Early-Life Sublethal Exposure to Thiacloprid Alters Adult Honeybee Gut Microbiota
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Sample Collection
2.3. DNA Extraction and Sequencing
2.4. Data Analyses
2.5. Statistics
3. Results
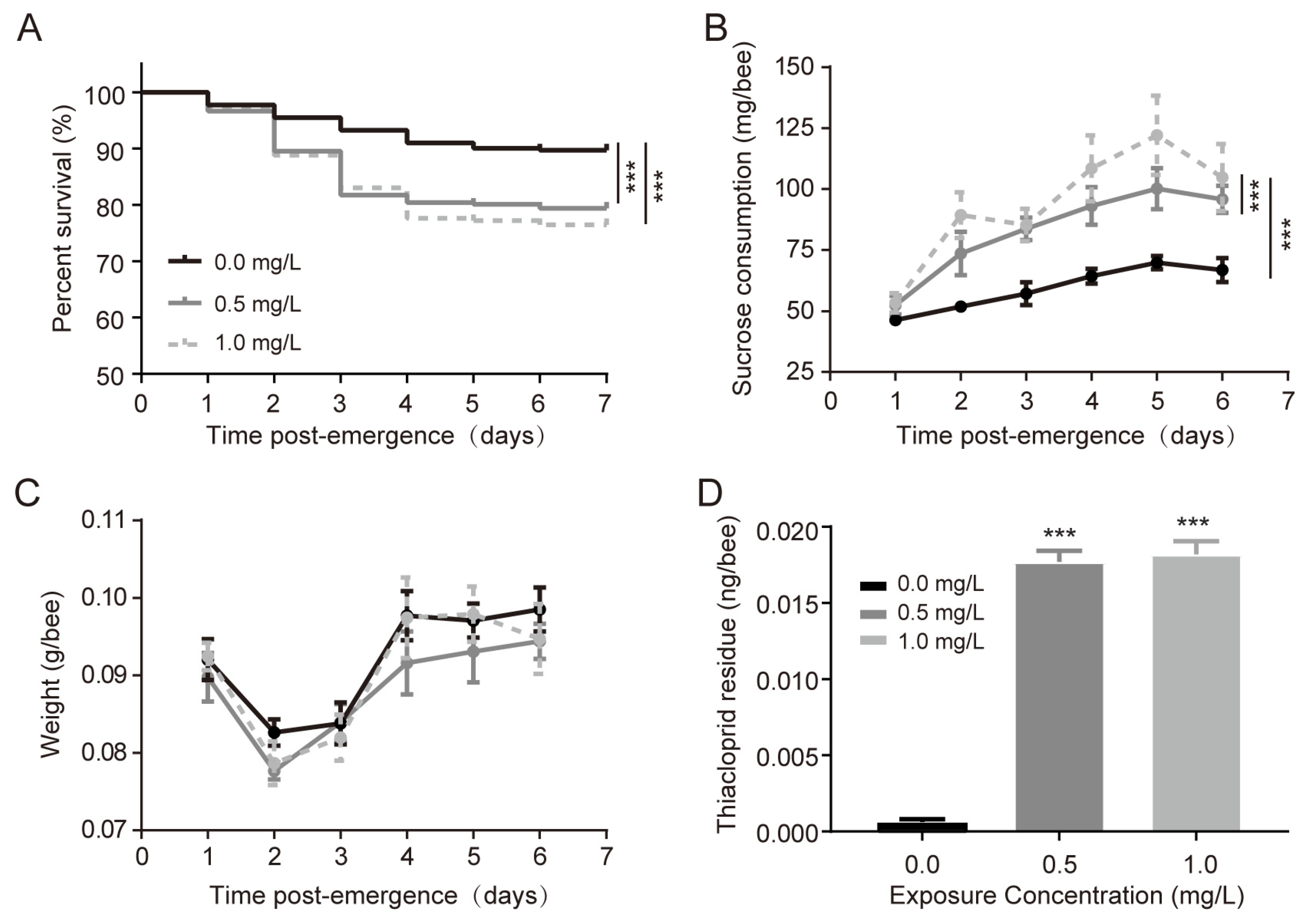
3.1. Thiacloprid Larval Exposure Affects the Physical Status of Adult Honeybees
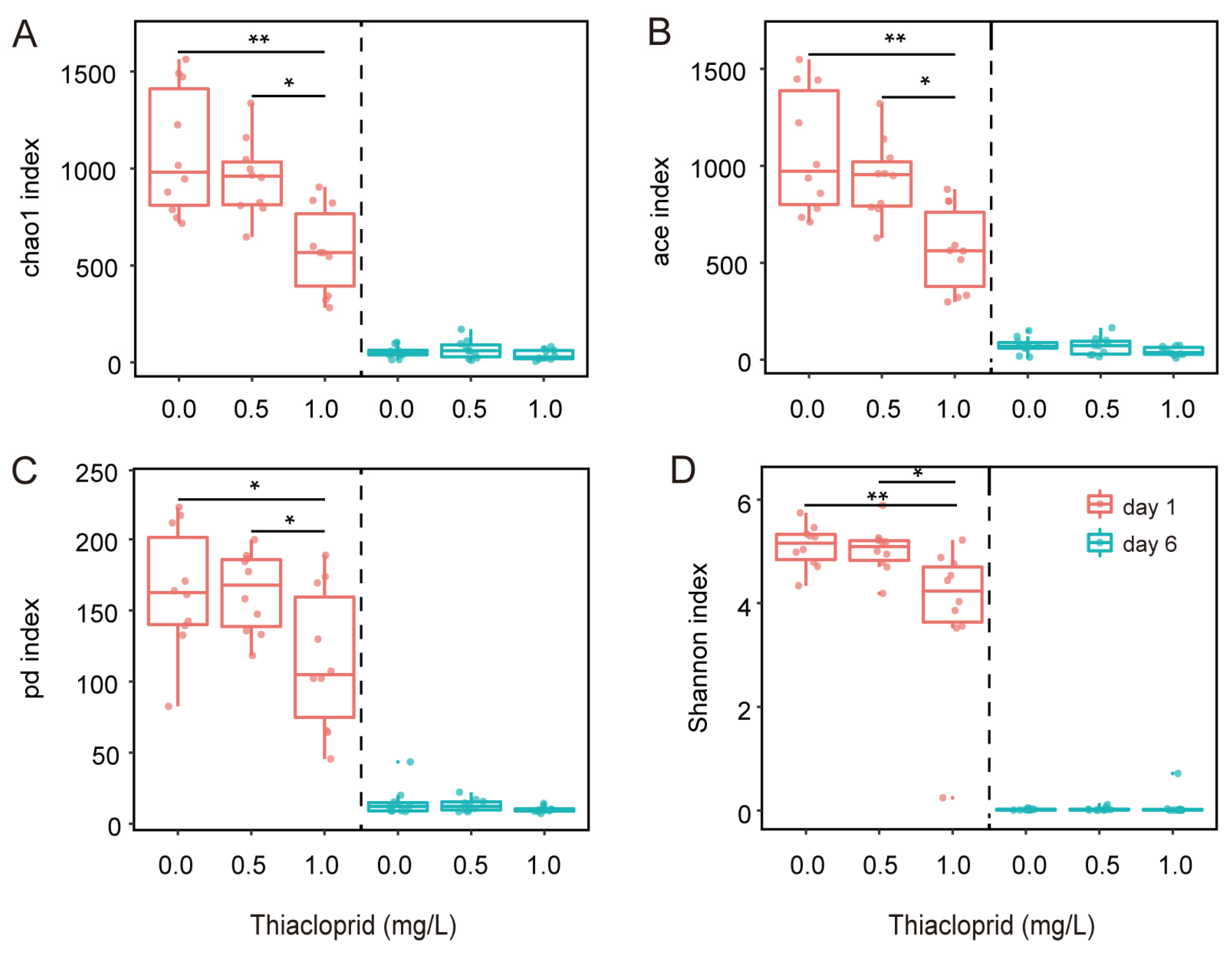
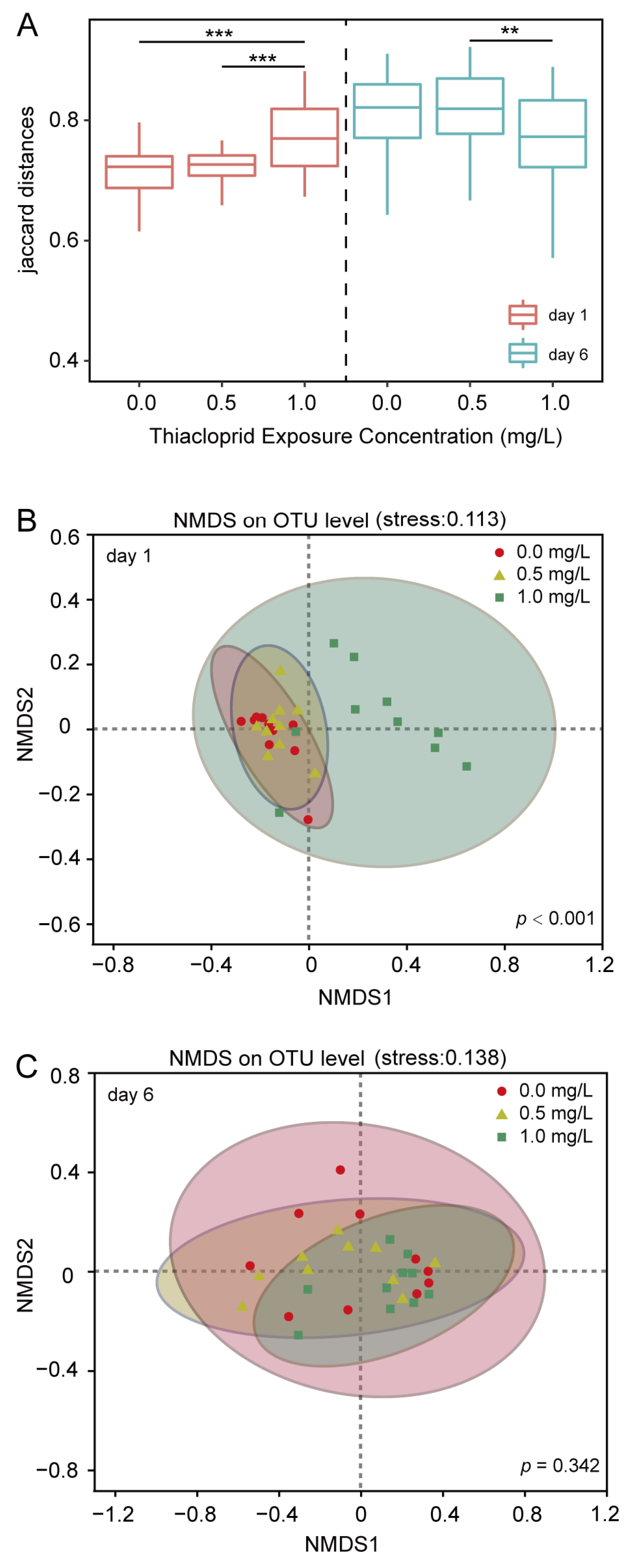
3.2. Thiacloprid Modulates the Diversity of Adult Honeybee Gut Microbiota
3.3. Factors Influencing the Early Establishment of Gut Microbiota in Adult Honeybees
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hung, K.L.J.; Kingston, J.M.; Albrecht, M.; Holway, D.A.; Kohn, J.R. The worldwide importance of honey bees as pollinators in natural habitats. Proc. R. Soc. B Biol. Sci. 2018, 285, 8. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Chen, C.; Zhang, H.; Lin, F.; He, H.; Sun, G. The toxicity effects of abamectin on honeybees (Apis mellifera L.). Asian J. Ecotoxicol. 2017, 12, 174–182. [Google Scholar] [CrossRef]
- Decourtye, A.; Devillers, J. Ecotoxicity of Neonicotinoid Insecticides to Bees; Springer: Berlin, Germany, 2010; pp. 85–95. [Google Scholar]
- Rundlof, M.; Andersson, G.K.S.; Bommarco, R.; Fries, I.; Hederstrom, V.; Herbertsson, L.; Jonsson, O.; Klatt, B.K.; Pedersen, T.R.; Yourstone, J.; et al. Seed coating with a neonicotinoid insecticide negatively affects wild bees. Nature 2015, 521, 77–80. [Google Scholar] [CrossRef]
- Tsvetkov, N.; Samson-Robert, O.; Sood, K.; Patel, H.S.; Malena, D.A.; Gajiwala, P.H.; Maciukiewicz, P.; Fournier, V.; Zayed, A. Chronic exposure to neonicotinoids reduces honey bee health near corn crops. Science 2017, 356, 1395–1397. [Google Scholar] [CrossRef]
- Li, B.; Ke, L.; Li, A.R.; Diao, Q.-Y.; Wang, Q.; Liu, Y.-J. Exposure of larvae to sublethal thiacloprid delays bee development and affects transcriptional responses of newly emerged honey bees. Front. Insect Sci. 2022, 2, 844957. [Google Scholar] [CrossRef]
