Identification of Site in the UTY Gene as Safe Harbor Locus on the Y Chromosome of Pig
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmid Construction
2.2. Cell Culture and Transfection
2.3. In Vitro Enzyme Cleavage
2.4. Off-Target Analysis of sgRNA
2.5. Screening of Monoc Slonal Cells and Junction PCR
2.6. Western Blot Analysis
2.7. Quantitative RT-PCR
3. Results
3.1. Identification of UTY Expression in Different Tissues
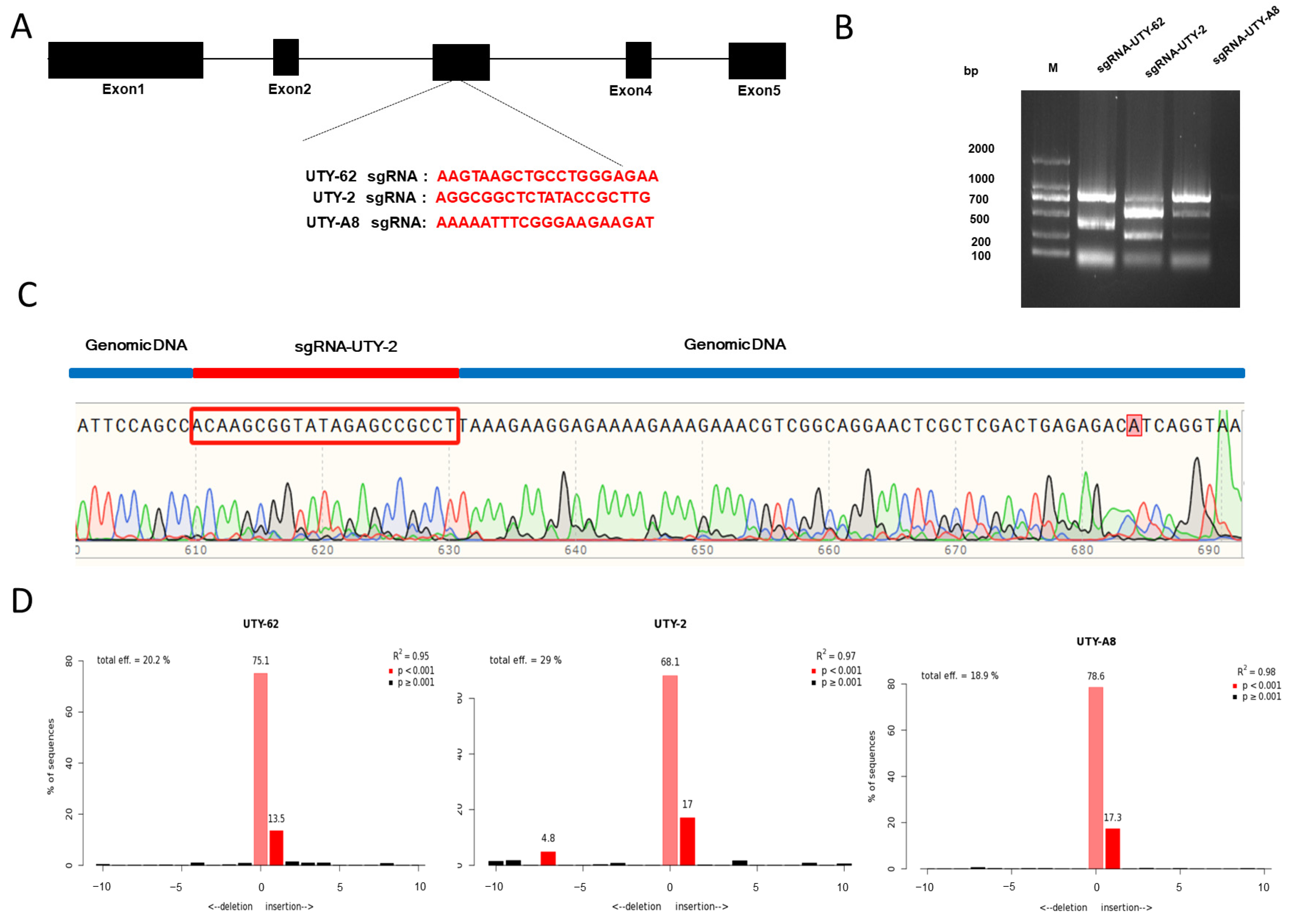
3.2. The Screening of sgRNA for Porcine UTY Gene Editing
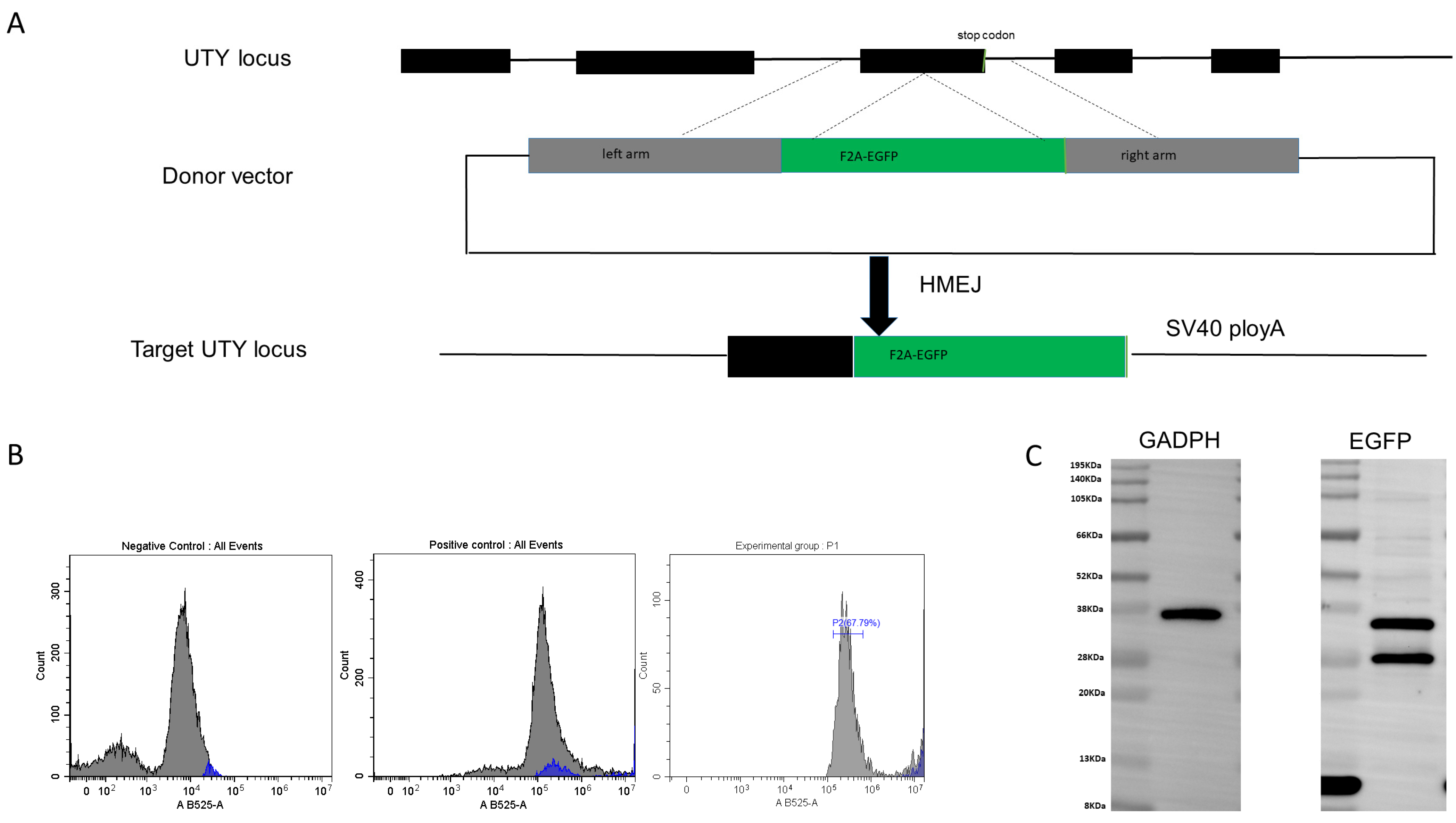
3.3. Establishment of a Reporter Gene System for Genome Editing in Pig Genome
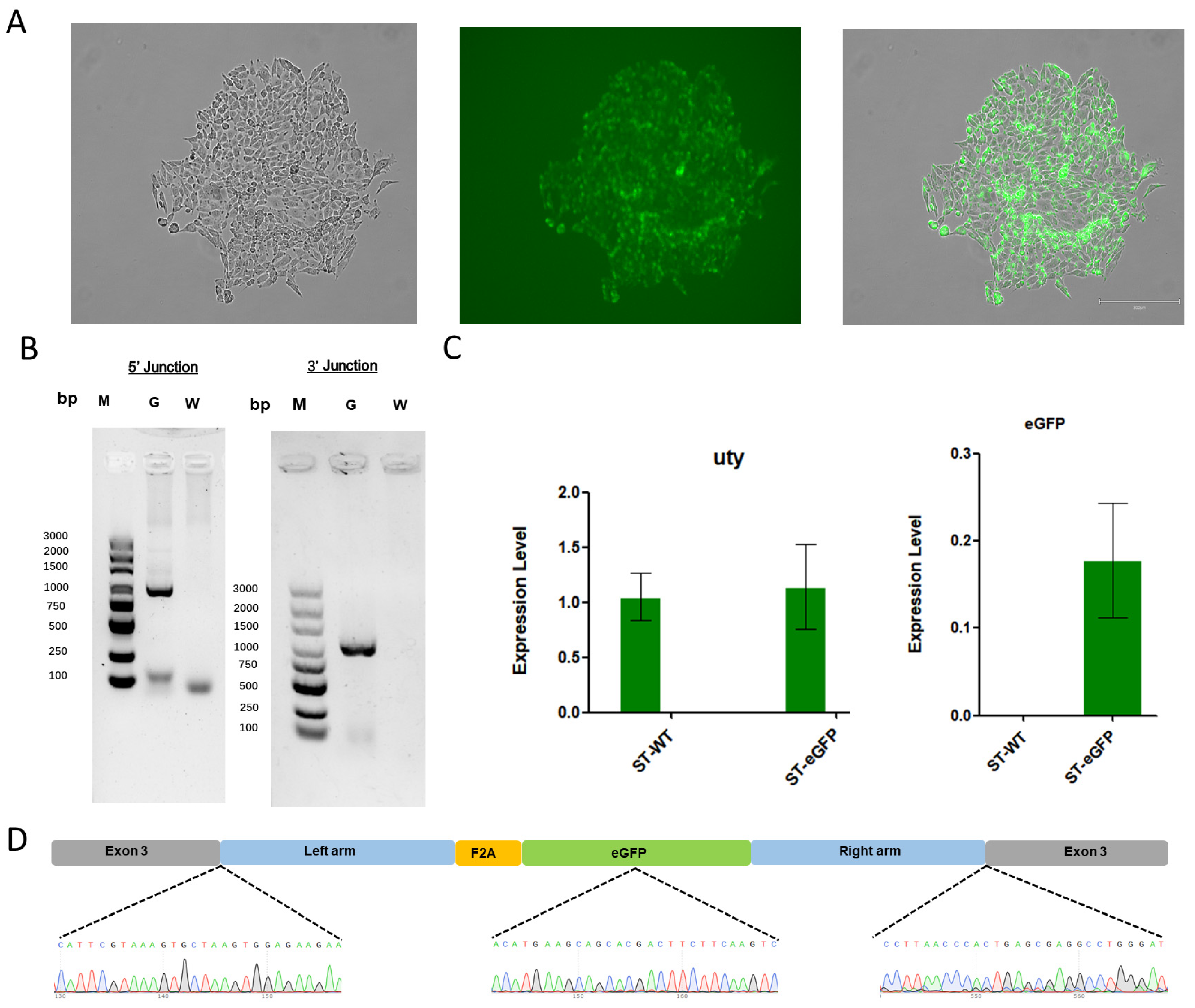
3.4. Analysis of Exogenous Gene Knock-In in the Pig Genome
4. Discussion
5. Patent
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Montgomery, R.A.; Stern, J.M.; Lonze, B.E.; Tatapudi, V.S.; Mangiola, M.; Wu, M.; Weldon, E.; Lawson, N.; Deterville, C.; Dieter, R.A.; et al. Results of Two Cases of Pig-to-Human Kidney Xenotransplantation. N. Engl. J. Med. 2022, 386, 1889–1898. [Google Scholar] [CrossRef]
- Moazami, N.; Stern, J.M.; Khalil, K.; Kim, J.I.; Narula, N.; Mangiola, M.; Weldon, E.P.; Kagermazova, L.; James, L.; Lawson, N.; et al. Pig-to-human heart xenotransplantation in two recently deceased human recipients. Nat. Med. 2023, 29, 1989–1997. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wang, T.; Yao, L.; Liu, B.; Teng, C.; Ouyang, H. Classical swine fever virus replicated poorly in cells from MxA transgenic pigs. BMC Vet. Res. 2016, 12, 169. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Yang, L.; Zhang, Y.; Xiao, W.; Wang, Z.; Tang, X.; Ouyang, H.; Pang, D. Current Status of Genetically Modified Pigs That Are Resistant to Virus Infection. Viruses 2022, 14, 417. [Google Scholar] [CrossRef] [PubMed]
