Molecular Mapping and Transfer of Quantitative Trait Loci (QTL) for Sheath Blight Resistance from Wild Rice Oryza nivara to Cultivated Rice (Oryza sativa L.)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Population Development
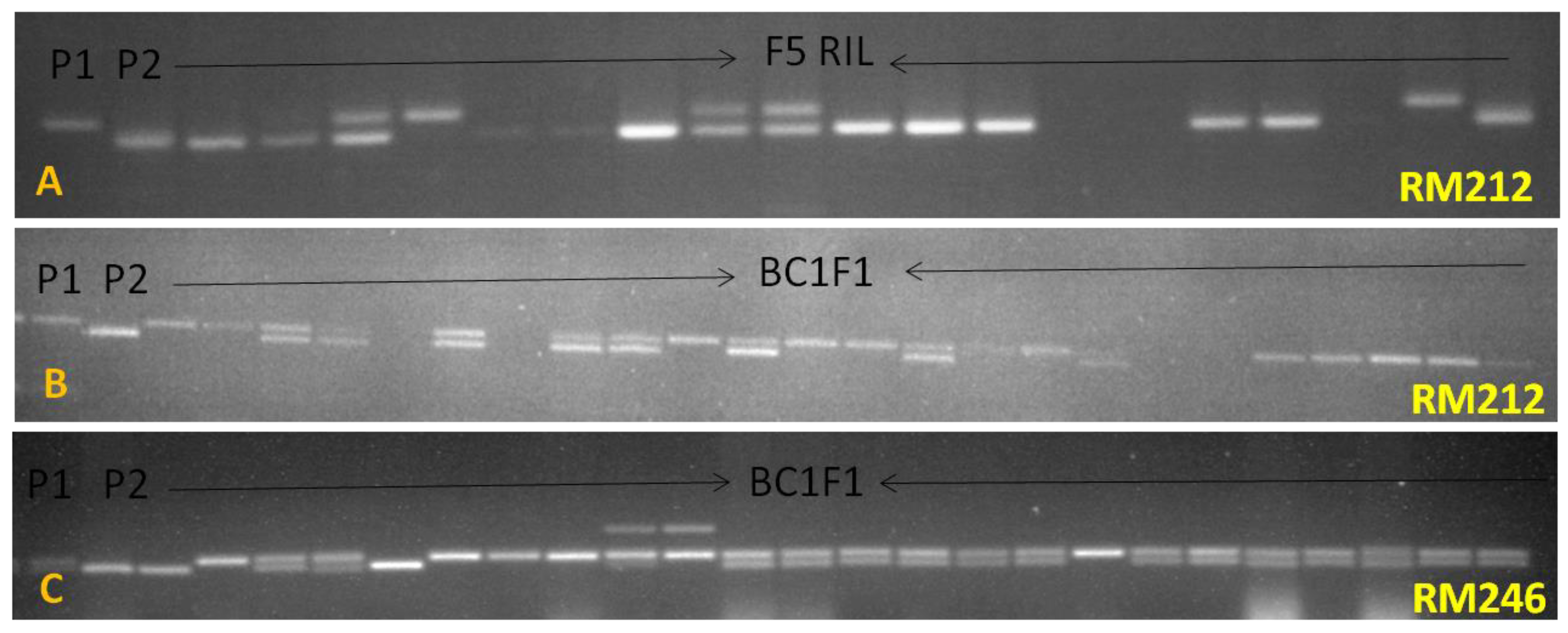
2.2. Development of Backcross Populations
2.3. Preparation of Pure Culture and Disease Assessment
2.4. DNA Extraction and SNP Genotyping
2.5. QTL Mapping
2.6. Statistical Analysis
2.6.1. Phenotypic Data Analysis
2.6.2. SNP Data Analysis
2.6.3. Linkage and QTL Mapping
2.7. Identification of the Linked Marker with Shb Resistance
2.8. Identification of Candidate Genes
3. Results
3.1. Frequency Distribution of ShB Disease in the F2 and F2:3 Populations
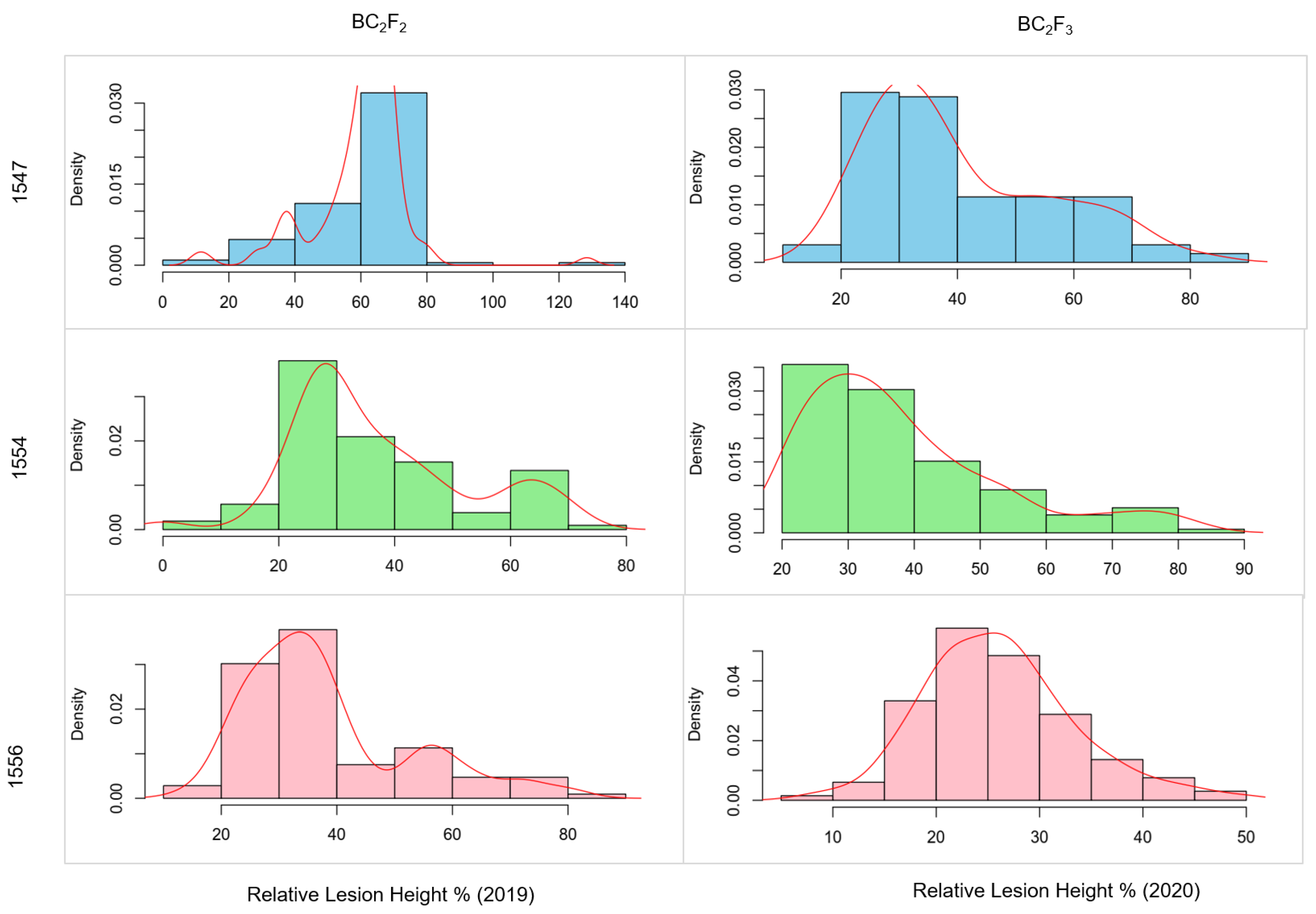
3.2. Identification of Promising Lines in the BC2F2 and BC2F3 Populations
3.3. Genotyping of the F2 Population Using ddRAD-seq
3.4. Linkage and QTL Mapping
3.5. Identification of the Linked Markers for MAS
3.6. Identification of Candidate Genes Underlying ShB Resistance QTL
4. Discussion
4.1. O. nivara as a Potential Donor for Resistance and Productivity Traits
4.2. ShB Resistance QTLs
4.3. Candidate Genes Confer Resistance to R. solani
4.4. Breeding Applications for Developing ShB-Resistant Rice Cultivars
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zheng, A.; Lin, R.; Zhang, D.; Qin, P.; Xu, L.; Ai, P.; Ding, L.; Wang, Y.; Chen, Y.; Liu, Y.; et al. The evolution and pathogenic mechanisms of the rice sheath blight pathogen. Nat. Commun. 2013, 4, 1424. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.K.; Islam, M.R.; Sundaram, R.M.; Bhuiyan, M.A.R.; Wickneswari, R. Introgression of the QTL qSB11-1TT conferring sheath blight resistance in rice (Oryza sativa) into an elite variety, UKMRC2, and evaluation of its backcross-derived plants. Front. Plant Sci. 2023, 13, 981345. [Google Scholar] [CrossRef] [PubMed]
- Rao, T.B.; Chopperla, R.; Prathi, N.B.; Balakrishnan, M.; Prakasam, V.; Laha, G.S.; Balachandran, S.M.; Mangrauthia, S.K. A Comprehensive gene expression profile of pectin degradation enzymes reveals the molecular events during cell wall degradation and pathogenesis of rice sheath blight pathogen Rhizoctonia solani AG1-IA. J. Fungi 2020, 6, 71. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhang, F.; Pinson, S.R.M.; Edwards, J.D.; Jackson, A.K.; Xia, X.; Eizenga, G.C. Assessment of rice sheath blight resistance including associations with plant architecture, as revealed by genome-wide association studies. Rice 2022, 15, 31. [Google Scholar] [CrossRef] [PubMed]
- Vijay Krishna Kumar, K.; Reddy, M.S.; Kloepper, J.W.; Lawrence, K.S.; Groth, D.E.; Miller, M.E. Sheath blight disease of rice (Oryza Sativa L.)—An Overview. Biosci. Biotechnol. Res. Asia 2009, 6, 465–480. [Google Scholar]
- Wu, W.; Huang, J.; Cui, K.; Nie, L.; Wang, Q.; Yang, F.; Shah, F.; Yao, F.; Peng, S. Sheath blight reduces stem breaking resistance and increases lodging susceptibility of rice plants. Field Crops Res. 2012, 128, 101–108. [Google Scholar] [CrossRef]
- Goad, D.M.; Jia, Y.; Gibbons, A.; Liu, Y.; Gealy, D.; Caicedo, A.L.; Olsen, K.M. Identification of novel QTL conferring sheath blight resistance in two weedy rice mapping populations. Rice 2020, 13, 21. [Google Scholar] [CrossRef] [PubMed]
- Savary, S.; Castilla, N.P.; Elazegui, F.A.; McLaren, C.G.; Ynalvez, M.A.; Teng, P.S. Direct and indirect effects of nitrogen supply and disease source structure on rice sheath blight spread. Phytopathology 1995, 85, 959–965. [Google Scholar] [CrossRef]
