A Closer Look into White Adipose Tissue Biology and the Molecular Regulation of Stem Cell Commitment and Differentiation
Abstract
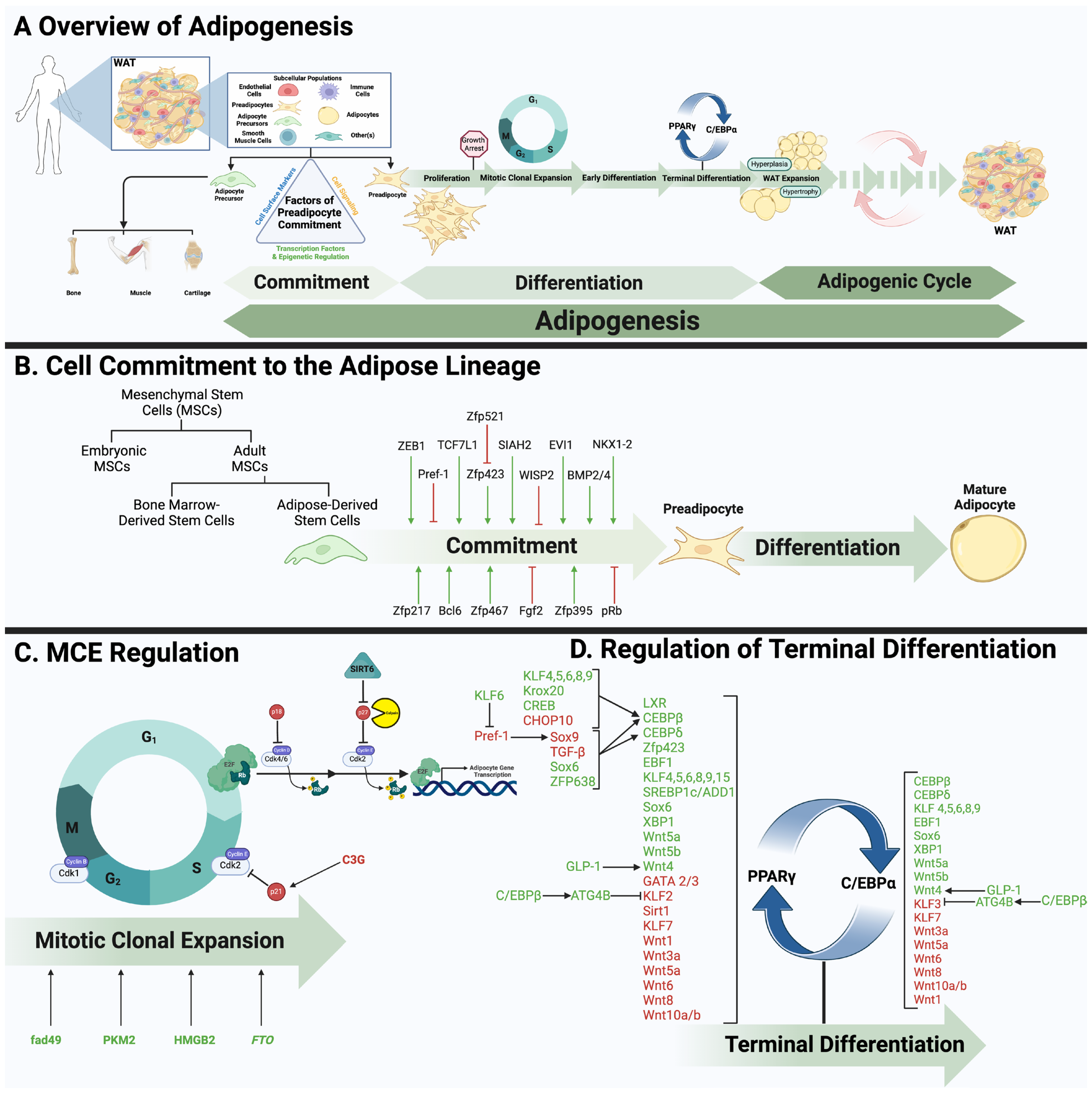
1. Introduction: An Overview of Adipose Tissue Biology and Function
2. WAT Development
2.1. Cellular Hierarchy
2.2. Commitment
2.3. Adipose Tissue Expansion
2.4. Adipose Stromal Cells
| Markers/Regulators | Overall Effect on WAT Development | Contribution | Commitment vs. Differentiation | Reference |
|---|---|---|---|---|
| Bcl6 | Promotes | Regulator | Commitment | [100] |
| BMP2/4 | Promotes | Regulator | Commitment | [121,172] |
| EVI1 | Promotes | Regulator | Commitment, Differentiation | [101,102,103] |
| Fgf2 | Inhibits | Regulator | Commitment, Differentiation | [173] |
| NKX1-2 | Promotes | Regulator | Commitment, Differentiation | [115] |
| Pref-1 | Inhibits | Marker, Regulator | Commitment, Differentiation | [91] |
| Regulator | Commitment | [93] | ||
| pRb | Inhibits | Regulator | Commitment, Differentiation | [98,99,174] |
| SIAH2 | Promotes | Regulator | Commitment | [95,96] |
| TCF7L1 | Promotes | Regulator | Commitment | [97] |
| ZEB1 | Promotes | Regulator | Commitment, Differentiation | [114] |
| Zfp217 | Promotes | Regulator | Commitment, Differentiation | [104,105,106] |
| Zfp395 | Promotes | Regulator | Commitment, Differentiation | [107] |
| Zfp423 | Promotes | Marker | Commitment | [109] |
| Regulator | Commitment | [108] | ||
| Zfp467 | Promotes | Regulator | Commitment, Differentiation | [111,123,124] |
| Zfp521 | Inhibits | Regulator | Commitment | [113] |
| WISP2 | Inhibits | Regulator | Commitment, Differentiation | [122] |
3. WAT Transcription
3.1. Mitotic Clonal Expansion (MCE)
3.2. PPARγ
3.3. C/EBPα
3.4. Additional Modulators
| Transcription Factors/Regulators | Primary Effect | Target(s) | Reference(s) |
|---|---|---|---|
| ATG4B | Inhibits | KLF2,3 | [264] |
| C/EBPα | Promotes | Sirt1 | [236] |
| C/EBPβ | Promotes | PPARγ, C/EBPα | [225,238] |
| C/EBPδ | |||
| CHOP10 | Inhibits | C/EBPβ | [265] |
| CREB | Promotes | C/EBPβ | [266] |
| EBF1 | Promotes | PPARγ, C/EBPα | [202] |
| GATA2/3 | Inhibits | PPARγ | [267] |
| GLP-1 | Promotes | PPARγ, C/EBPα | [268] |
| Krox20 | Promotes | C/EBPβ | [269] |
| LXR | Promotes | PPARγ | [270] |
| Sirt1 | Inhibits | PPARγ | [234,271] |
| Sox6 | Promotes | PPARγ, C/EBPα, β, δ | [272] |
| SREBP1c/ADD1 | Promotes | PPARγ | [273] |
| TGF-β | Inhibits | C/EBPβ, δ | [274] |
| Wnt1 | Inhibits | PPARγ, C/EBPα | [275] |
| Wnt3a | [276] | ||
| Wnt4 | Promotes | [258,268] | |
| Wnt5a | Promotes | [258] | |
| Wnt5a | Inhibits | [259] | |
| Wnt5b | Promotes | [262,277] | |
| Wnt6 | Inhibits | [278] | |
| Wnt8 | [279] | ||
| Wnt10a/b | [278,280] | ||
| XBP1 | Promotes | PPARγ, C/EBPα | [281,282] |
| Zfp423 | Promotes | PPARγ | [108] |
| Zfp638 | Promotes | C/EBPβ, δ | [283] |
4. Future Directions
