Genotype-by-Environment Interactions in Nonalcoholic Fatty Liver Disease and Chronic Illness among Mexican Americans: The Role of Acculturation Stress
Abstract
1. Background
2. Materials and Methods
2.1. Statistical Analysis
2.2. Genotype-by Environment (G×E) Interaction Model for Continuous Environments
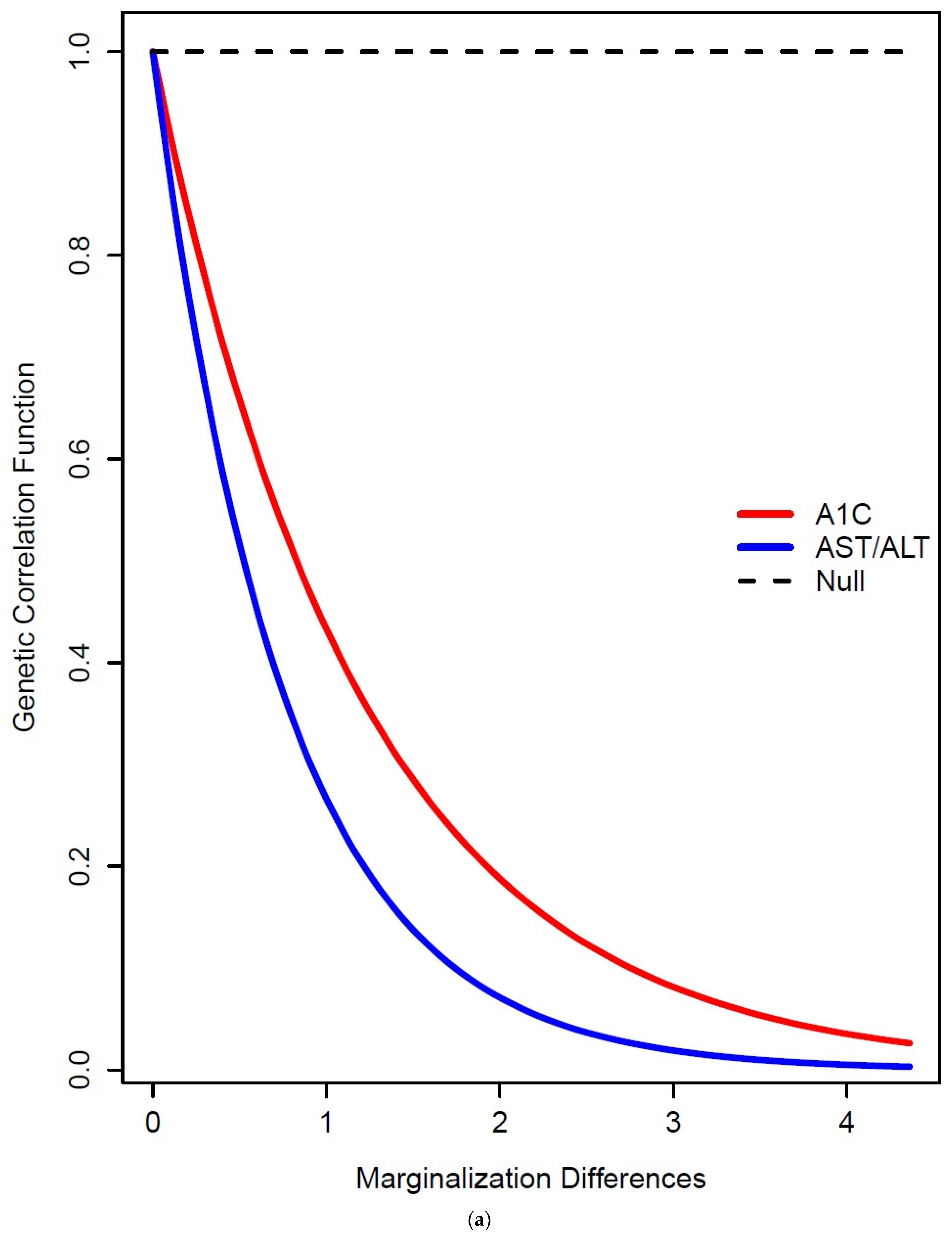
3. Results
4. Discussion
5. Limitations
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wanat, K.A.; Kovarik, C.L.; Shuman, S.; Whitaker, R.C.; Foster, G.D.; O’Brien, M.J. The association between obesity and health-related quality of life among urban Latinos. Ethn. Dis. 2014, 24, 14–18. [Google Scholar] [PubMed]
- Manusov, E.G.; Diego, V.P.; Smith, J.; Garza, J.R., 2nd; Lowdermilk, J.; Blangero, J.; Williams-Blangero, S.; Fernandez, F. UniMóvil: A Mobile Health Clinic Providing Primary Care to the Colonias of the Rio Grande Valley, South Texas. Front. Public Health 2019, 7, 215. [Google Scholar] [CrossRef] [PubMed]
- Manusov, E.G.; Diego, V.P.; Abrego, E.; Herklotz, K.; Almeida, M.; Mao, X.; Laston, S.; Blangero, J.; Blangero, S. Gene-by-Environment Interaction in Non-Alcoholic Fatty Liver Disease and Depression: The Role of Hepatic Transaminases. Med. Res. Arch. 2023, 11, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Manusov, E.; Reininger, B.; Ulloa-Aguirre, A.; Williams-Blangero, S. Editorial: The biology and management of chronic diseases in Mexican Americans. Front. Med. 2022, 9, 1095203. [Google Scholar] [CrossRef] [PubMed]
- Heredia, N.I.; Xu, T.; Lee, M.; McNeill, L.H.; Reininger, B.M. The Neighborhood Environment and Hispanic/Latino Health. Am. J. Health Promot. 2022, 36, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Vilar-Gomez, E.; Vuppalanchi, R.; Mladenovic, A.; Samala, N.; Gawrieh, S.; Newsome, P.N.; Chalasani, N. Prevalence of High-risk Nonalcoholic Steatohepatitis (NASH) in the United States: Results From NHANES 2017–2018. Clin. Gastroenterol. Hepatol. 2023, 21, 115–124. [Google Scholar] [CrossRef]
- Makri, E.; Goulas, A.; Polyzos, S.A. Epidemiology, Pathogenesis, Diagnosis and Emerging Treatment of Nonalcoholic Fatty Liver Disease. Arch. Med. Res. 2021, 52, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Sabotta, C.M.; Kwan, S.Y.; Petty, L.E.; Below, J.E.; Joon, A.; Wei, P.; Fisher-Hoch, S.P.; McCormick, J.B.; Beretta, L. Genetic variants associated with circulating liver injury markers in Mexican Americans, a population at risk for non-alcoholic fatty liver disease. Front. Genet. 2022, 13, 995488. [Google Scholar] [CrossRef] [PubMed]
- Young, K.A.; Palmer, N.D.; Fingerlin, T.E.; Langefeld, C.D.; Norris, J.M.; Wang, N.; Xiang, A.H.; Guo, X.; Williams, A.H.; Chen, Y.I.; et al. Genome-Wide Association Study Identifies Loci for Liver Enzyme Concentrations in Mexican Americans: The GUARDIAN Consortium. Obesity 2019, 27, 1331–1337. [Google Scholar] [CrossRef] [PubMed]