- Molla, K.A.; Karmakar, S.; Molla, J.; Bajaj, P.; Varshney, R.K.; Datta, S.K.; Datta, K. Understanding sheath blight resistance in rice: The road behind and the road ahead. Plant Biotechnol. J. 2020, 18, 895–915. [Google Scholar] [CrossRef]
- Zeng, Y.X.; Ji, Z.J.; Ma, L.Y.; Li, X.M.; Yang, C.D. Advances in mapping loci conferring resistance to rice sheath blight and mining Rhizoctonia solani resistant resources. Rice Sci. 2011, 18, 56–66. [Google Scholar] [CrossRef]
- Singh, P.; Mazumdar, P.; Harikrishna, J.A.; Babu, S. Sheath blight of rice: A review and identification of priorities for future research. Planta 2019, 250, 1387–1407. [Google Scholar] [CrossRef] [PubMed]
- Srinivasachary; Willocquet, L.; Savary, S. Resistance to rice sheath blight (Rhizoctonia Solani Kühn) [(teleomorph: Thanatephorus cucumeris (A.B. Frank) Donk.] Disease: Current status and perspectives. Euphytica 2011, 178, 1–22. [Google Scholar] [CrossRef]
- Zou, J.H.; Pan, X.B.; Chen, Z.X.; Xu, J.Y.; Lu, J.F.; Zhai, W.X.; Zhu, L.H. Mapping quantitative trait loci controlling sheath blight resistance in two rice cultivars (Oryza Sativa L.). Theor. Appl. Genet. 2000, 101, 569–573. [Google Scholar] [CrossRef]
- Li, D.; Li, S.; Wei, S.; Sun, W. Strategies to manage rice sheath blight: Lessons from interactions between rice and Rhizoctonia Solani. Rice 2021, 14, 21. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Pinson, S.R.M.; Marchetti, M.A.; Stansel, J.W.; Park, W.D. Characterization of quantitative trait loci (QTLs) in cultivated rice contributing to field resistance to sheath blight (Rhizoctonia Solani). Theor. Appl. Genet. 1995, 91, 382–388. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.P.; Xing, Y.Z.; Chen, Z.X.; Gu, S.L.; Pan, X.B.; Chen, X.L.; Zhang, Q.F. Mapping QTLs for horizontal resistance to sheath blight in an elite rice restorer line, Minghui 63. Acta Genet. Sin. 2002, 29, 622–626. [Google Scholar] [PubMed]
- Pinson, S.R.M.; Capdevielle, F.M.; Oard, J.H. Confirming QTLs and finding additional loci conditioning sheath blight resistance in rice using recombinant inbred lines. Crop Sci. 2005, 45, 503–510. [Google Scholar] [CrossRef]
- Tan, C.X.; Ji, X.M.; Yang, Y.; Pan, X.Y.; Zuo, S.M.; Zhang, Y.F.; Zou, J.H.; Chen, Z.X.; Zhu, L.H.; Pan, X.B. Identification and marker-assisted selection of two major quantitative genes controlling rice sheath blight resistance in backcross generations. Acta Genet. Sin. 2005, 32, 399–405. [Google Scholar] [PubMed]
- Sharma, A.; McClung, A.M.; Pinson, S.R.M.; Kepiro, J.L.; Shank, A.R.; Tabien, R.E.; Fjellstrom, R. Genetic mapping of sheath blight resistance QTLs within tropical japonica rice cultivars. Crop Sci. 2009, 49, 256–264. [Google Scholar] [CrossRef]
- Channamallikarjuna, V.; Sonah, H.; Prasad, M.; Rao, G.J.N.; Chand, S.; Upreti, H.C.; Singh, N.K.; Sharma, T.R. Identification of major quantitative trait loci qSBR11-1 for sheath blight resistance in rice. Mol. Breed. 2010, 25, 155–166. [Google Scholar] [CrossRef]
- Silva, J.; Scheffler, B.; Sanabria, Y.; de Guzman, C.; Galam, D.; Farmer, A.; Woodward, J.; May, G.; Oard, J. Identification of candidate genes in rice for resistance to sheath blight disease by whole genome sequencing. Theor. Appl. Genet. 2012, 124, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Zuo, S.; Yin, Y.; Pan, C.; Chen, Z.; Zhang, Y.; Gu, S.; Zhu, L.; Pan, X. Fine mapping of qSB-11LE, the QTL that confers partial resistance to rice sheath blight. Theor. Appl. Genet. 2013, 126, 1257–1272. [Google Scholar] [CrossRef] [PubMed]