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Azeez, O.I.; Meintjes, R.; Chamunorwa, J.P. Fat body, fat pad and adipose tissues in invertebrates and vertebrates: The nexus. Lipids Health Dis. 2014, 13, 71. [Google Scholar] [CrossRef]
- Ottaviani, E.; Malagoli, D.; Franceschi, C. The evolution of the adipose tissue: A neglected enigma. Gen. Comp. Endocrinol. 2011, 174, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Chusyd, D.E.; Wang, D.; Huffman, D.M.; Nagy, T.R. Relationships between Rodent White Adipose Fat Pads and Human White Adipose Fat Depots. Front. Nutr. 2016, 3, 10. [Google Scholar] [CrossRef] [PubMed]
- Wronska, A.; Kmiec, Z. Structural and biochemical characteristics of various white adipose tissue depots. Acta Physiol. 2012, 205, 194–208. [Google Scholar] [CrossRef] [PubMed]
- Martins, T.; Castro-Ribeiro, C.; Lemos, S.; Ferreira, T.; Nascimento-Gonçalves, E.; Rosa, E.; Oliveira, P.A.; Antunes, L.M. Murine Models of Obesity. Obesities 2022, 2, 127–147. [Google Scholar] [CrossRef]
- Giordano, A.; Cinti, F.; Canese, R.; Carpinelli, G.; Colleluori, G.; Di Vincenzo, A.; Palombelli, G.; Severi, I.; Moretti, M.; Redaelli, C.; et al. The Adipose Organ Is a Unitary Structure in Mice and Humans. Biomedicines 2022, 10, 2275. [Google Scholar] [CrossRef] [PubMed]
- Cinti, S. The Adipose Organ. In Adipose Tissue and Adipokines in Health and Disease; Humana Press: Totowa, NJ, USA, 2007; pp. 3–19. [Google Scholar]
- Mandarim-de-Lacerda, C.; Sol, M.; Vásquez, B.; Aguila, M. Mice as an Animal Model for the Study of Adipose Tissue and Obesity. Int. J. Morphol. 2021, 39, 1521–1528. [Google Scholar] [CrossRef]
- Börgeson, E.; Boucher, J.; Hagberg, C.E. Of mice and men: Pinpointing species differences in adipose tissue biology. Front. Cell Dev. Biol. 2022, 10, 1003118. [Google Scholar] [CrossRef] [PubMed]
- Richard, M.A.; Pallubinsky, H.; Blondin, D.P. Functional characterization of human brown adipose tissue metabolism. Biochem. J. 2020, 477, 1261–1286. [Google Scholar] [CrossRef] [PubMed]
- Kiefer, F.W.; Cohen, P.; Plutzky, J. Fifty shades of brown: Perivascular fat, thermogenesis, and atherosclerosis. Circulation 2012, 126, 1012–1015. [Google Scholar] [CrossRef] [PubMed]
- Ziqubu, K.; Dludla, P.V.; Mthembu, S.X.H.; Nkambule, B.B.; Mabhida, S.E.; Jack, B.U.; Nyambuya, T.M.; Mazibuko-Mbeje, S.E. An insight into brown/beige adipose tissue whitening, a metabolic complication of obesity with the multifactorial origin. Front. Endocrinol. 2023, 14, 1114767. [Google Scholar] [CrossRef] [PubMed]
- Maurer, S.; Harms, M.; Boucher, J. The colorful versatility of adipocytes: White-to-brown transdifferentiation and its therapeutic potential in humans. FEBS J. 2021, 288, 3628–3646. [Google Scholar] [CrossRef] [PubMed]
- Shinde, A.B.; Song, A.; Wang, Q.A. Brown Adipose Tissue Heterogeneity, Energy Metabolism, and Beyond. Front. Endocrinol. 2021, 12, 651763. [Google Scholar] [CrossRef] [PubMed]