- Brandt, A.; Gorenflo, A.; Siede, R.; Meixner, M.; Buchler, R. The neonicotinoids thiacloprid, imidacloprid, and clothianidin affect the immunocompetence of honey bees (Apis mellifera L.). J. Insect Physiol. 2016, 86, 40–47. [Google Scholar] [CrossRef]
- Di Prisco, G.; Cavaliere, V.; Annoscia, D.; Varricchio, P.; Caprio, E.; Nazzi, F.; Gargiulo, G.; Pennacchio, F. Neonicotinoid clothianidin adversely affects insect immunity and promotes replication of a viral pathogen in honey bees. Proc. Natl. Acad. Sci. USA 2013, 110, 18466–18471. [Google Scholar] [CrossRef]
- Williams, G.R.; Troxler, A.; Retschnig, G.; Roth, K.; Yanez, O.; Shutler, D.; Neumann, P.; Gauthier, L. Neonicotinoid pesticides severely affect honey bee queens. Sci. Rep. 2015, 5, 8. [Google Scholar] [CrossRef]
- Tosi, S.; Burgio, G.; Nieh, J.C. A common neonicotinoid pesticide, thiamethoxam, impairs honey bee flight ability. Sci. Rep. 2017, 7, 8. [Google Scholar] [CrossRef]
- Stanley, D.A.; Smith, K.E.; Raine, N.E. Bumblebee learning and memory is impaired by chronic exposure to a neonicotinoid pesticide. Sci. Rep. 2015, 5, 10. [Google Scholar] [CrossRef]
- Eiri, D.M.; Nieh, J.C. A nicotinic acetylcholine receptor agonist affects honey bee sucrose responsiveness and decreases waggle dancing. J. Exp. Biol. 2016, 219, 2081. [Google Scholar] [CrossRef] [PubMed]
- Iwasa, T.; Motoyama, N.; Ambrose, J.T.; Roe, R.M. Mechanism for the differential toxicity of neonicotinoid insecticides in the honey bee, Apis mellifera. Crop Prot. 2004, 23, 371–378. [Google Scholar] [CrossRef]
- Manjon, C.; Troczka, B.J.; Zaworra, M.; Beadle, K.; Randall, E.; Hertlein, G.; Singh, K.S.; Zimmer, C.T.; Homem, R.A.; Lueke, B.; et al. Unravelling the molecular determinants of bee sensitivity to neonicotinoid insecticides. Curr. Biol. 2018, 28, 1137–1143. [Google Scholar] [CrossRef]
- Christen, V.; Bachofer, S.; Fent, K. Binary mixtures of neonicotinoids show different transcriptional changes than single neonicotinoids in honeybees (Apis mellifera). Environ. Pollut. 2017, 220, 1264–1270. [Google Scholar] [CrossRef] [PubMed]