- Zuo, S.; Zhang, Y.; Yin, Y.; Li, G.; Zhang, G.; Wang, H.; Chen, Z.; Pan, X. Fine-Mapping of qSB-9 TQ, a gene conferring major quantitative resistance to rice sheath blight. Mol. Breed. 2014, 34, 2191–2203. [Google Scholar] [CrossRef]
- Yadav, S.; Anuradha, G.; Kumar, R.R.; Vemireddy, L.R.; Sudhakar, R.; Donempudi, K.; Venkata, D.; Jabeen, F.; Narasimhan, Y.K.; Marathi, B.; et al. Identification of QTLs and possible candidate genes conferring sheath blight resistance in rice (Oryza sativa L.). Springerplus 2015, 4, 175. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Jia, Y.; Correa-Victoria, F.J.; Prado, G.A.; Yeater, K.M.; McClung, A.; Correll, J.C. Mapping quantitative trait loci responsible for resistance to sheath blight in rice. Phytopathology 2009, 99, 1078–1084. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Feng, Z.; Kang, H.; Zhao, J.; Chen, T.; Li, Q.; Gong, H.; Zhang, Y.; Chen, X.; Pan, X.; et al. Identification of new resistance loci against sheath blight disease in rice through genome-wide association study. Rice Sci. 2019, 26, 21–31. [Google Scholar] [CrossRef]
- Zhang, F.; Zeng, D.; Zhang, C.S.; Lu, J.L.; Chen, T.J.; Xie, J.P.; Zhou, Y.L. Genome-wide association analysis of the genetic basis for sheath blight resistance in rice. Rice 2019, 12, 93. [Google Scholar] [CrossRef] [PubMed]
- Oreiro, E.G.; Grimares, E.K.; Atienza-Grande, G.; Quibod, I.L.; Roman-Reyna, V.; Oliva, R. Genome-wide associations and transcriptional profiling reveal ros regulation as one underlying mechanism of sheath blight resistance in rice. Mol. Plant-Microbe Interact. 2020, 33, 212–222. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Shu, X.; Jing, X.; Jiao, C.; Chen, L.; Zhang, J.; Ma, L.; Jiang, Y.; Yamamoto, N.; Li, S.; et al. Identification of rice (Oryza sativa L.) genes involved in sheath blight resistance via a genome-wide association study. Plant Biotechnol. J. 2021, 19, 1553–1566. [Google Scholar] [CrossRef]
- Kumar, K.; Sarao, P.S.; Bhatia, D.; Neelam, K.; Kaur, A.; Mangat, G.S.; Brar, D.S.; Singh, K. High-resolution genetic mapping of a novel brown planthopper resistance locus, Bph34 in Oryza sativa L. X Oryza nivara (Sharma & Shastry) derived interspecific F2 population. Theor. Appl. Genet. 2018, 131, 1163–1171. [Google Scholar] [CrossRef]
- Kumar, K.; Neelam, K.; Singh, G.; Mathan, J.; Ranjan, A.; Brar, D.S.; Singh, K. Production and cytological characterization of a synthetic amphiploid derived from a cross between Oryza sativa and Oryza punctata. Genome 2019, 62, 705–714. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, S.K.; Neelam, K.; Jain, J.; Kaur, R.; Pannu, P.P.S.; Lenka, S.K.; Lore, J.S.; Singh, K. Identification of promising resistance sources against sheath blight from the annual wild species of rice Oryza nivara (Sharma et Shastry). Plant Genet. Resour. Characterisation Util. 2019, 17, 554–558. [Google Scholar] [CrossRef]
- Lore, J.S.; Hunjan, M.S.; Singh, P.; Willocquet, L.; Sri, S.; Savary, S. Phenotyping of partial physiological resistance to rice sheath blight. J. Phytopathol. 2013, 161, 224–229. [Google Scholar] [CrossRef]
- Sharma, N.R.; Teng, P.S. Comparison of rice sheath blight (ShB) assessment methods. Int. Rice Res. Newsl. 1990, 15, 20–21. [Google Scholar]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Van Os, H.; Stam, P.; Visser, R.G.F.; Van Eck, H.J. RECORD: A novel method for ordering loci on a genetic linkage map. Theor. Appl. Genet. 2005, 112, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Churchill, G.A.; Doerge, R.W. Empirical threshold values for quantitative trait mapping. Genetics 1994, 138, 963–971. [Google Scholar] [CrossRef]