- Younossi, Z.M.; Golabi, P.; de Avila, L.; Paik, J.M.; Srishord, M.; Fukui, N.; Qiu, Y.; Burns, L.; Afendy, A.; Nader, F. The global epidemiology of NAFLD and NASH in patients with type 2 diabetes: A systematic review and meta-analysis. J. Hepatol. 2019, 71, 793–801. [Google Scholar] [CrossRef] [PubMed]
- Stefan, N.; Cusi, K. A global view of the interplay between non-alcoholic fatty liver disease and diabetes. Lancet Diabetes Endocrinol. 2022, 10, 284–296. [Google Scholar] [CrossRef] [PubMed]
- Tanase, D.M.; Gosav, E.M.; Costea, C.F.; Ciocoiu, M.; Lacatusu, C.M.; Maranduca, M.A.; Ouatu, A.; Floria, M. The Intricate Relationship between Type 2 Diabetes Mellitus (T2DM), Insulin Resistance (IR), and Nonalcoholic Fatty Liver Disease (NAFLD). J. Diabetes Res. 2020, 2020, 3920196. [Google Scholar] [CrossRef] [PubMed]
- Muzica, C.M.; Sfarti, C.; Trifan, A.; Zenovia, S.; Cuciureanu, T.; Nastasa, R.; Huiban, L.; Cojocariu, C.; Singeap, A.M.; Girleanu, I.; et al. Nonalcoholic Fatty Liver Disease and Type 2 Diabetes Mellitus: A Bidirectional Relationship. Can. J. Gastroenterol. Hepatol. 2020, 2020, 6638306. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Lim, L.K.E.; Ng, C.H.; Tan, D.J.H.; Lim, W.H.; Ho, C.S.H.; Tan, E.X.X.; Sanyal, A.J.; Muthiah, M.D. Is Fatty Liver Associated With Depression? A Meta-Analysis and Systematic Review on the Prevalence, Risk Factors, and Outcomes of Depression and Non-alcoholic Fatty Liver Disease. Front. Med. 2021, 8, 691696. [Google Scholar] [CrossRef] [PubMed]
- Soto-Angona, Ó.; Anmella, G.; Valdés-Florido, M.J.; De Uribe-Viloria, N.; Carvalho, A.F.; Penninx, B.; Berk, M. Non-alcoholic fatty liver disease (NAFLD) as a neglected metabolic companion of psychiatric disorders: Common pathways and future approaches. BMC Med. 2020, 18, 261. [Google Scholar] [CrossRef] [PubMed]
- Manusov, E.G.; Diego, V.P.; Sheikh, K.; Laston, S.; Blangero, J.; Williams-Blangero, S. Non-alcoholic Fatty Liver Disease and Depression: Evidence for Genotype × Environment Interaction in Mexican Americans. Front. Psychiatry 2022, 13, 936052. [Google Scholar] [CrossRef] [PubMed]
- Trépo, E.; Valenti, L. Update on NAFLD genetics: From new variants to the clinic. J. Hepatol. 2020, 72, 1196–1209. [Google Scholar] [CrossRef] [PubMed]
- Locke, A.E.; Kahali, B.; Berndt, S.I.; Justice, A.E.; Pers, T.H.; Day, F.R.; Powell, C.; Vedantam, S.; Buchkovich, M.L.; Yang, J.; et al. Genetic studies of body mass index yield new insights for obesity biology. Nature 2015, 518, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Suppli, N.P.; Andersen, K.K.; Agerbo, E.; Rajagopal, V.M.; Appadurai, V.; Coleman, J.R.I.; Breen, G.; Bybjerg-Grauholm, J.; Bækvad-Hansen, M.; Pedersen, C.B.; et al. Genome-wide by Environment Interaction Study of Stressful Life Events and Hospital-Treated Depression in the iPSYCH2012 Sample. Biol. Psychiatry Glob. Open Sci. 2022, 2, 400–410. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.Z.; Song, X.S.; Ma, J.S. Gene × environment interaction in major depressive disorder. World J. Clin. Cases 2021, 9, 9368–9375. [Google Scholar] [CrossRef] [PubMed]