- Tomizawa, M.; Casida, J.E. Neonicotinoid insecticide toxicology: Mechanisms of selective action. Annu. Rev. Pharmacol. Toxicol. 2005, 45, 247–268. [Google Scholar] [CrossRef] [PubMed]
- Sabree, Z.L.; Hansen, A.K.; Moran, N.A. Independent studies using deep sequencing resolve the same set of core bacterial species dominating gut communities of honey bees. PLoS ONE 2012, 7, e41250. [Google Scholar] [CrossRef]
- Powell, J.E.; Martinson, V.G.; Urban-Mead, K.; Moran, N.A. Routes of acquisition of the gut microbiota of the honey bee Apis mellifera. Appl. Environ. Microbiol. 2014, 80, 7378–7387. [Google Scholar] [CrossRef]
- Zheng, H.; Powell, J.E.; Steele, M.I.; Dietrich, C.; Moran, N.A. Honeybee gut microbiota promotes host weight gain via bacterial metabolism and hormonal signaling. Proc. Natl. Acad. Sci. USA 2017, 114, 4775–4780. [Google Scholar] [CrossRef]
- Zheng, H.; Perreau, J.; Powell, J.E.; Han, B.; Zhang, Z.; Kwong, W.K.; Tringe, S.G.; Moran, N.A. Division of labor in honey bee gut microbiota for plant polysaccharide digestion. Proc. Natl. Acad. Sci. USA 2019, 116, 25909–25916. [Google Scholar] [CrossRef]
- Engel, P.; Moran, N.A. Functional and evolutionary insights into the simple yet specific gut microbiota of the honey bee from metagenomic analysis. Gut Microbes 2013, 4, 60–65. [Google Scholar] [CrossRef]
- Kwong, W.K.; Mancenido, A.L.; Moran, N.A. Immune system stimulation by the native gut microbiota of honey bees. R. Soc. Open Sci. 2017, 4, 170003. [Google Scholar] [CrossRef] [PubMed]
- Motta, E.V.S.; Moran, N.A. The honeybee microbiota and its impact on health and disease. Nat. Rev. Microbiol. 2024, 22, 122–137. [Google Scholar] [CrossRef] [PubMed]
- Motta, E.V.S.; Raymann, K.; Moran, N.A. Glyphosate perturbs the gut microbiota of honey bees. Proc. Natl. Acad. Sci. USA 2018, 115, 10305–10310. [Google Scholar] [CrossRef] [PubMed]