- Jin, J.; Huang, W.; Gao, J.P.; Yang, J.; Shi, M.; Zhu, M.Z.; Luo, D.; Lin, H.X. Genetic control of rice plant architecture under domestication. Nat. Genet. 2008, 40, 1365–1369. [Google Scholar] [CrossRef]
- Li, C.; Zhou, A.; Sang, T. Genetic analysis of rice domestication syndrome with the wild annual species, Oryza nivara. New Phytol. 2006, 170, 185–194. [Google Scholar] [CrossRef]
- Zhu, Z.; Tan, L.; Fu, Y.; Liu, F.; Cai, H.; Xie, D.; Wu, F.; Wu, J.; Matsumoto, T.; Sun, C. Genetic control of inflorescence architecture during rice domestication. Nat. Commun. 2013, 4, 2200. [Google Scholar] [CrossRef] [PubMed]
- Cheema, K.K.; Grewal, N.K.; Vikal, Y.; Sharma, R.; Lore, J.S.; Das, A.; Bhatia, D.; Mahajan, R.; Gupta, V.; Bharaj, T.S.; et al. A novel bacterial blight resistance gene from Oryza nivara mapped to 38 Kb region on chromosome 4L and transferred to Oryza sativa L. Genet. Res. 2008, 90, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Neelam, K.; Lore, J.S.; Kaur, K.; Pathania, S.; Kumar, K.; Sahi, G.; Mangat, G.S.; Singh, K. Identification of resistance sources in wild species of rice against two recently evolved pathotypes of Xanthomonas Oryzae pv. Oryzae. Plant Genet. Resour. Characterisation Util. 2017, 15, 558–562. [Google Scholar] [CrossRef]
- Sarao, P.S.; Sahi, G.K.; Neelam, K.; Mangat, G.S.; Patra, B.C.; Singh, K. Donors for resistance to brown planthopper Nilaparvata lugens (Stål) from wild rice species. Rice Sci. 2016, 23, 219–224. [Google Scholar] [CrossRef]
- Kumar, K.; Kaur, P.; Kishore, A.; Vikal, Y.; Singh, K.; Neelam, K. Recent advances in genomics-assisted breeding of brown planthopper (Nilaparvata lugens) resistance in rice (Oryza sativa). Plant Breed. 2020, 139, 1052–1066. [Google Scholar] [CrossRef]
- Bhatia, D.; Joshi, S.; Das, A.; Vikal, Y.; Sahi, G.K.; Neelam, K.; Kaur, K.; Singh, K. Introgression of yield component traits in rice (Oryza sativa ssp. indica) through interspecific hybridization. Crop Sci. 2017, 57, 1557–1573. [Google Scholar] [CrossRef]
- Eizenga, G.C.; Prasad, B.; Jackson, A.K.; Jia, M.H. Identification of rice sheath blight and blast quantitative trait loci in two different O. sativa/O. nivara advanced backcross populations. Mol. Breed. 2013, 31, 889–907. [Google Scholar] [CrossRef]
- Kumar, K.; Neelam, K.; Bhatia, D.; Kaur, R.; Khanna, R.; Mangat, G.S.; Vikal, Y.; Singh, K. High resolution genetic mapping and identification of a candidate gene(s) for the purple sheath color and plant height in an interspecific F2 population derived from Oryza nivara Sharma & Shastry × Oryza sativa L. cross. Genet. Resour. Crop Evol. 2020, 67, 97–105. [Google Scholar] [CrossRef]
- Eizenga, G.C.; Neves, P.C.F.; Bryant, R.J.; Agrama, H.A.; Mackill, D.J. Evaluation of a M-202 × Oryza nivara advanced backcross mapping population for seedling vigor, yield components and quality. Euphytica 2016, 208, 157–171. [Google Scholar] [CrossRef]
- Mallikarjuna Swamy, B.P.; Kaladhar, K.; Reddy, G.A.S.H.O.K.; Viraktamath, B.C.; Sarla, N. Mapping and introgression of qtl for yield and related traits in two backcross populations derived from Oryza sativa cv. Swarna and two accessions of O. nivara. J. Genet. 2014, 93, 643–654. [Google Scholar] [CrossRef]