- Glahn, D.C.; Curran, J.E.; Winkler, A.M.; Carless, M.A.; Kent, J.W., Jr.; Charlesworth, J.C.; Johnson, M.P.; Göring, H.H.; Cole, S.A.; Dyer, T.D.; et al. High dimensional endophenotype ranking in the search for major depression risk genes. Biol. Psychiatry 2012, 71, 6–14. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Zhang, W.; Hu, Y.; Chen, Y.; Shi, J. Association between nonalcoholic fatty liver disease and depression: A systematic review and meta-analysis of observational studies. J. Affect. Disord. 2022, 301, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Albuquerque, D.; Nóbrega, C.; Manco, L.; Padez, C. The contribution of genetics and environment to obesity. Br. Med. Bull. 2017, 123, 159–173. [Google Scholar] [CrossRef] [PubMed]
- Virolainen, S.J.; VonHandorf, A.; Viel, K.; Weirauch, M.T.; Kottyan, L.C. Gene-environment interactions and their impact on human health. Genes Immun. 2023, 24, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Diego, V.P.; Manusov, E.G.; Mao, X.; Curran, J.E.; Göring, H.; Almeida, M.; Mahaney, M.C.; Peralta, J.M.; Blangero, J.; Williams-Blangero, S. Genotype-by-socioeconomic status interaction influences heart disease risk scores and carotid artery thickness in Mexican Americans: The predominant role of education in comparison to household income and socioeconomic index. Front. Genet. 2023, 14, 1132110. [Google Scholar] [CrossRef] [PubMed]
- Arya, R.; Farook, V.S.; Fowler, S.P.; Puppala, S.; Chittoor, G.; Resendez, R.G.; Mummidi, S.; Vanamala, J.; Almasy, L.; Curran, J.E.; et al. Genetic and environmental (physical fitness and sedentary activity) interaction effects on cardiometabolic risk factors in Mexican American children and adolescents. Genet. Epidemiol. 2018, 42, 378–393. [Google Scholar] [CrossRef] [PubMed]
- Choh, A.C.; Lee, M.; Kent, J.W.; Diego, V.P.; Johnson, W.; Curran, J.E.; Dyer, T.D.; Bellis, C.; Blangero, J.; Siervogel, R.M.; et al. Gene-by-age effects on BMI from birth to adulthood: The Fels Longitudinal Study. Obesity 2014, 22, 875–881. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Lopez, O.; Martinez-Lopez, E.; Roman, S.; Fierro, N.A.; Panduro, A. Genetic, metabolic and environmental factors involved in the development of liver cirrhosis in Mexico. World J. Gastroenterol. 2015, 21, 11552–11566. [Google Scholar] [CrossRef] [PubMed]
- Barton, J.; Perlmeter, E.R.; Blum, E.S.; Marquez, R.R. Las Colonias in th 21st Century: Progress along the Texas-Mexico Border. Available online: https://www.dallasfed.org/~/media/microsites/cd/colonias/index.html (accessed on 3 January 2024).