- Pallio, G. New Insights into Adipose Tissue Metabolic Function and Dysfunction. Int. J. Mol. Sci. 2023, 24, 9953. [Google Scholar] [CrossRef] [PubMed]
- Cinti, S. White, brown, beige and pink: A rainbow in the adipose organ. Curr. Opin. Endocr. Metab. Res. 2019, 4, 29–36. [Google Scholar] [CrossRef]
- Fève, B.; Cintid, S.; Beaupère, C.; Vatier, C.; Vigouroux, C.; Vali, A.; Capeau, J.; Grosfed, A.; Moldes, M. Pink adipose tissue: A paradigm of adipose tissue plasticity. Ann. Endocrinol. 2024, 85, 248–251. [Google Scholar] [CrossRef] [PubMed]
- Aparisi Gómez, M.P.; Ayuso Benavent, C.; Simoni, P.; Aparisi, F.; Guglielmi, G.; Bazzocchi, A. Fat and bone: The multiperspective analysis of a close relationship. Quant. Imaging Med. Surg. 2020, 10, 1614–1635. [Google Scholar] [CrossRef] [PubMed]
- Lecka-Czernik, B.; Baroi, S.; Stechschulte, L.A.; Chougule, A.S. Marrow Fat-a New Target to Treat Bone Diseases? Curr. Osteoporos. Rep. 2018, 16, 123–129. [Google Scholar] [CrossRef]
- Scheller, E.L.; Cawthorn, W.P.; Burr, A.A.; Horowitz, M.C.; MacDougald, O.A. Marrow Adipose Tissue: Trimming the Fat. Trends Endocrinol. Metab. 2016, 27, 392–403. [Google Scholar] [CrossRef] [PubMed]
- Machado, S.A.; Pasquarelli-do-Nascimento, G.; da Silva, D.S.; Farias, G.R.; de Oliveira Santos, I.; Baptista, L.B.; Magalhães, K.G. Browning of the white adipose tissue regulation: New insights into nutritional and metabolic relevance in health and diseases. Nutr. Metab. 2022, 19, 61. [Google Scholar] [CrossRef]
- Bartness, T.J.; Ryu, V. Neural control of white, beige and brown adipocytes. Int. J. Obes. Suppl. 2015, 5, S35–S39. [Google Scholar] [CrossRef] [PubMed]
- Cedikova, M.; Kripnerová, M.; Dvorakova, J.; Pitule, P.; Grundmanova, M.; Babuska, V.; Mullerova, D.; Kuncova, J. Mitochondria in White, Brown, and Beige Adipocytes. Stem. Cells Int. 2016, 2016, 6067349. [Google Scholar] [CrossRef] [PubMed]
- Yuko, O.O.; Saito, M. Brown Fat as a Regulator of Systemic Metabolism beyond Thermogenesis. Diabetes Metab. J. 2021, 45, 840–852. [Google Scholar] [CrossRef]
- Ikeda, K.; Yamada, T. UCP1 Dependent and Independent Thermogenesis in Brown and Beige Adipocytes. Front. Endocrinol. 2020, 11, 498. [Google Scholar] [CrossRef] [PubMed]
- Rui, L. Brown and Beige Adipose Tissues in Health and Disease. Compr. Physiol. 2017, 7, 1281–1306. [Google Scholar] [CrossRef] [PubMed]
- Amri, E.Z. Beige or brite adipocytes of the adipose organ: Link with white and brown adipocytes. Ann. Endocrinol. 2024, 85, 253–254. [Google Scholar] [CrossRef] [PubMed]