- Dolgin, E. First GM pigs for allergies. Could xenotransplants be next? Nat. Biotechnol. 2021, 39, 397–400. [Google Scholar] [CrossRef]
- Cohen, J. Meat from gene-edited pigs could hit the market. Science 2024, 383, 940–941. [Google Scholar] [CrossRef]
- Bibikova, M.; Beumer, K.; Trautman, J.K.; Carroll, D. Enhancing Gene Targeting with Designed Zinc Finger Nucleases. Science 2003, 300, 764. [Google Scholar] [CrossRef] [PubMed]
- Moehle, E.A.; Rock, J.M.; Lee, Y.-L.; Jouvenot, Y.; DeKelver, R.C.; Gregory, P.D.; Urnov, F.D.; Holmes, M.C. Targeted gene addition into a specified location in the human genome using designed zinc finger nucleases. Proc. Natl. Acad. Sci. USA 2007, 104, 3055–3060. [Google Scholar] [CrossRef] [PubMed]
- Hockemeyer, D.; Wang, H.; Kiani, S.; Lai, C.S.; Gao, Q.; Cassady, J.P.; Cost, G.J.; Zhang, L.; Santiago, Y.; Miller, J.C.; et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat. Biotechnol. 2011, 29, 731–734. [Google Scholar] [CrossRef]
- Christian, M.; Cermak, T.; Doyle, E.L.; Schmidt, C.; Zhang, F.; Hummel, A.; Bogdanove, A.J.; Voytas, D.F. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 2010, 186, 757–761. [Google Scholar] [CrossRef]
- Yang, H.; Wang, H.; Shivalila, C.S.; Cheng, A.W.; Shi, L.; Jaenisch, R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 2013, 154, 1370–1379. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Maresca, M.; Lin, V.G.; Guo, N.; Yang, Y. Obligate ligation-gated recombination (ObLiGaRe): Custom-designed nuclease-mediated targeted integration through nonhomologous end joining. Genome Res. 2013, 23, 539–546. [Google Scholar] [CrossRef]