- Alarcón, R.D.; Parekh, A.; Wainberg, M.L.; Duarte, C.S.; Araya, R.; Oquendo, M.A. Hispanic immigrants in the USA: Social and mental health perspectives. Lancet Psychiatry 2016, 3, 860–870. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.A.; Becker, H.A.; García, A.A.; Velasquez, M.M.; Tanaka, H.; Winter, M.A.; Perkison, W.B.; Brown, E.L.; Aguilar, D.; Hanis, C.L. Acculturation, Dietary Behaviors, and Macronutrient Intake Among Mexican Americans With Prediabetes: The Starr County Diabetes Prevention Initiative. Sci. Diabetes Self Manag. Care 2023, 49, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Nelson, T.; Perez, A.; Alcaraz, J.; Talavera, G.; McCarthy, J.J. Family History of Diabetes, Acculturation, and the Metabolic Syndrome among Mexican Americans: Proyecto SALSA. Metab. Syndr. Relat. Disord. 2007, 5, 262–269. [Google Scholar] [CrossRef] [PubMed]
- Cuellar, I.; Arnold, B.; Maldonado, R. Acculturation Rating Scale for Mexican Americans-II: A Revision of the Original ARSMA Scale. Hisp. J. Behav. Sci. 1995, 17, 275–304. [Google Scholar] [CrossRef]
- Jimenez, D.E.; Gray, H.L.; Cucciare, M.; Kumbhani, S.; Gallagher-Thompson, D. Using the Revised Acculturation Rating Scale for Mexican Americans (ARSMA-II) with Older Adults. Hisp. Health Care Int. 2010, 8, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Suarez, L.; Pulley, L. Comparing acculturation scales and their relationship to cancer screening among older Mexican-American women. J. Natl. Cancer Inst. Monogr. 1995, 18, 4–47. [Google Scholar]
- Hull, S.C.; Glanz, K.; Steffen, A.; Wilfond, B.S. Recruitment approaches for family studies: Attitudes of index patients and their relatives. IRB Ethics Hum. Res. 2004, 26, 12–17. [Google Scholar] [CrossRef]
- Eddowes, P.J.; Sasso, M.; Allison, M.; Tsochatzis, E.; Anstee, Q.M.; Sheridan, D.; Guha, I.N.; Cobbold, J.F.; Deeks, J.J.; Paradis, V.; et al. Accuracy of FibroScan Controlled Attenuation Parameter and Liver Stiffness Measurement in Assessing Steatosis and Fibrosis in Patients With Nonalcoholic Fatty Liver Disease. Gastroenterology 2019, 156, 1717–1730. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, M.S.; Vuppalanchi, R.; Van Natta, M.L.; Hallinan, E.; Kowdley, K.V.; Abdelmalek, M.; Neuschwander-Tetri, B.A.; Loomba, R.; Dasarathy, S.; Brandman, D.; et al. Vibration-Controlled Transient Elastography to Assess Fibrosis and Steatosis in Patients With Nonalcoholic Fatty Liver Disease. Clin. Gastroenterol. Hepatol. 2019, 17, 156–163.e152. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Kim, D.; Cho, Y. Exploratory Factor Analysis of the Beck Anxiety Inventory and the Beck Depression Inventory-II in a Psychiatric Outpatient Population. J. Korean Med. Sci. 2018, 33, e128. [Google Scholar] [CrossRef] [PubMed]
- Penley, J.A.; Wiebe, J.S.; Nwosu, A. Psychometric properties of the Spanish Beck Depression Inventory-II in a medical sample. Psychol. Assess. 2003, 15, 569–577. [Google Scholar] [CrossRef] [PubMed]