- Rouze, R.; Mone, A.; Delbac, F.; Belzunces, L.; Blot, N. The Honeybee Gut Microbiota Is Altered after Chronic Exposure to Different Families of Insecticides and Infection by Nosema ceranae. Microbes Environ. 2019, 34, 226–233. [Google Scholar] [CrossRef] [PubMed]
- Dai, P.; Yan, Z.; Ma, S.; Yang, Y.; Wang, Q.; Hou, C.; Wu, Y.; Liu, Y.; Diao, Q. The herbicide glyphosate negatively affects midgut bacterial communities and survival of honey bee during larvae reared in vitro. J. Agric. Food Chem. 2018, 66, 7786–7793. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.J.; Qiao, N.H.; Diao, Q.Y.; Jing, Z.W.; Vukanti, R.; Dai, P.L.; Ge, Y. Thiacloprid exposure perturbs the gut microbiota and reduces the survival status in honeybees. J. Hazard. Mater. 2020, 389, 11. [Google Scholar] [CrossRef] [PubMed]
- Ellegaard, K.M.; Engel, P. Genomic diversity landscape of the honey bee gut microbiota. Nat. Commun. 2019, 10, 13. [Google Scholar] [CrossRef] [PubMed]
- Schmehl, D.R.; Tome, H.V.V.; Mortensen, A.N.; Martins, G.F.; Ellis, J.D. Protocol for the in vitro rearing of honey bee (Apis mellifera L.) workers. J. Apic. Res. 2016, 55, 113–129. [Google Scholar] [CrossRef]
- Alkassab, A.T.; Kunz, N.; Bischoff, G.; Luken, D.; Janke, M.; Wallner, K.; Kirchner, W.H.; Pistorius, J. Large-scale study investigating the effects of a tank mixture containing thiacloprid-prochloraz on honey bees (Apis mellifera). Chemosphere 2023, 313, 137396. [Google Scholar] [CrossRef]
- Ebeling, J.; Pieper, F.; Gobel, J.; Knispel, H.; McCarthy, M.; Goncalves, M.; Turner, M.; Merrill, A.R.; Genersch, E. Anti-virulence strategy against the honey bee pathogenic bacterium paenibacillus larvae via small molecule inhibitors of the bacterial toxin Plx2A. Toxins 2021, 13, 607. [Google Scholar] [CrossRef]
- Amsterdam, A.; Sadler, K.C.; Lai, K.; Farrington, S.; Bronson, R.T.; Lees, J.A.; Hopkins, N. Many ribosomal protein genes are cancer genes in zebrafish. PLoS Biol. 2004, 2, 690–698. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Feng, J.; Shi, Z.; Zhou, X.; Yuan, M.; Tao, X.; Hale, L.; Yuan, T.; Wang, J.; Qin, Y.; et al. Climate warming leads to divergent succession of grassland microbial communities. Nat. Clim. Change 2018, 8, 813–818. [Google Scholar] [CrossRef]