- Trayhurn, P.; Beattie, J.H. Physiological role of adipose tissue: White adipose tissue as an endocrine and secretory organ. Proc. Nutr. Soc. 2001, 60, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Richard, A.J.; White, U.; Elks, C.M.; Stephens, J.M. Adipose Tissue: Physiology to Metabolic Dysfunction. In Endotext; Feingold, K.R., Anawalt, B., Blackman, M.R., Boyce, A., Chrousos, G., Corpas, E., de Herder, W.W., Dhatariya, K., Dungan, K., Hofland, J., et al., Eds.; MDText.com, Inc.: South Dartmouth, MA, USA, 2000. [Google Scholar]
- Lee, Y.H.; Mottillo, E.P.; Granneman, J.G. Adipose tissue plasticity from WAT to BAT and in between. Biochim. Biophys. Acta 2014, 1842, 358–369. [Google Scholar] [CrossRef]
- Cao, W.; Daniel, K.W.; Robidoux, J.; Puigserver, P.; Medvedev, A.V.; Bai, X.; Floering, L.M.; Spiegelman, B.M.; Collins, S. p38 mitogen-activated protein kinase is the central regulator of cyclic AMP-dependent transcription of the brown fat uncoupling protein 1 gene. Mol. Cell. Biol. 2004, 24, 3057–3067. [Google Scholar] [CrossRef] [PubMed]
- Fenzl, A.; Kiefer, F.W. Brown adipose tissue and thermogenesis. Horm. Mol. Biol. Clin. Investig. 2014, 19, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.M.; Sanchez-Gurmaches, J.; Guertin, D.A. Brown Adipose Tissue Development and Metabolism. In Brown Adipose Tissue; Springer: Cham, Switzerland, 2019; Volume 251, pp. 3–36. [Google Scholar] [CrossRef]
- Brondani, L.A.; Assmann, T.S.; Duarte, G.C.; Gross, J.L.; Canani, L.H.; Crispim, D. The role of the uncoupling protein 1 (UCP1) on the development of obesity and type 2 diabetes mellitus. Arq. Bras. Endocrinol. Metabol. 2012, 56, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Cannon, B.; Nedergaard, J. What Ignites UCP1? Cell Metab. 2017, 26, 697–698. [Google Scholar] [CrossRef] [PubMed]
- Gagelin, A.; Largeau, C.; Masscheleyn, S.; Piel, M.S.; Calderón-Mora, D.; Bouillaud, F.; Hénin, J.; Miroux, B. Molecular determinants of inhibition of UCP1-mediated respiratory uncoupling. Nat. Commun. 2023, 14, 2594. [Google Scholar] [CrossRef] [PubMed]
- Cannon, B.; Nedergaard, J. Brown adipose tissue: Function and physiological significance. Physiol. Rev. 2004, 84, 277–359. [Google Scholar] [CrossRef] [PubMed]
- Azzu, V.; Brand, M.D. The on-off switches of the mitochondrial uncoupling proteins. Trends Biochem. Sci. 2010, 35, 298–307. [Google Scholar] [CrossRef] [PubMed]
- Ricquier, D. Uncoupling protein 1 of brown adipocytes, the only uncoupler: A historical perspective. Front. Endocrinol. 2011, 2, 85. [Google Scholar] [CrossRef] [PubMed]
- Saito, M.; Matsushita, M.; Yoneshiro, T.; Okamatsu-Ogura, Y. Brown Adipose Tissue, Diet-Induced Thermogenesis, and Thermogenic Food Ingredients: From Mice to Men. Front. Endocrinol. 2020, 11, 222. [Google Scholar] [CrossRef] [PubMed]
- Scheel, A.K.; Espelage, L.; Chadt, A. Many Ways to Rome: Exercise, Cold Exposure and Diet—Do They All Affect BAT Activation and WAT Browning in the Same Manner? Int. J. Mol. Sci. 2022, 23, 4759. [Google Scholar] [CrossRef]