- Wiebe, J.S.; Penley, J.A. A psychometric comparison of the Beck Depression Inventory-II in English and Spanish. Psychol. Assess. 2005, 17, 481–485. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.P.; Gorenstein, C. Psychometric properties of the Beck Depression Inventory-II: A comprehensive review. Braz. J. Psychiatry 2013, 35, 416–431. [Google Scholar] [CrossRef] [PubMed]
- Eilenberg, M.; Munda, P.; Stift, J.; Langer, F.B.; Prager, G.; Trauner, M.; Staufer, K. Accuracy of non-invasive liver stiffness measurement and steatosis quantification in patients with severe and morbid obesity. Hepatobiliary Surg. Nutr. 2021, 10, 610–622. [Google Scholar] [CrossRef] [PubMed]
- Newsome, P.N.; Sasso, M.; Deeks, J.J.; Paredes, A.; Boursier, J.; Chan, W.K.; Yilmaz, Y.; Czernichow, S.; Zheng, M.H.; Wong, V.W.; et al. FibroScan-AST (FAST) score for the non-invasive identification of patients with non-alcoholic steatohepatitis with significant activity and fibrosis: A prospective derivation and global validation study. Lancet Gastroenterol. Hepatol. 2020, 5, 362–373. [Google Scholar] [CrossRef] [PubMed]
- Oeda, S.; Takahashi, H.; Imajo, K.; Seko, Y.; Kobayashi, T.; Ogawa, Y.; Moriguchi, M.; Yoneda, M.; Anzai, K.; Irie, H.; et al. Diagnostic accuracy of FibroScan-AST score to identify non-alcoholic steatohepatitis with significant activity and fibrosis in Japanese patients with non-alcoholic fatty liver disease: Comparison between M and XL probes. Hepatol. Res. 2020, 50, 831–839. [Google Scholar] [CrossRef] [PubMed]
- Cassinotto, C.; Boursier, J.; de Ledinghen, V.; Lebigot, J.; Lapuyade, B.; Cales, P.; Hiriart, J.B.; Michalak, S.; Bail, B.L.; Cartier, V.; et al. Liver stiffness in nonalcoholic fatty liver disease: A comparison of supersonic shear imaging, FibroScan, and ARFI with liver biopsy. Hepatology 2016, 63, 1817–1827. [Google Scholar] [CrossRef] [PubMed]
- Mikolasevic, I.; Orlic, L.; Franjic, N.; Hauser, G.; Stimac, D.; Milic, S. Transient elastography (FibroScan((R))) with controlled attenuation parameter in the assessment of liver steatosis and fibrosis in patients with nonalcoholic fatty liver disease—Where do we stand? World J. Gastroenterol. 2016, 22, 7236–7251. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Liu, Y.; Cao, Z.; Wang, L.; Li, Z.; Sheng, Z.; Cai, W.; Wang, H.; Bao, S.; Xie, Q. Comparison of FibroTouch and FibroScan for staging fibrosis in chronic liver disease: Single-center prospective study. Dig. Liver Dis. 2019, 51, 1323–1329. [Google Scholar] [CrossRef] [PubMed]
- Grob, S.R.; Suter, F.; Katzke, V.; Rohrmann, S. The Association between Liver Enzymes and Mortality Stratified by Non-Alcoholic Fatty Liver Disease: An Analysis of NHANES III. Nutrients 2023, 15, 3063. [Google Scholar] [CrossRef]
- Ghaddar, S.; Brown, C.J.; Pagán, J.A.; Díaz, V. Acculturation and healthy lifestyle habits among Hispanics in United States-Mexico border communities. Rev. Panam. Salud Publica 2010, 28, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Delaney, C.L.; Spaccarotella, K.; Quick, V.; Byrd-Bredbenner, C. A Comparison of Weight-Related Behaviors of Hispanic Mothers and Children by Acculturation Level. Int. J. Environ. Res. Public Health 2021, 18, 503. [Google Scholar] [CrossRef]