- Ning, D.; Deng, Y.; Tiedje, J.M.; Zhou, J. A general framework for quantitatively assessing ecological stochasticity. Proc. Natl. Acad. Sci. USA 2019, 116, 16892–16898. [Google Scholar] [CrossRef] [PubMed]
- Tosi, S.; Sfeir, C.; Carnesecchi, E.; vanEngelsdorp, D.; Chauzat, M.P. Lethal, sublethal, and combined effects of pesticides on bees: A meta-analysis and new risk assessment tools. Sci. Total Environ. 2022, 844, 12. [Google Scholar] [CrossRef] [PubMed]
- Klingelhofer, D.; Braun, M.; Bruggmann, D.; Groneberg, D.A. Neonicotinoids: A critical assessment of the global research landscape of the most extensively used insecticide. Environ. Res. 2022, 213, 13. [Google Scholar] [CrossRef] [PubMed]
- Weitekamp, C.A.; Koethe, R.W.; Lehmann, D.M. A comparison of pollen and syrup exposure routes in bombus impatiens (Hymenoptera: Apidae) microcolonies: Implications for pesticide risk assessment. Environ. Entomol. 2022, 51, 613–620. [Google Scholar] [CrossRef] [PubMed]
- Siede, R.; Faust, L.; Meixner, M.D.; Maus, C.; Grunewald, B.; Buchler, R. Performance of honey bee colonies under a long-lasting dietary exposure to sublethal concentrations of the neonicotinoid insecticide thiacloprid. Pest Manag. Sci. 2017, 73, 1334–1344. [Google Scholar] [CrossRef] [PubMed]
- Hamdi, C.; Balloi, A.; Essanaa, J.; Crotti, E.; Gonella, E.; Raddadi, N.; Ricci, I.; Boudabous, A.; Borin, S.; Manino, A.; et al. Gut microbiome dysbiosis and honeybee health. J. Appl. Entomol. 2011, 135, 524–533. [Google Scholar] [CrossRef]
- Zhu, L.; Qi, S.; Xue, X.; Niu, X.; Wu, L. Nitenpyram disturbs gut microbiota and influences metabolic homeostasis and immunity in honey bee (Apis mellifera L.). Environ. Pollut. 2020, 258, 113671. [Google Scholar] [CrossRef]
- Aufauvre, J.; Misme-Aucouturier, B.; Vigues, B.; Texier, C.; Delbac, F.; Blot, N. Transcriptome Analyses of the Honeybee Response to Nosema ceranae and Insecticides. PLoS ONE 2014, 9, 12. [Google Scholar] [CrossRef]