- Balakrishnan, D.; Subrahmanyam, D.; Badri, J.; Raju, A.K.; Rao, Y.V.; Beerelli, K.; Mesapogu, S.; Surapaneni, M.; Ponnuswamy, R.; Padmavathi, G.; et al. Genotype × environment interactions of yield traits in backcross introgression lines derived from Oryza sativa cv. Swarna/Oryza nivara. Front. Plant Sci. 2016, 7, 1530. [Google Scholar] [CrossRef] [PubMed]
- Balakrishnan, D.; Surapaneni, M.; Yadavalli, V.R.; Addanki, K.R.; Mesapogu, S.; Beerelli, K.; Neelamraju, S. Detecting CSSLs and yield QTLs with additive, epistatic and QTL×environment interaction effects from Oryza sativa × O. nivara IRGC81832 cross. Sci. Rep. 2020, 10, 7766. [Google Scholar] [CrossRef] [PubMed]
- Haritha, G.; Swamy, B.P.M.; Naik, M.L.; Jyothi, B.; Divya, B.; Malathi, S.; Sarla, N. Yield traits and associated marker segregation in elite introgression lines derived from O. Sativa × O. nivara. Rice Sci. 2018, 25, 19–31. [Google Scholar] [CrossRef]
- Surapaneni, M.; Balakrishnan, D.; Mesapogu, S.; Addanki, K.R.; Yadavalli, V.R.; Tripura Venkata, V.G.N.; Neelamraju, S. Identification of major effect QTLs for agronomic traits and cssls in rice from Swarna/Oryza nivara derived backcross inbred lines. Front. Plant Sci. 2017, 8, 1027. [Google Scholar] [CrossRef] [PubMed]
- Bal, A.; Samal, P.; Chakraborti, M.; Mukherjee, A.K.; Ray, S.; Molla, K.A.; Behera, L.; Samal, R.; Sarangi, S.; Sahoo, P.; et al. Stable Quantitative Trait Locus (QTL) for Sheath Blight Resistance from Rice Cultivar CR 1014. Euphytica 2020, 216, 182. [Google Scholar] [CrossRef]
- Nelson, J.C.; Oard, J.H.; Groth, D.; Utomo, H.S.; Jia, Y.; Liu, G.; Moldenhauer, K.A.K.; Correa-Victoria, F.J.; Fjellstrom, R.G.; Scheffler, B.; et al. Sheath-blight resistance QTLs in japonica rice germplasm. Euphytica 2012, 184, 23–34. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, L.; Fu, D.; Lou, Q.; Mei, H.; Xiong, L.; Li, M.; Xu, X.; Mei, X.; Luo, L. Dissection of additive, epistatic effect and qtl × environment interaction of quantitative trait loci for sheath blight resistance in rice. Hereditas 2014, 151, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.Q.; Zhu, J.; He, C.X.; Benmoussa, M.; Wu, P. Quantitative trait loci analysis for the developmental behavior of tiller number in rice (Oryza Sativa L.). Theor. Appl. Genet. 1998, 97, 267–274. [Google Scholar] [CrossRef]
- Yan, J.; Zhu, J.; He, C.; Benmoussa, M.; Wu, P. Molecular marker-assisted dissection of genotype x environment interaction for plant type traits in rice (Oryza Sativa L.). Crop Sci. 1999, 39, 538–544. [Google Scholar] [CrossRef]
- Hittalmani, S.; Shashidhar, H.E.; Bagali, P.G.; Huang, N.; Sidhu, J.S.; Singh, V.P.; Khush, G.S. Molecular mapping of quantitative trait loci for plant growth, yield and yield related traits across three diverse locations in a doubled haploid rice population. Euphytica 2002, 125, 207–214. [Google Scholar] [CrossRef]
- Hua, J.P.; Xing, Y.Z.; Xu, C.G.; Sun, X.L.; Yu, S.B.; Zhang, Q. Genetic dissection of an elite rice hybrid revealed that heterozygotes are not always advantageous for performance. Genetics 2002, 162, 1885–1895. [Google Scholar] [CrossRef] [PubMed]
- Cui, K.H.; Peng, S.B.; Xing, Y.Z.; Yu, S.B.; Xu, C.G.; Zhang, Q. Molecular dissection of the genetic relationships of source, sink and transport tissue with yield traits in rice. Theor. Appl. Genet. 2003, 106, 649–658. [Google Scholar] [CrossRef]
- Cho, Y.G.; Kang, H.J.; Lee, J.S.; Lee, Y.T.; Lim, S.J.; Gauch, H.; Eun, M.Y.; McCouch, S.R. Identification of quantitative trait loci in rice for yield, yield components, and agronomic traits across years and locations. Crop Sci. 2007, 47, 2403–2417. [Google Scholar] [CrossRef]