- McIntire, R.K.; Scalzo, L.; Doran, C.; Bucher, K.; Juon, H.S. Acculturation and Hypertension Diagnoses Among Hispanics in California. J. Racial Ethn. Health Disparities 2022, 9, 946–953. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Rhodes, L.; McArdle, C.E.; Rao, H.; Wang, Y.; Martinez-Miller, E.E.; Ward, J.B.; Cai, J.; Sofer, T.; Isasi, C.R.; North, K.E. A Gene-Acculturation Study of Obesity Among US Hispanic/Latinos: The Hispanic Community Health Study/Study of Latinos. Psychosom. Med. 2023, 85, 358–365. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Fulda, K.G.; Tao, M.H. Association between acculturation and metabolic syndrome in Hispanic adults mediated by fruits intake. Public Health Nutr. 2021, 24, 6472–6476. [Google Scholar] [CrossRef] [PubMed]
- Isasi, C.R.; Ayala, G.X.; Sotres-Alvarez, D.; Madanat, H.; Penedo, F.; Loria, C.M.; Elder, J.P.; Daviglus, M.L.; Barnhart, J.; Siega-Riz, A.M.; et al. Is acculturation related to obesity in Hispanic/Latino adults? Results from the Hispanic community health study/study of Latinos. J. Obes. 2015, 2015, 186276. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, C.M.; Moreno, O.; Garcia-Rodriguez, I.; Fuentes, L.; Nelson, T. The Hispanic Paradox: A Moderated Mediation Analysis of Health Conditions, Self-Rated Health, and Mental Health among Mexicans and Mexican Americans. Health Psychol. Behav. Med. 2022, 10, 180–198. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Guarda, R.M.; McCabe, B.E.; Nagy, G.A.; Stafford, A.M.; Matos, L.; Lu, M.; Felsman, I.; Rocha-Goldberg, P.; Cervantes, R.C. Acculturative Stress, Resilience, and a Syndemic Factor Among Latinx Immigrants. Nurs. Res. 2023, 72, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Ray, M.; Wallace, M.K.; Grayson, S.C.; Cummings, M.H.; Davis, J.A.; Scott, J.; Belcher, S.M.; Davis, T.S.; Conley, Y.P. Epigenomic Links Between Social Determinants of Health and Symptoms: A Scoping Review. Biol. Res. Nurs. 2023, 25, 404–416. [Google Scholar] [CrossRef] [PubMed]
- Balakrishnan, M.; Kanwal, F.; El-Serag, H.B.; Thrift, A.P. Acculturation and Nonalcoholic Fatty Liver Disease Risk Among Hispanics of Mexican Origin: Findings From the National Health and Nutrition Examination Survey. Clin. Gastroenterol. Hepatol. 2017, 15, 310–312. [Google Scholar] [CrossRef]
- Kallwitz, E.R.; Daviglus, M.L.; Allison, M.A.; Emory, K.T.; Zhao, L.; Kuniholm, M.H.; Chen, J.; Gouskova, N.; Pirzada, A.; Talavera, G.A.; et al. Prevalence of suspected nonalcoholic fatty liver disease in Hispanic/Latino individuals differs by heritage. Clin. Gastroenterol. Hepatol. 2015, 13, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, A.; Villavicencio, E.A.; Vogel, R.M.; Pace, T.W.; Ruiz, J.M.; Alkhouri, N.; Garcia, D.O. The association between perceived stress, acculturation, and non-alcoholic fatty liver disease in Mexican-origin adults in Southern Arizona. Prev. Med. Rep. 2023, 32, 102147. [Google Scholar] [CrossRef] [PubMed]