- Jurado-Fasoli, L.; Merchan-Ramirez, E.; Martinez-Tellez, B.; Acosta, F.M.; Sanchez-Delgado, G.; Amaro-Gahete, F.J.; Munoz Hernandez, V.; Martinez-Avila, W.D.; Ortiz-Alvarez, L.; Xu, H.; et al. Association between dietary factors and brown adipose tissue volume/(18)F-FDG uptake in young adults. Clin. Nutr. 2021, 40, 1997–2008. [Google Scholar] [CrossRef] [PubMed]
- Saito, M. Brown adipose tissue as a therapeutic target for human obesity. Obes. Res. Clin. Pract. 2013, 7, e432–e438. [Google Scholar] [CrossRef]
- Johnson, S.B.; Tomer, A.; Cunningham, W.R.; Henretta, J.C. Adherence in childhood diabetes: Results of a confirmatory factor analysis. Health Psychol. 1990, 9, 493–501. [Google Scholar] [CrossRef] [PubMed]
- Scheele, C.; Wolfrum, C. Brown Adipose Crosstalk in Tissue Plasticity and Human Metabolism. Endocr. Rev. 2020, 41, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Kulterer, O.C.; Herz, C.T.; Prager, M.; Schmöltzer, C.; Langer, F.B.; Prager, G.; Marculescu, R.; Kautzky-Willer, A.; Hacker, M.; Haug, A.R.; et al. Brown Adipose Tissue Prevalence Is Lower in Obesity but Its Metabolic Activity Is Intact. Front. Endocrinol. 2022, 13, 858417. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Delgado, G.; Martinez-Tellez, B.; Acosta, F.M.; Virtue, S.; Vidal-Puig, A.; Gil, A.; Llamas-Elvira, J.M.; Ruiz, J.R. Brown Adipose Tissue Volume and Fat Content Are Positively Associated with Whole-Body Adiposity in Young Men—Not in Women. Diabetes 2021, 70, 1473–1485. [Google Scholar] [CrossRef] [PubMed]
- Dewal, R.S.; Yang, F.T.; Baer, L.A.; Vidal, P.; Hernandez-Saavedra, D.; Seculov, N.P.; Ghosh, A.; Noé, F.; Togliatti, O.; Hughes, L.; et al. Transplantation of committed pre-adipocytes from brown adipose tissue improves whole-body glucose homeostasis. iScience 2024, 27, 108927. [Google Scholar] [CrossRef] [PubMed]
- White, J.D.; Dewal, R.S.; Stanford, K.I. The beneficial effects of brown adipose tissue transplantation. Mol. Asp. Med. 2019, 68, 74–81. [Google Scholar] [CrossRef]
- Warrier, M.; Paules, E.M.; Silva-Gomez, J.; Friday, W.B.; Bramlett, F.; Kim, H.; Zhang, K.; Trujillo-Gonzalez, I. Homocysteine-induced endoplasmic reticulum stress activates FGF21 and is associated with browning and atrophy of white adipose tissue in Bhmt knockout mice. Heliyon 2023, 9, e13216. [Google Scholar] [CrossRef] [PubMed]
- Alipoor, E.; Hosseinzadeh-Attar, M.J.; Rezaei, M.; Jazayeri, S.; Chapman, M. White adipose tissue browning in critical illness: A review of the evidence, mechanisms and future perspectives. Obes. Rev. 2020, 21, e13085. [Google Scholar] [CrossRef] [PubMed]
- McClave, S.A.; Martindale, R.G. Browning of white adipose tissue may be an appropriate adaptive response to critical illness. J. Parenter. Enter. Nutr. 2024, 48, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Patsouris, D.; Qi, P.; Abdullahi, A.; Stanojcic, M.; Chen, P.; Parousis, A.; Amini-Nik, S.; Jeschke, M.G. Burn Induces Browning of the Subcutaneous White Adipose Tissue in Mice and Humans. Cell Rep. 2015, 13, 1538–1544. [Google Scholar] [CrossRef] [PubMed]