- Qi, Y.; Zhang, Y.; Zhang, F.; Baller, J.A.; Cleland, S.C.; Ryu, Y.; Starker, C.G.; Voytas, D.F. Increasing frequencies of site-specific mutagenesis and gene targeting in Arabidopsis by manipulating DNA repair pathways. Genome Res. 2013, 23, 547–554. [Google Scholar] [CrossRef]
- Nakade, S.; Tsubota, T.; Sakane, Y.; Kume, S.; Sakamoto, N.; Obara, M.; Daimon, T.; Sezutsu, H.; Yamamoto, T.; Sakuma, T.; et al. Microhomology-mediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9. Nat. Commun. 2014, 5, 5560. [Google Scholar] [CrossRef]
- Yao, X.; Wang, X.; Hu, X.; Liu, Z.; Liu, J.; Zhou, H.; Shen, X.; Wei, Y.; Huang, Z.; Ying, W.; et al. Homology-mediated end joining-based targeted integration using CRISPR/Cas9. Cell Res. 2017, 27, 801–814. [Google Scholar] [CrossRef] [PubMed]
- Zambrowicz, B.P.; Imamoto, A.; Fiering, S.; Herzenberg, L.A.; Kerr, W.G.; Soriano, P. Disruption of overlapping transcripts in the ROSA beta geo 26 gene trap strain leads to widespread expression of beta-galactosidase in mouse embryos and hematopoietic cells. Proc. Natl. Acad. Sci. USA 1997, 94, 3789–3794. [Google Scholar] [CrossRef]
- Papapetrou, E.P.; Lee, G.; Malani, N.; Setty, M.; Riviere, I.; Tirunagari, L.M.; Kadota, K.; Roth, S.L.; Giardina, P.; Viale, A.; et al. Genomic safe harbors permit high beta-globin transgene expression in thalassemia induced pluripotent stem cells. Nat. Biotechnol. 2011, 29, 73–78. [Google Scholar] [CrossRef]
- Li, X.; Yang, Y.; Bu, L.; Guo, X.; Tang, C.; Song, J.; Fan, N.; Zhao, B.; Ouyang, Z.; Liu, Z.; et al. Rosa26-targeted swine models for stable gene over-expression and Cre-mediated lineage tracing. Cell Res. 2014, 24, 501–504. [Google Scholar] [CrossRef]
- Ruan, J.; Li, H.; Xu, K.; Wu, T.; Wei, J.; Zhou, R.; Liu, Z.; Mu, Y.; Yang, S.; Ouyang, H.; et al. Highly efficient CRISPR/Cas9-mediated transgene knockin at the H11 locus in pigs. Sci. Rep. 2015, 5, 14253. [Google Scholar] [CrossRef]