- Tomeno, W.; Kawashima, K.; Yoneda, M.; Saito, S.; Ogawa, Y.; Honda, Y.; Kessoku, T.; Imajo, K.; Mawatari, H.; Fujita, K.; et al. Non-alcoholic fatty liver disease comorbid with major depressive disorder: The pathological features and poor therapeutic efficacy. J. Gastroenterol. Hepatol. 2015, 30, 1009–1014. [Google Scholar] [CrossRef] [PubMed]
- Shea, S.; Lionis, C.; Kite, C.; Atkinson, L.; Chaggar, S.S.; Randeva, H.S.; Kyrou, I. Non-Alcoholic Fatty Liver Disease (NAFLD) and Potential Links to Depression, Anxiety, and Chronic Stress. Biomedicines 2021, 9, 1697. [Google Scholar] [CrossRef] [PubMed]
- Serafica, R.; Lekhak, N.; Bhatta, T. Acculturation, acculturative stress and resilience among older immigrants in United States. Int. Nurs. Rev. 2019, 66, 442–448. [Google Scholar] [CrossRef] [PubMed]
- Liem, A.; Renzaho, A.M.N.; Hannam, K.; Lam, A.I.F.; Hall, B.J. Acculturative stress and coping among migrant workers: A global mixed-methods systematic review. Appl. Psychol. Health Well Being 2021, 13, 491–517. [Google Scholar] [CrossRef] [PubMed]
- Bekteshi, V.; Kang, S.W. Contextualizing acculturative stress among Latino immigrants in the United States: A systematic review. Ethn. Health 2020, 25, 897–914. [Google Scholar] [CrossRef] [PubMed]
- Korbecki, J.; Bobiński, R.; Dutka, M. Self-regulation of the inflammatory response by peroxisome proliferator-activated receptors. Inflamm. Res. 2019, 68, 443–458. [Google Scholar] [CrossRef] [PubMed]
- Lommel, L.L.; Thompson, L.; Chen, J.L.; Waters, C.; Carrico, A. Acculturation, Inflammation, and Self-rated Health in Mexican American Immigrants. J. Immigr. Minor. Health 2019, 21, 1052–1060. [Google Scholar] [CrossRef] [PubMed]
- Polacchini, A.; Girardi, D.; Falco, A.; Zanotta, N.; Comar, M.; De Carlo, N.A.; Tongiorgi, E. Distinct CCL2, CCL5, CCL11, CCL27, IL-17, IL-6, BDNF serum profiles correlate to different job-stress outcomes. Neurobiol. Stress. 2018, 8, 82–91. [Google Scholar] [CrossRef]
- Sohal, A.; Chaudhry, H.; Kowdley, K.V. Genetic Markers Predisposing to Nonalcoholic Steatohepatitis. Clin. Liver Dis. 2023, 27, 333–352. [Google Scholar] [CrossRef] [PubMed]


| Trait | Females | Males | p-Value for Difference | ||
|---|---|---|---|---|---|
| Mean (N = 406) | S.D. | Mean (N = 141) | S.D. | ||
| Age | 44.13 | 14.61 | 46.18 | 15.69 | 0.1739 |
| Marginalization | 24.21 | 10.96 | 24.43 | 12.02 | 0.8543 |
| BDI-II | 6.79 | 8.14 | 4.59 | 6.51 | 1.4 × 10−3 |
| BMI | 32.49 | 7.07 | 32.25 | 6.97 | 0.7272 |
| HbA1C | 6.60 | 1.96 | 6.62 | 2.13 | 0.9318 |
| AST | 19.91 | 15.78 | 25.69 | 19.83 | 2.0 × 10−3 |
| ALT | 22.30 | 23.73 | 33.35 | 31.98 | 1.2 × 10−4 |
| AST/ALT | 1.16 | 0.65 | 0.99 | 0.63 | 6.4 × 10−3 |