- Abdullahi, A.; Chen, P.; Stanojcic, M.; Sadri, A.R.; Coburn, N.; Jeschke, M.G. IL-6 Signal From the Bone Marrow is Required for the Browning of White Adipose Tissue Post Burn Injury. Shock 2017, 47, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Heier, C.; Meng, X.; Bakiri, L.; Pototschnig, I.; Tang, Z.; Schauer, S.; Baumgartner, V.J.; Grabner, G.F.; Schabbauer, G.; et al. An immune-sympathetic neuron communication axis guides adipose tissue browning in cancer-associated cachexia. Proc. Natl. Acad. Sci. USA 2022, 119, e2112840119. [Google Scholar] [CrossRef] [PubMed]
- Kir, S.; Spiegelman, B.M. Cachexia & Brown Fat: A Burning Issue in Cancer. Trends Cancer 2016, 2, 461–463. [Google Scholar] [CrossRef] [PubMed]
- Ayalon, I.; Shen, H.; Williamson, L.; Stringer, K.; Zingarelli, B.; Kaplan, J.M. Sepsis Induces Adipose Tissue Browning in Nonobese Mice But Not in Obese Mice. Shock 2018, 50, 557–564. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Davis, X.; Lahni, P.; Stuck, J.; Williamson, L.; Kaplan, J. Obesity protects against sepsis-induced and norepinephrine-induced white adipose tissue browning. Am. J. Physiol. Endocrinol. Metab. 2021, 321, E433–E442. [Google Scholar] [CrossRef]
- Pinckard, K.M.; Stanford, K.I. The Heartwarming Effect of Brown Adipose Tissue. Mol. Pharmacol. 2022, 102, 460–471. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, Z.; Song, Y.; Xie, H.; Dong, M. An update on brown adipose tissue and obesity intervention: Function, regulation and therapeutic implications. Front. Endocrinol. 2022, 13, 1065263. [Google Scholar] [CrossRef] [PubMed]
- Ahmadian, M.; Duncan, R.E.; Jaworski, K.; Sarkadi-Nagy, E.; Sul, H.S. Triacylglycerol metabolism in adipose tissue. Future Lipidol. 2007, 2, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.T.; Hochfeld, W.E.; Myburgh, R.; Pepper, M.S. Adipocyte and adipogenesis. Eur. J. Cell Biol. 2013, 92, 229–236. [Google Scholar] [CrossRef]
- Liao, X.; Zhou, H.; Deng, T. The composition, function, and regulation of adipose stem and progenitor cells. J. Genet. Genom. 2022, 49, 308–315. [Google Scholar] [CrossRef] [PubMed]
- Pyrina, I.; Chung, K.J.; Michailidou, Z.; Koutsilieris, M.; Chavakis, T.; Chatzigeorgiou, A. Fate of Adipose Progenitor Cells in Obesity-Related Chronic Inflammation. Front. Cell Dev. Biol. 2020, 8, 644. [Google Scholar] [CrossRef] [PubMed]
- Rodeheffer, M.S.; Birsoy, K.; Friedman, J.M. Identification of white adipocyte progenitor cells in vivo. Cell 2008, 135, 240–249. [Google Scholar] [CrossRef] [PubMed]
- Berry, R.; Rodeheffer, M.S. Characterization of the adipocyte cellular lineage in vivo. Nat. Cell Biol. 2013, 15, 302–308. [Google Scholar] [CrossRef] [PubMed]