- Ma, L.; Wang, Y.; Wang, H.; Hu, Y.; Chen, J.; Tan, T.; Hu, M.; Liu, X.; Zhang, R.; Xing, Y.; et al. Screen and Verification for Transgene Integration Sites in Pigs. Sci. Rep. 2018, 8, 7433. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Tu, Z.; Liu, Z.; Fan, N.; Yang, H.; Yang, S.; Yang, W.; Zhao, Y.; Ouyang, Z.; Lai, C.; et al. A Huntingtin Knockin Pig Model Recapitulates Features of Selective Neurodegeneration in Huntington’s Disease. Cell 2018, 173, 989–1002. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Xiong, Y.; Zhao, C.; Xie, S.; Li, C.; Li, X.; Liu, X.; Li, K.; Zhao, S.; Ruan, J. Identification of Glyceraldehyde-3-Phosphate Dehydrogenase Gene as an Alternative Safe Harbor Locus in Pig Genome. Genes 2019, 10, 660. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Han, X.; Zhang, J.; Zhao, G.; Wang, Z.; Zhuang, R.; Nie, X.; Xie, S.; Li, C.; Li, X.; et al. Identification of ACTB Gene as a Potential Safe Harbor Locus in Pig Genome. Mol. Biotechnol. 2020, 62, 589–597. [Google Scholar] [CrossRef] [PubMed]
- Douglas, C.; Maciulyte, V.; Zohren, J.; Snell, D.M.; Mahadevaiah, S.K.; Ojarikre, O.A.; Ellis, P.J.I.; Turner, J.M.A. CRISPR-Cas9 effectors facilitate generation of single-sex litters and sex-specific phenotypes. Nat. Commun. 2021, 12, 6926. [Google Scholar] [CrossRef] [PubMed]
- Shpargel, K.B.; Sengoku, T.; Yokoyama, S.; Magnuson, T. UTX and UTY Demonstrate Histone Demethylase-Independent Function in Mouse Embryonic Development. PLoS Genet. 2012, 8, e1002964. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Tan, C.; Wang, F.; Wang, Y.; Zhou, R.; Cui, D.; You, W.; Zhao, H.; Ren, J.; Feng, B. Knock-in of large reporter genes in human cells via CRISPR/Cas9-induced homology-dependent and independent DNA repair. Nucleic Acids Res. 2016, 44, e85. [Google Scholar] [CrossRef] [PubMed]