- Li, Z.K.; Yu, S.B.; Lafitte, H.R.; Huang, N.; Courtois, B.; Hittalmani, S.; Vijayakumar, C.H.M.; Liu, G.F.; Wang, G.C.; Shashidhar, H.E.; et al. QTL X environment interactions in rice. I. Heading date and plant height. Theor. Appl. Genet. 2003, 108, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Pinson, S.R.M.; Paterson, A.H.; Park, W.D.; Stansel, J.W. Genetics of hybrid sterility and hybrid breakdown in an intersubspecific rice (Oryza Sativa L.) population. Genetics 1997, 145, 1139–1148. [Google Scholar] [CrossRef]
- Kamoshita, A.; Zhang, J.; Siopongco, J.; Sarkarung, S.; Nguyen, H.T.; Wade, L.J. Effects of phenotyping environment on identification of quantitative trait loci for rice root morphology under anaerobic conditions. Crop Sci. 2002, 42, 255–265. [Google Scholar] [CrossRef]
- Dong, Y.; Kamiunten, H.; Ogawa, T.; Tsuzuki, E.; Terao, H.; Lin, D.; Matsuo, M. Mapping of QTLs for leaf developmental behavior in rice (Oryza Sativa L.). Euphytica 2004, 138, 169–175. [Google Scholar] [CrossRef]
- Jiang, L.; Zhang, W.; Xia, Z.; Jiang, G.; Qian, Q.; Li, A.; Cheng, Z.; Zhu, L.; Mao, L.; Zhai, W. A paracentric inversion suppresses genetic recombination at the FON3 locus with breakpoints corresponding to sequence gaps on rice chromosome 11L. Mol. Genet. Genomics 2007, 277, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Afzal, A.J.; Wood, A.J.; Lightfoot, D.A. Plant receptor-like serine threonine kinases: Roles in signaling and plant defense. Mol. Plant-Microbe Interact. 2008, 21, 507–517. [Google Scholar] [CrossRef]
- Bajsa, J.; Pan, Z.; Duke, S.O. Serine/threonine protein phosphatases: Multi-purpose enzymes in control of defense mechanisms. Plant Signal. Behav. 2011, 6, 1921–1925. [Google Scholar] [CrossRef]
- Aggarwal, S.K.; Malik, P.; Neelam, K.; Kumar, K.; Kaur, R.; Lore, J.S.; Singh, K. Genome-wide association mapping for identification of sheath blight resistance loci from wild rice Oryza Rufipogon. Euphytica 2022, 218, 144. [Google Scholar] [CrossRef]
- Büschges, R.; Hollricher, K.; Panstruga, R.; Simons, G.; Wolter, M.; Frijters, A.; Van Daelen, R.; Van der Lee, T.; Diergaarde, P.; Groenendijk, J.; et al. The barley Mlo gene: A novel control element of plant pathogen resistance. Cell 1997, 88, 695–705. [Google Scholar] [CrossRef] [PubMed]
- Datta, K.; Velazhahan, R.; Oliva, N.; Ona, I.; Mew, T.; Khush, G.S.; Muthukrishnan, S.; Datta, S.K. Over-Expression of the Cloned Rice Thaumatin-like Protein (PR-5) Gene in transgenic rice plants enhances environmental friendly resistance to Rhizoctonia Solani causing sheath blight disease. Theor. Appl. Genet. 1999, 98, 1138–1145. [Google Scholar] [CrossRef]
- Kalpana, K.; Maruthasalam, S.; Rajesh, T.; Poovannan, K.; Kumar, K.K.; Kokiladevi, E.; Raja, J.A.J.; Sudhakar, D.; Velazhahan, R.; Samiyappan, R.; et al. Engineering sheath blight resistance in elite indica rice cultivars using genes encoding defense proteins. Plant Sci. 2006, 170, 203–215. [Google Scholar] [CrossRef]
- Chen, Z.X.; Zhang, Y.F.; Feng, F.; Feng, M.H.; Jiang, W.; Ma, Y.Y.; Pan, C.H.; Hua, H.L.; Li, G.S.; Pan, X.B.; et al. Improvement of japonica rice resistance to sheath blight by pyramiding qSB-9TQ and qSB-7TQ. Filed Crop. Res. 2014, 161, 118–127. [Google Scholar] [CrossRef]
- Ramalingam, J.; Raveendra, C.; Savitha, P.; Vidya, V.; Chaithra, T.L.; Velprabakaran, S.; Saraswathi, R.; Ramanathan, A.; Arumugam Pillai, M.P.; Arumugachamy, S.; et al. Gene pyramiding for achieving enhanced resistance to bacterial blight, blast, and sheath blight diseases in rice. Front. Plant Sci. 2020, 11, 591457. [Google Scholar] [CrossRef]