- Corvera, S. Cellular Heterogeneity in Adipose Tissues. Annu. Rev. Physiol. 2021, 83, 257–278. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, A.; Guo, J.; Banyard, D.A.; Fadavi, D.; Toranto, J.D.; Wirth, G.A.; Paydar, K.Z.; Evans, G.R.; Widgerow, A.D. Stromal vascular fraction: A regenerative reality? Part 1: Current concepts and review of the literature. J. Plast. Reconstr. Aesthet. Surg. 2016, 69, 170–179. [Google Scholar] [CrossRef] [PubMed]
- Sebo, Z.L.; Rodeheffer, M.S. Assembling the adipose organ: Adipocyte lineage segregation and adipogenesis in vivo. Development 2019, 146, dev172098. [Google Scholar] [CrossRef] [PubMed]
- Billon, N.; Iannarelli, P.; Monteiro, M.C.; Glavieux-Pardanaud, C.; Richardson, W.D.; Kessaris, N.; Dani, C.; Dupin, E. The generation of adipocytes by the neural crest. Development 2007, 134, 2283–2292. [Google Scholar] [CrossRef] [PubMed]
- Takashima, Y.; Era, T.; Nakao, K.; Kondo, S.; Kasuga, M.; Smith, A.G.; Nishikawa, S. Neuroepithelial cells supply an initial transient wave of MSC differentiation. Cell 2007, 129, 1377–1388. [Google Scholar] [CrossRef] [PubMed]
- Morikawa, S.; Mabuchi, Y.; Niibe, K.; Suzuki, S.; Nagoshi, N.; Sunabori, T.; Shimmura, S.; Nagai, Y.; Nakagawa, T.; Okano, H.; et al. Development of mesenchymal stem cells partially originate from the neural crest. Biochem. Biophys. Res. Commun. 2009, 379, 1114–1119. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.; Kim, H.; Elkabetz, Y.; Al Shamy, G.; Panagiotakos, G.; Barberi, T.; Tabar, V.; Studer, L. Isolation and directed differentiation of neural crest stem cells derived from human embryonic stem cells. Nat. Biotechnol. 2007, 25, 1468–1475. [Google Scholar] [CrossRef] [PubMed]
- Vodyanik, M.A.; Yu, J.; Zhang, X.; Tian, S.; Stewart, R.; Thomson, J.A.; Slukvin, I.I. A mesoderm-derived precursor for mesenchymal stem and endothelial cells. Cell Stem Cell 2010, 7, 718–729. [Google Scholar] [CrossRef] [PubMed]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.; Krause, D.; Deans, R.; Keating, A.; Prockop, D.; Horwitz, E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef] [PubMed]
- Horwitz, E.M.; Le Blanc, K.; Dominici, M.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.C.; Deans, R.J.; Krause, D.S.; Keating, A. Clarification of the nomenclature for MSC: The International Society for Cellular Therapy position statement. Cytotherapy 2005, 7, 393–395. [Google Scholar] [CrossRef] [PubMed]
- Viswanathan, S.; Shi, Y.; Galipeau, J.; Krampera, M.; Leblanc, K.; Martin, I.; Nolta, J.; Phinney, D.G.; Sensebe, L. Mesenchymal stem versus stromal cells: International Society for Cell & Gene Therapy (ISCT®) Mesenchymal Stromal Cell committee position statement on nomenclature. Cytotherapy 2019, 21, 1019–1024. [Google Scholar] [CrossRef] [PubMed]