| CAP | 284.24 | 64.94 | 289.64 | 59.21 | 0.4919 |
| kPa | 6.64 | 4.49 | 7.02 | 4.11 | 0.4808 |
| FAST | 0.14 | 0.20 | 0.22 | 0.26 | 8.4 × 10−3 |
| Trait | Heritability | Standard Error | p-Value |
|---|---|---|---|
| HbA1C | 0.52 | 0.11 | 2.5 × 10−6 |
| Marginalization | 0.30 | 0.08 | 3.8 × 10−5 |
| AST | 0.25 | 0.14 | 2.0 × 10−2 |
| ALT | 0.41 | 0.13 | 6.9 × 10−3 |
| AST/ALT | 0.27 | 0.10 | 1.9 × 10−3 |
| BDI-II | 0.36 | 0.10 | 1.5 × 10−5 |
| BMI | 0.55 | 0.11 | 8.0 × 10−7 |
| CAP | 0.34 | 0.10 | 3.6 × 10−4 |
| FAST | 0.35 | 0.13 | 2.3 × 10−3 |
| kPa | 0.33 | 0.13 | 4.3 × 10−3 |
| Trait | Model | Ln Likelihood | Chi-Square | p-Value |
|---|---|---|---|---|
| HbA1c | Polygenic | −261.838 | 9.72868 | 4.8 × 10−3 |
| Reduced G×E | −256.973 | |||
| AST | Polygenic | −261.088 | 2.503294 | 0.199821 |
| Full G×E | −259.836 | |||
| ALT | Polygenic | −264.444 | 2.952168 | 0.157147 |
| Full G×E | −262.968 | |||
| AST/ALT | Polygenic | −265.081 | 14.65585 | 3.9 × 10−4 |
| Full G×E | −257.753 | |||
| BDI-II | Polygenic | −247.356 | 20.96177 | 1.6 × 10−5 |
| Full G×E | −236.876 | |||
| BMI | Polygenic | −260.745 | 12.40074 | 2.03 × 10−3 |
| Reduced G×E | −254.545 | |||
| CAP | Polygenic | −261.838 | 9.72868 | 7.7 × 10−3 |
| Reduced G×E | −256.973 | |||
| FAST | Polygenic | −226.611 | 1.992278 | 0.263703 |
| Full G×E | −225.615 | |||
| kPa | Polygenic | −275.19 | 0.375754 | 0.684301 |
| Full G×E | −275.002 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manusov, E.G.; Diego, V.P.; Almeida, M.; Ortiz, D.; Curran, J.E.; Galan, J.; Leandro, A.C.; Laston, S.; Blangero, J.; Williams-Blangero, S. Genotype-by-Environment Interactions in Nonalcoholic Fatty Liver Disease and Chronic Illness among Mexican Americans: The Role of Acculturation Stress. Genes 2024, 15, 1006. https://doi.org/10.3390/genes15081006
Manusov EG, Diego VP, Almeida M, Ortiz D, Curran JE, Galan J, Leandro AC, Laston S, Blangero J, Williams-Blangero S. Genotype-by-Environment Interactions in Nonalcoholic Fatty Liver Disease and Chronic Illness among Mexican Americans: The Role of Acculturation Stress. Genes. 2024; 15(8):1006. https://doi.org/10.3390/genes15081006
Chicago/Turabian StyleManusov, Eron G., Vincent P. Diego, Marcio Almeida, David Ortiz, Joanne E. Curran, Jacob Galan, Ana C. Leandro, Sandra Laston, John Blangero, and Sarah Williams-Blangero. 2024. "Genotype-by-Environment Interactions in Nonalcoholic Fatty Liver Disease and Chronic Illness among Mexican Americans: The Role of Acculturation Stress" Genes 15, no. 8: 1006. https://doi.org/10.3390/genes15081006
APA StyleManusov, E. G., Diego, V. P., Almeida, M., Ortiz, D., Curran, J. E., Galan, J., Leandro, A. C., Laston, S., Blangero, J., & Williams-Blangero, S. (2024). Genotype-by-Environment Interactions in Nonalcoholic Fatty Liver Disease and Chronic Illness among Mexican Americans: The Role of Acculturation Stress. Genes, 15(8), 1006. https://doi.org/10.3390/genes15081006







