, D.; Xiao, Y.; Liu, G.; Zheng, Y.; Wu, Q.; Liao, M.; Lv, M.; et al. An MPS-Based 50plex Microhaplotype Assay for Forensic DNA Analysis. Genes 2023, 14, 865. [Google Scholar] [CrossRef] [PubMed]
- LI, Y.; Wang, H.; Li, C.; Guo, M.; Wei, J.; Zhang, Y.; Huang, R.; Chen, Z.; Du, X. Population Genetic Structure Analysis of 3 Outbred Mice Populations from National Rodent Seed Center Using 45 loci of Single Nucleotide Polymorphism. Lab. Anim. Sci. 2018, 35, 1–7. [Google Scholar]
- Porras-Hurtado, L.; Ruiz, Y.; Santos, C.; Phillips, C.; Carracedo, A.; Lareu, M.V. An overview of STRUCTURE: Applications, parameter settings, and supporting software. Front. Genet. 2013, 4, 98. [Google Scholar] [CrossRef]
- Oldoni, F.; Yoon, L.; Wootton, S.C.; Lagacé, R.; Kidd, K.K.; Podini, D. Population genetic data of 74 microhaplotypes in four major U.S. population groups. Forensic Sci. Int. Genet. 2020, 49, 102398. [Google Scholar] [CrossRef]
- Wang, X.; Shi, W.; Zhao, S.; Gong, D.; Li, S.; Hu, C.; Chen, Z.J.; Li, Y.; Yan, J. Whole exome sequencing in unexplained recurrent miscarriage families identified novel pathogenic genetic causes of euploid miscarriage. Hum. Reprod. 2023, 38, 1003–1018. [Google Scholar] [CrossRef]
- Zhang, Q.; Qi, Y.; Wang, S.; Zhao, F.; Zou, L.; Zhou, Q.; Geng, P.; Hong, Y.; Yang, H.; Luo, Q.; et al. Identification and in vitro functional assessment of 10 CYP2C9 variants found in Chinese Han subjects. Front. Endocrinol. 2023, 14, 1139805. [Google Scholar] [CrossRef]
- Kidd, K.K.; Pakstis, A.J.; Speed, W.C.; Lagacé, R.; Chang, J.; Wootton, S.; Haigh, E.; Kidd, J.R. Current sequencing technology makes microhaplotypes a powerful new type of genetic marker for forensics. Forensic Sci. Int. Genet. 2014, 12, 215–224. [Google Scholar] [CrossRef]