- Wang, G.L.; Yue, W.L.; Liu, Y.; Li, F.; Xiong, M.H.; Zhang, H. Biodegradation of the neonicotinoid insecticide Acetamiprid by bacterium Pigmentiphaga sp strain AAP-1 isolated from soil. Bioresour. Technol. 2013, 138, 359–368. [Google Scholar] [CrossRef]
- Zhou, G.C.; Wang, Y.; Zhai, S.; Ge, F.; Liu, Z.H.; Dai, Y.J.; Yuan, S.; Hou, J.Y. Biodegradation of the neonicotinoid insecticide thiamethoxam by the nitrogen-fixing and plant-growth-promoting rhizobacterium Ensifer adhaerens strain TMX-23. Appl. Microbiol. Biotechnol. 2013, 97, 4065–4074. [Google Scholar] [CrossRef]
- Martinson, V.G.; Moy, J.; Moran, N.A. Establishment of characteristic gut bacteria during development of the honeybee worker. Appl. Environ. Microbiol. 2012, 78, 2830–2840. [Google Scholar] [CrossRef] [PubMed]
- Kwong, W.K.; Moran, N.A. Gut microbial communities of social bees. Nat. Rev. Microbiol. 2016, 14, 374–384. [Google Scholar] [CrossRef]
- Suchail, S.; Debrauwer, L.; Belzunces, L.P. Metabolism of imidacloprid in Apis mellifera. Pest Manag. Sci. 2004, 60, 291–296. [Google Scholar] [CrossRef]
- Christen, V.; Krebs, J.; Bünter, I.; Fent, K. Biopesticide spinosad induces transcriptional alterations in genes associated with energy production in honey bees (Apis mellifera) at sublethal concentrations. J. Hazard. Mater. 2019, 378, 120736. [Google Scholar] [CrossRef]
- Castelli, L.; Branchiccela, B.; Romero, H.; Zunino, P.; Antunez, K. Seasonal dynamics of the honey bee gut microbiota in colonies under subtropical climate seasonal dynamics of honey bee gut microbiota. Microb. Ecol. 2022, 83, 492–500. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Jing, Z.; Diao, Q.; He, J.Z.; Liu, Y.J. Host species and geography differentiate honeybee gut bacterial communities by changing the relative contribution of community assembly processes. mBio 2021, 12, e0075121. [Google Scholar] [CrossRef] [PubMed]