- Wang, Y.; Pinson, S.R.M.; Fjellstrom, R.G.; Tabien, R.E. Phenotypic gain from introgression of two QTL, qSB9-2 and qSB12-1, for rice sheath blight resistance. Mol. Breed. 2012, 30, 293–303. [Google Scholar] [CrossRef]




| Seasons | Traits | LGs | QTLs | Left Marker | Right Marker | Genetic Distance (cM) | LOD Score | Additive | R2 |
|---|---|---|---|---|---|---|---|---|---|
| 2017 | Plant Height | 1 | qPH1.1 | rs.Chr1.37378469 | rs.Chr1.37464136 | 1321.1–1324.9 | 13.0331 | 13.923 | 0.173 |
| 4 | qPH4.1 | rs.Chr4.13005766 | rs.Chr4.13217468 | 1160.8–1164.7 | 2.9013 | 1.6064 | 0.2412 | ||
| 6 | qPH6.1 | rs.Chr6.12096188 | rs.Chr6.11979786 | 499.3–510.9 | 2.8491 | 6.2268 | 0.0502 | ||
| 8 | qPH8.1 | rs.Chr8.17265874 | rs.Chr8.15842297 | 487.0–490.9 | 2.9012 | −5.2969 | 0.1062 | ||
| Lesion Height | 2 | qLH2.1 | rs.Chr2.35457758 | rs.Chr2.35457823 | 0–2.7 | 3.3578 | −1.9855 | 0.1602 | |
| 3 | qLH3.1 | rs.Chr3.2694761 | rs.Chr3.3801316 | 605.7–626.6 | 2.7423 | −0.8633 | 0.11 | ||
| 7 | qLH7.1 | rs.Chr7.17816084 | rs.Chr7.17848951 | 378.5–386.6 | 3.4374 | 2.1383 | 0.1834 | ||
| 11 | qLH11.1 | rs.Chr11.27787652 | rs.Chr11.28970389 | 31.5–34.8 | 2.8294 | −2.0404 | 0.2308 | ||
| 11 | qLH11.2 | rs.Chr11.26522147 | rs.Chr11.26053892 | 90.5–98.5 | 2.863 | 1.8248 | 0.1896 | ||
| Relative Lesion Height | 1 | qRLH1.1 | rs.Chr1.38966610 | rs.Chr1.38764373 | 1345.9–1353.8 | 9.9124 | −7.0955 | 0.1667 | |
| 1 | qRLH1.2 | rs.Chr1.39839247 | rs.Chr1.39839342 | 1442.4–1443.5 | 3.3124 | 0.6929 | 0.1001 | ||
| 6 | qRLH6.1 | rs.Chr6.28594729 | rs.Chr6.29343276 | 927.3–932.9 | 3.4633 | 1.7665 | 0.1161 | ||
| 8 | qRLH8.1 | rs.Chr8.17265874 | rs.Chr8.15842297 | 487.0–490.9 | 3.4087 | 3.4381 | 0.1458 | ||
| 2018 | Plant Height | 1 | qPH1.2 | rs.Chr1.35874500 | rs.Chr1.37378469 | 1315.6–1321.1 | 3.1704 | 12.997 | 0.2974 |
| 2 | qPH2.1 | rs.Chr2.22628796 | rs.Chr2.22747179 | 624.4–632.5 | 3.1502 | −7.2949 | 0.0745 | ||
| Lesion Height | 3 | qLH3.2 | rs.Chr3.2694761 | rs.Chr3.3801316 | 605.7–626.6 | 4.9772 | −1.6642 | 0.1768 | |
| Relative Lesion Height | 1 | qRLH1.3 | rs.Chr1.614777 | rs.Chr1.649072 | 307.7–326.3 | 4.7678 | 2.4154 | 0.4805 | |
| 1 | qRLH1.4 | rs.Chr1.649072 | rs.Chr1.613037 | 326.3–338.2 | 3.7844 | 2.1927 | 0.4468 | ||
| 1 | qRLH1.5 | rs.Chr1.38678740 | rs.Chr1.38601901 | 1364.3–1367.5 | 11.8044 | −9.9129 | 0.2236 | ||
| 3 | qRLH3.1 | rs.Chr3.2801043 | rs.Chr3.3215296 | 566.3–573.8 | 2.672 | −3.6511 | 0.1088 | ||
| Mean | Plant Height | 1 | qPH1.3 | rs.Chr1.35874500 | rs.Chr1.37378469 | 1315.6–1321.1 | 4.0694 | 7.5129 | 0.3769 |
| 2 | qPH2.2 | rs.Chr2.28243124 | rs.Chr2.28243186 | 531.4–532.6 | 4.0487 | −4.5695 | 0.1224 | ||
| Lesion Height | 3 | qLH3.2 | rs.Chr3.2801043 | rs.Chr3.3215296 | 566.3–573.8 | 2.9809 | −2.4302 | 0.1453 | |
| 11 | qLH11.3 | rs.Chr11.27721783 | rs.Chr11.27721767 | 43.6–44.1 | 3.4717 | −2.2544 | 0.0482 | ||
| Relative Lesion Height | 1 | qRLH1.6 | rs.Chr1.614777 | rs.Chr1.649072 | 307.7–326.3 | 4.4345 | 3.3726 | 0.3992 | |
| 1 | qRLH1.7 | rs.Chr1.649072 | rs.Chr1.613037 | 326.3–338.2 | 3.116 | 4.4559 | 0.4052 | ||
| 1 | qRLH1.8 | rs.Chr1.35880614 | rs.Chr1.36300744 | 1328.8–1336.4 | 7.1119 | −6.4116 | 0.1372 | ||
| 2 | qRLH2.1 | rs.Chr2.20104645 | rs.Chr2.19443601 | 762.9–771.0 | 4.0941 | 2.9825 | 0.0595 | ||
| 11 | qRLH11.1 | rs.Chr11.26611565 | rs.Chr11.26522147 | 83.7–90.5 | 2.7475 | 0.6426 | 0.047 |
| QTL | Locus Id | Encoded Protein | |
|---|---|---|---|