- Andrews, K.R.; Hunter, S.S.; Torrevillas, B.K.; Céspedes, N.; Garrison, S.M.; Strickland, J.; Wagers, D.; Hansten, G.; New, D.D.; Fagnan, M.W.; et al. A new mouse SNP genotyping assay for speed congenics: Combining flexibility, affordability, and power. BMC Genom. 2021, 22, 378. [Google Scholar] [CrossRef]
- Wang, F.-Q.; Fan, X.-C.; Zhang, Y.; Sun, L.; Liu, C.-H.; Jiang, J.-F. Establishment and application of an SNP molecular identification system for grape cultivars. J. Integr. Agric. 2022, 21, 1044–1057. [Google Scholar] [CrossRef]
- Petkov, P.M.; Ding, Y.; Cassell, M.A.; Zhang, W.; Wagner, G.; Sargent, E.E.; Asquith, S.; Crew, V.; Johnson, K.A.; Robinson, P.; et al. An efficient SNP system for mouse genome scanning and elucidating strain relationships. Genome Res. 2004, 14, 1806–1811. [Google Scholar] [CrossRef]
- Li, Y.; Chen, Z.; Liu, J.; Li, C.; Liu, X.; Lv, J.; Guo, M.; Du, X. Genetic Mornitoring for Inbred Mice with Single Nucleotide Polymorphisms. Lab. Anim. Comp. Med. 2018, 38, 16–21. [Google Scholar]
- Wei, J.; Wang, H.; Zhou, J.; Zhao, L.; Li, H.; Yue, B. Application of SNP Markers in Identification of Inbred Mice Strains and Primer Sequences. CN115976226A, 18 April 2023. [Google Scholar]


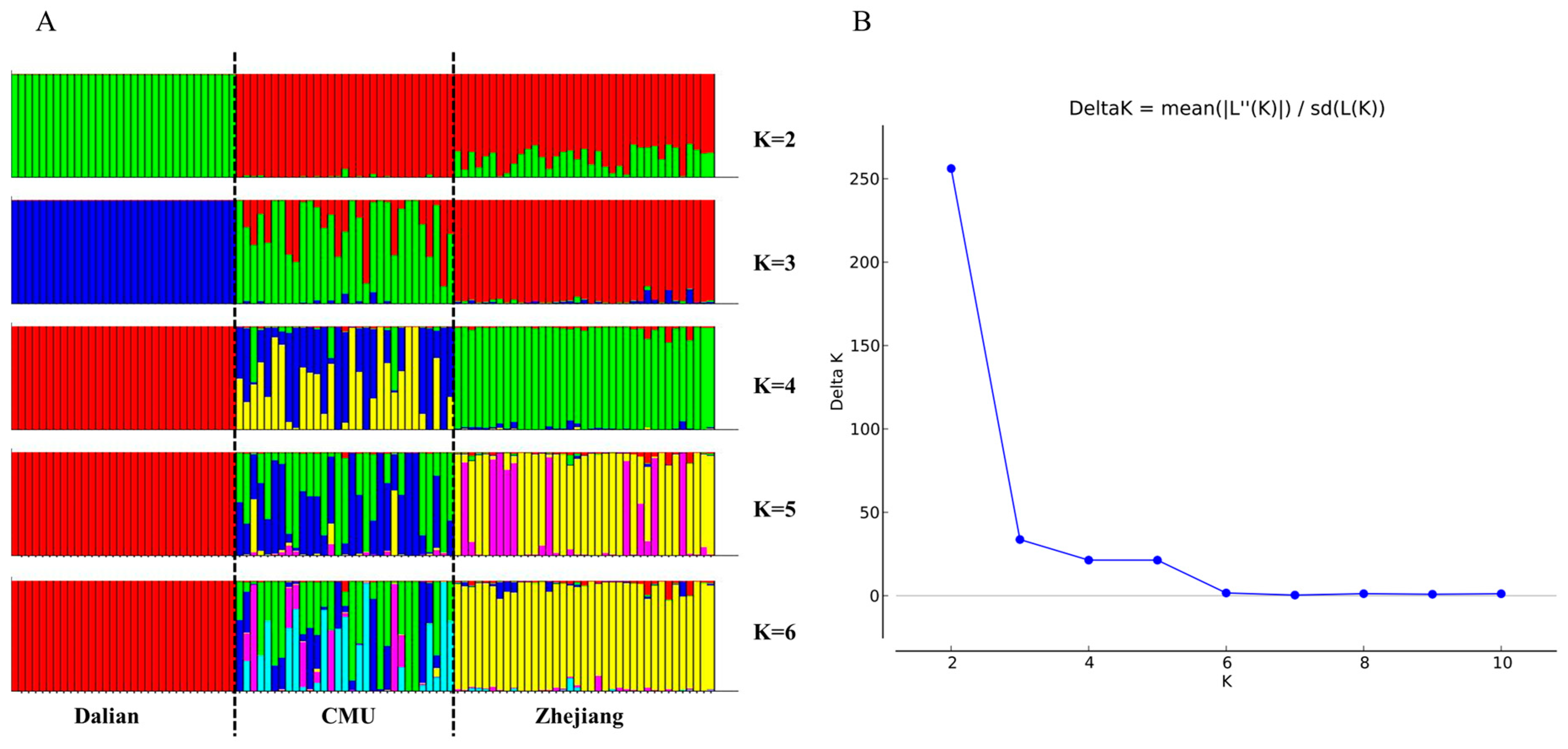
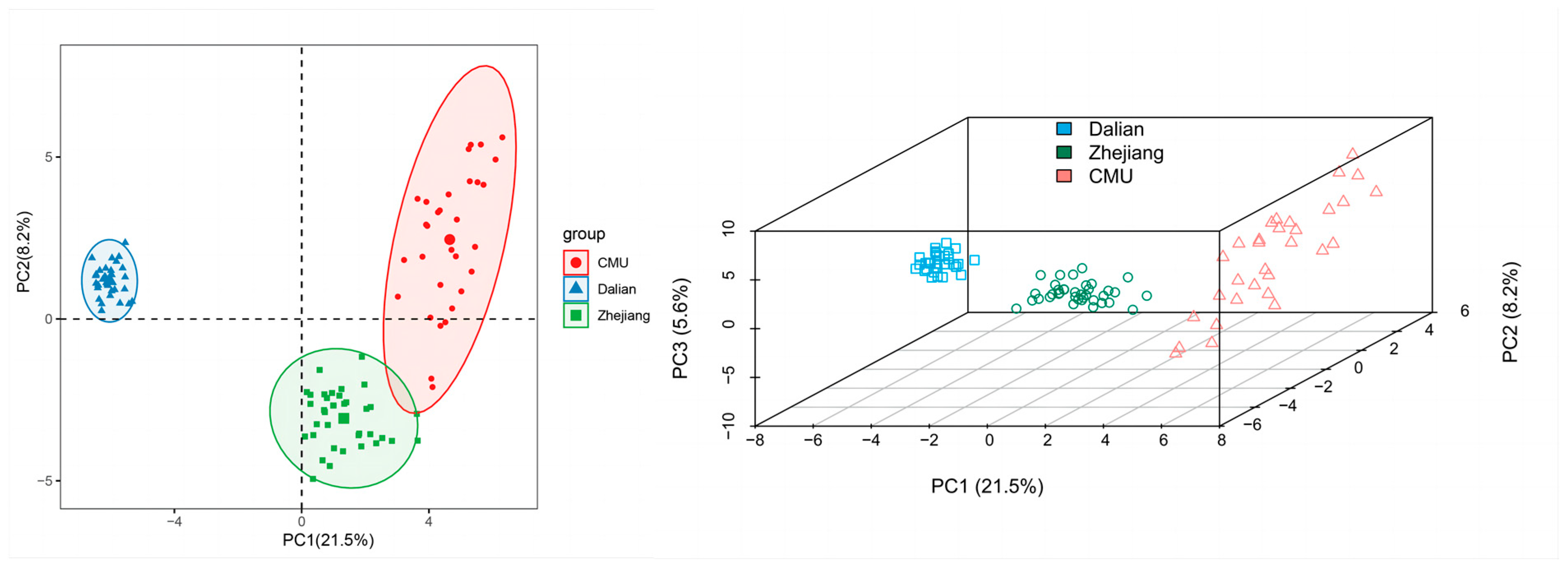
| Mongolian Gerbils | Na | ne | I | Obs Hom | Obs Het | Exp Hom | Exp Het | Ave Het | Percentage of Polymorphic Loci |
|---|---|---|---|---|---|---|---|---|---|
| CMU | 1.9903 | 1.6953 | 0.5753 | 0.6019 | 0.3981 | 0.6002 | 0.3998 | 0.3934 | 99.03% |