- Sadelain, M.; Papapetrou, E.P.; Bushman, F.D. Safe harbours for the integration of new DNA in the human genome. Nat. Rev. Cancer 2012, 12, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.R.; Maguire, S.; Davis, L.A.; Alexander, M.; Yang, F.; Chandran, S.; ffrench-Constant, C.; Pedersen, R.A. Robust, persistent transgene expression in human embryonic stem cells is achieved with AAVS1-targeted integration. Stem Cells 2008, 26, 496–504. [Google Scholar] [CrossRef]
- Xiang, G.; Ren, J.; Hai, T.; Fu, R.; Yu, D.; Wang, J.; Li, W.; Wang, H.; Zhou, Q. Editing porcine IGF2 regulatory element improved meat production in Chinese Bama pigs. Cell. Mol. Life Sci. 2018, 75, 4619–4628. [Google Scholar] [CrossRef]
- Xie, Z.; Pang, D.; Yuan, H.; Jiao, H.; Lu, C.; Wang, K.; Yang, Q.; Li, M.; Chen, X.; Yu, T.; et al. Genetically modified pigs are protected from classical swine fever virus. PLoS Pathog. 2018, 14, e1007193. [Google Scholar] [CrossRef]
- Wang, H.; Shen, L.; Chen, J.; Liu, X.; Tan, T.; Hu, Y.; Bai, X.; Li, Y.; Tian, K.; Li, N.; et al. Deletion of CD163 Exon 7 Confers Resistance to Highly Pathogenic Porcine Reproductive and Respiratory Viruses on Pigs. Int. J. Biol. Sci. 2019, 15, 1993–2005. [Google Scholar] [CrossRef]
- Pacesa, M.; Pelea, O.; Jinek, M. Past, present, and future of CRISPR genome editing technologies. Cell 2024, 187, 1076–1100. [Google Scholar] [CrossRef]
- Xiong, Y.; Zhuang, R.; Zhao, G.; Liu, Y.; Su, Y.; Wang, W.; Xi, X.; Yang, Y.; Han, X.; Xie, S.; et al. Identification of the CKM Gene as a Potential Muscle-Specific Safe Harbor Locus in Pig Genome. Genes 2022, 13, 921. [Google Scholar] [CrossRef]