| qRLH1.1 (202.2 kb) | rs.Chr1.38966610 rs.Chr1.38764373 | LOC_Os01g66820 | Inactive receptor kinase |
| LOC_Os01g66830 | Pectinacetylesterase domain protein | ||
| LOC_Os01g66840 | Pectinacetylesterase domain protein | ||
| LOC_Os01g66850 | Pectinacetylesterase domain protein | ||
| LOC_Os01g66860 | Serine/threonine-protein kinase receptor | ||
| LOC_Os01g66890 | BTBZ1—Bric-a-Brac, Tramtrack | ||
| LOC_Os01g66920 | Ser/Thr protein phosphatase protein | ||
| LOC_Os01g66970 | Zinc finger, C3HC4 type domain protein | ||
| LOC_Os01g67030 | Auxin-responsive protein | ||
| qRLH1.2 (0.096 kb) | rs.Chr1.39839247 rs.Chr1.39839342 | LOC_Os01g68589 | LTPL39—Protease inhibitor/seed storage/LTP family protein precursor, expressed |
| qRLH1.5 (76.84 kb) | rs.Chr1.38678740 rs.Chr1.38601901 | LOC_Os01g66490 | No apical meristem protein |
| LOC_Os01g66510 | MLO domain containing protein | ||
| LOC_Os01g66520 | serine/threonine-protein kinase RIO-like | ||
| qRLH1.8 (420.1 kb) | rs.Chr1.35880614 rs.Chr1.36300744 | LOC_01g62030 | Disease resistance-responsive protein |
| LOC_01g62080 | Serine/threonine-protein kinase | ||
| LOC_01g62190 | ZOS1-15—C2H2 zinc finger protein | ||
| LOC_01g62260 | Thaumatin, putative, expressed | ||
| LOC_01g62410 | MYB family transcription factor | ||
| LOC_01g62460 | ZOS1-16—C2H2 zinc finger protein | ||
| LOC_01g62480 | Laccase precursor protein | ||
| LOC_01g62490 | Laccase precursor protein | ||
| LOC_01g62510 | WRKY119 | ||
| LOC_01g62514 | WRKY56 | ||
| LOC_01g62570 | ATP/GTP/Ca++ binding protein | ||
| LOC_01g62584 | Peptidase aspartic family protein | ||
| LOC_01g62600 | Laccase precursor protein | ||
| LOC_01g62610 | Peptidyl-prolyl cis-trans isomerase | ||
| LOC_01g62620 | DHHC zinc finger domain protein | ||
| LOC_01g62630 | Aspartic proteinase nepenthesin | ||
| LOC_01g62640 | Zinc finger, C3HC4 type domain protein | ||
| LOC_01g62650 | Pumilio-family RNA binding protein | ||
| LOC_01g62660 | MYB family transcription factor | ||
| LOC_01g62670 | avr9/Cf-9 rapidly elicited protein |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Neelam, K.; Aggarwal, S.K.; Kumari, S.; Kumar, K.; Kaur, A.; Babbar, A.; Lore, J.S.; Kaur, R.; Khanna, R.; Vikal, Y.; et al. Molecular Mapping and Transfer of Quantitative Trait Loci (QTL) for Sheath Blight Resistance from Wild Rice Oryza nivara to Cultivated Rice (Oryza sativa L.). Genes 2024, 15, 919. https://doi.org/10.3390/genes15070919
Neelam K, Aggarwal SK, Kumari S, Kumar K, Kaur A, Babbar A, Lore JS, Kaur R, Khanna R, Vikal Y, et al. Molecular Mapping and Transfer of Quantitative Trait Loci (QTL) for Sheath Blight Resistance from Wild Rice Oryza nivara to Cultivated Rice (Oryza sativa L.). Genes. 2024; 15(7):919. https://doi.org/10.3390/genes15070919
Chicago/Turabian StyleNeelam, Kumari, Sumit Kumar Aggarwal, Saundarya Kumari, Kishor Kumar, Amandeep Kaur, Ankita Babbar, Jagjeet Singh Lore, Rupinder Kaur, Renu Khanna, Yogesh Vikal, and et al. 2024. "Molecular Mapping and Transfer of Quantitative Trait Loci (QTL) for Sheath Blight Resistance from Wild Rice Oryza nivara to Cultivated Rice (Oryza sativa L.)" Genes 15, no. 7: 919. https://doi.org/10.3390/genes15070919
APA StyleNeelam, K., Aggarwal, S. K., Kumari, S., Kumar, K., Kaur, A., Babbar, A., Lore, J. S., Kaur, R., Khanna, R., Vikal, Y., & Singh, K. (2024). Molecular Mapping and Transfer of Quantitative Trait Loci (QTL) for Sheath Blight Resistance from Wild Rice Oryza nivara to Cultivated Rice (Oryza sativa L.). Genes, 15(7), 919. https://doi.org/10.3390/genes15070919