- Cuesta-Mate, A.; Renelies-Hamilton, J.; Kryger, P.; Jensen, A.B.; Sinotte, V.M.; Poulsen, M. Resistance and vulnerability of honeybee (Apis mellifera) gut bacteria to commonly used pesticides. Front. Microbiol. 2021, 12, 16. [Google Scholar] [CrossRef]
- Ricigliano, V.A.; Williams, S.T.; Oliver, R. Effects of different artificial diets on commercial honey bee colony performance, health biomarkers, and gut microbiota. BMC Vet. Res. 2022, 18, 14. [Google Scholar] [CrossRef]
- Brochet, S.; Quinn, A.; At Mars, R.; Neuschwander, N.; Sauer, U.; Engel, P. Niche partitioning facilitates coexistence of closely related honey bee gut bacteria. eLife 2021, 10, 23. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wu, Y.; Long, H.; Ma, X.; Shariati, K.; Webb, J.; Guo, L.; Pan, Y.; Ma, M.; Deng, C.; et al. Global honeybee health decline factors and potential conservation techniques. Food Secur. 2023, 15, 855–875. [Google Scholar] [CrossRef]

| CAP1 | CAP2 | r2 | p-Values | |
|---|---|---|---|---|
| thiacloprid level | 0.119 | −0.993 | 0.699 | 0.001 |
| age | 0.989 | 0.150 | 0.967 | 0.001 |
| thiacloprid | 0.104 | −0.995 | 0.300 | 0.001 |
| weight | 1.000 | 0.009 | 0.071 | 0.136 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, B.; Chen, X.; Ke, L.; Dai, P.; Ge, Y.; Liu, Y.-J. Early-Life Sublethal Exposure to Thiacloprid Alters Adult Honeybee Gut Microbiota. Genes 2024, 15, 1001. https://doi.org/10.3390/genes15081001
Li B, Chen X, Ke L, Dai P, Ge Y, Liu Y-J. Early-Life Sublethal Exposure to Thiacloprid Alters Adult Honeybee Gut Microbiota. Genes. 2024; 15(8):1001. https://doi.org/10.3390/genes15081001
Chicago/Turabian StyleLi, Bin, Xiasang Chen, Li Ke, Pingli Dai, Yuan Ge, and Yong-Jun Liu. 2024. "Early-Life Sublethal Exposure to Thiacloprid Alters Adult Honeybee Gut Microbiota" Genes 15, no. 8: 1001. https://doi.org/10.3390/genes15081001
APA StyleLi, B., Chen, X., Ke, L., Dai, P., Ge, Y., & Liu, Y.-J. (2024). Early-Life Sublethal Exposure to Thiacloprid Alters Adult Honeybee Gut Microbiota. Genes, 15(8), 1001. https://doi.org/10.3390/genes15081001