| Zhejiang | 2.0000 | 1.6550 | 0.5594 | 0.6229 | 0.3771 | 0.6167 | 0.3833 | 0.3782 | 100% |
| Dalian | 1.5146 | 1.3408 | 0.2882 | 0.7770 | 0.2230 | 0.8014 | 0.1986 | 0.1955 | 51.46% |
| Number | Genotyping | Zhejiang | Dalian | CMU | p Value |
|---|---|---|---|---|---|
| MG15 | AA | 0.08 | 0.09 | 0.00 | <0.001 |
| AG | 0.35 | 0.47 | 0.00 | ||
| GG | 0.57 | 0.44 | 1.00 | ||
| MG89 | CC | 0.00 | 0.00 | 0.68 | <0.001 |
| CT | 0.05 | 0.00 | 0.32 | ||
| TT | 0.95 | 1.00 | 0.00 | ||
| MG126 | CC | 0.03 | 1.00 | 0.00 | <0.001 |
| CT | 0.09 | 0.00 | 0.00 | ||
| TT | 0.89 | 0.00 | 1.00 | ||
| MG159 | CC | 0.97 | 0.59 | 0.97 | <0.001 |
| TT | 0.03 | 0.41 | 0.03 | ||
| MG164 | AA | 0.32 | 0.97 | 0.74 | <0.001 |
| AC | 0.35 | 0.03 | 0.26 | ||
| CC | 0.32 | 0.00 | 0.00 | ||
| MG206 | AA | 0.22 | 0.97 | 0.39 | <0.001 |
| AG | 0.62 | 0.03 | 0.42 | ||
| GG | 0.16 | 0.00 | 0.19 | ||
| MG215 | CC | 0.03 | 0.00 | 0.90 | <0.001 |
| CT | 0.54 | 0.00 | 0.10 | ||
| TT | 0.43 | 1.00 | 0.00 |
| Number | CHROM | POS | Variant_Classification | REF | ALT | Inbred Cerebral Ischemia Line | Inbred Diabetes Line |
|---|---|---|---|---|---|---|---|
| MG37 | NW_018657859.1 | 1,081,095 | 5′Flank | C | G | G/G | C/C |
| MG46 | NW_018657888.1 | 735,326 | Missense_Mutation | A | G | A/A | G/G |
| MG47 | NW_018657888.1 | 1,469,235 | Nonstop_Mutation | A | T | T/T | A/A |
| MG49 | NW_018657893.1 | 1,416,535 | Missense_Mutation | C | T | C/C | T/T |
| MG54 | NW_018657915.1 | 154,939 | Intron | T | C | T/T | C/C |
| MG73 | NW_018658000.1 | 110,223 | Missense_Mutation | T | C | T/T | C/C |
| MG82 | NW_018658044.1 | 1,083,386 | Missense_Mutation | G | T | T/T | G/G |
| MG94 | NW_018658087.1 | 982,880 | Missense_Mutation | T | C | C/C | T/T |