- Gu, X.; Hu, X.; Wang, D.; Xu, Z.; Wang, F.; Li, D.; Li, G.-l.; Yang, H.; Li, H.; Zuo, E.; et al. Treatment of autosomal recessive hearing loss via in vivo CRISPR/Cas9-mediated optimized homology-directed repair in mice. Cell Res. 2022, 32, 699–702. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Liu, J.; Janssen, J.M.; Gonçalves, M.A.F.V. Precise homology-directed installation of large genomic edits in human cells with cleaving and nicking high-specificity Cas9 variants. Nucleic Acids Res. 2023, 51, 3465–3484. [Google Scholar] [CrossRef]
- Chen, X.; Du, J.; Yun, S.; Xue, C.; Yao, Y.; Rao, S. Recent advances in CRISPR-Cas9-based genome insertion technologies. Mol. Ther. Nucleic Acids 2024, 35, 102138. [Google Scholar] [CrossRef] [PubMed]

| Off-Target Gene | Sequence (5′-3′) | Indel |
|---|---|---|
| USPL1 | AGCCGTCT [TTATACAGCTTG] TGG | NO |
| MAST2 | CGGCGGGT [CTATACCGCGGG] CGG | NO |
| SPON1 | AAGCGGAT [CTGTGCCGCTTG] AGG | NO |
| LARP1 | AGGCGGGC [CTTTACCCCTTG] GGG | NO |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, X.; Yang, G.; Ji, P.; Liu, G.; Zhang, L. Identification of Site in the UTY Gene as Safe Harbor Locus on the Y Chromosome of Pig. Genes 2024, 15, 1005. https://doi.org/10.3390/genes15081005
Chen X, Yang G, Ji P, Liu G, Zhang L. Identification of Site in the UTY Gene as Safe Harbor Locus on the Y Chromosome of Pig. Genes. 2024; 15(8):1005. https://doi.org/10.3390/genes15081005
Chicago/Turabian StyleChen, Xiaomei, Guang Yang, Pengyun Ji, Guoshi Liu, and Lu Zhang. 2024. "Identification of Site in the UTY Gene as Safe Harbor Locus on the Y Chromosome of Pig" Genes 15, no. 8: 1005. https://doi.org/10.3390/genes15081005
APA StyleChen, X., Yang, G., Ji, P., Liu, G., & Zhang, L. (2024). Identification of Site in the UTY Gene as Safe Harbor Locus on the Y Chromosome of Pig. Genes, 15(8), 1005. https://doi.org/10.3390/genes15081005