| MG107 | NW_018658151.1 | 621,718 | 3′Flank | G | T | T/T | G/G |
| MG114 | NW_018658168.1 | 333,441 | 5′Flank | C | T | T/T | C/C |
| MG124 | NW_018658203.1 | 697,270 | Missense_Mutation | C | T | T/T | C/C |
| MG132 | NW_018658278.1 | 540,596 | Nonsense_Mutation | C | A | A/A | C/C |
| MG142 | NW_018658707.1 | 206,847 | Missense_Mutation | C | A | A/A | C/C |
| MG190 | NW_018661696.1 | 2303 | Missense_Mutation | A | G | G/G | A/A |
| MG219 | NW_018692212.1 | 1211 | 5′Flank | G | A | G/G | A/A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, Y.; Cui, Y.; Sun, M.; Zhu, X.; Zhang, Y.; Lu, J.; Li, C.; Lv, J.; Guo, M.; Liu, X.; et al. Establishment and Application of a Novel Genetic Detection Panel for SNPs in Mongolian Gerbils. Genes 2024, 15, 817. https://doi.org/10.3390/genes15060817
Guo Y, Cui Y, Sun M, Zhu X, Zhang Y, Lu J, Li C, Lv J, Guo M, Liu X, et al. Establishment and Application of a Novel Genetic Detection Panel for SNPs in Mongolian Gerbils. Genes. 2024; 15(6):817. https://doi.org/10.3390/genes15060817
Chicago/Turabian StyleGuo, Yafang, Yutong Cui, Minghe Sun, Xiao Zhu, Yilang Zhang, Jing Lu, Changlong Li, Jianyi Lv, Meng Guo, Xin Liu, and et al. 2024. "Establishment and Application of a Novel Genetic Detection Panel for SNPs in Mongolian Gerbils" Genes 15, no. 6: 817. https://doi.org/10.3390/genes15060817
APA StyleGuo, Y., Cui, Y., Sun, M., Zhu, X., Zhang, Y., Lu, J., Li, C., Lv, J., Guo, M., Liu, X., Chen, Z., Du, X., & Huo, X. (2024). Establishment and Application of a Novel Genetic Detection Panel for SNPs in Mongolian Gerbils. Genes, 15(6), 817. https://doi.org/10.3390/genes15060817