- Bianco, P.; Robey, P.G.; Simmons, P.J. Mesenchymal stem cells: Revisiting history, concepts, and assays. Cell Stem Cell 2008, 2, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, W.; Rubin, J.P.; Marra, K.G. Adipose-derived stem cells: Implications in tissue regeneration. World J. Stem Cells 2014, 6, 312–321. [Google Scholar] [CrossRef] [PubMed]
- Cawthorn, W.P.; Scheller, E.L.; MacDougald, O.A. Adipose tissue stem cells meet preadipocyte commitment: Going back to the future. J. Lipid Res. 2012, 53, 227–246. [Google Scholar] [CrossRef] [PubMed]
- Mildmay-White, A.; Khan, W. Cell Surface Markers on Adipose-Derived Stem Cells: A Systematic Review. Curr Stem Cell Res. Ther. 2017, 12, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Ferrero, R.; Rainer, P.; Deplancke, B. Toward a Consensus View of Mammalian Adipocyte Stem and Progenitor Cell Heterogeneity. Trends Cell Biol. 2020, 30, 937–950. [Google Scholar] [CrossRef] [PubMed]
- Merrick, D.; Sakers, A.; Irgebay, Z.; Okada, C.; Calvert, C.; Morley, M.P.; Percec, I.; Seale, P. Identification of a mesenchymal progenitor cell hierarchy in adipose tissue. Science 2019, 364, eaav2501. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Kim, S.; Li, L.; Chattopadhyay, S.; Rando, T.A.; Feldman, B.J. Identification of an adipose tissue-resident pro-preadipocyte population. Cell Rep. 2023, 42, 112440. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Liao, K. Analysis of gene expression profile during 3T3-L1 preadipocyte differentiation. Gene 2000, 251, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Burton, G.R.; Nagarajan, R.; Peterson, C.A.; McGehee, R.E., Jr. Microarray analysis of differentiation-specific gene expression during 3T3-L1 adipogenesis. Gene 2004, 329, 167–185. [Google Scholar] [CrossRef] [PubMed]
- Green, H.; Meuth, M. An established pre-adipose cell line and its differentiation in culture. Cell 1974, 3, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Ojeda, F.J.; Rupérez, A.I.; Gomez-Llorente, C.; Gil, A.; Aguilera, C.M. Cell Models and Their Application for Studying Adipogenic Differentiation in Relation to Obesity: A Review. Int. J. Mol. Sci. 2016, 17, 1040. [Google Scholar] [CrossRef] [PubMed]
- Hudak, C.S.; Sul, H.S. Pref-1, a gatekeeper of adipogenesis. Front. Endocrinol. 2013, 4, 79. [Google Scholar] [CrossRef] [PubMed]
- Smas, C.M.; Sul, H.S. Pref-1, a protein containing EGF-like repeats, inhibits adipocyte differentiation. Cell 1993, 73, 725–734. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Kim, K.A.; Kim, J.H.; Sul, H.S. Pref-1, a preadipocyte secreted factor that inhibits adipogenesis. J. Nutr. 2006, 136, 2953–2956. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.A.; Kim, J.H.; Wang, Y.; Sul, H.S. Pref-1 (preadipocyte factor 1) activates the MEK/extracellular signal-regulated kinase pathway to inhibit adipocyte differentiation. Mol. Cell. Biol. 2007, 27, 2294–2308. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sul, H.S. Pref-1 regulates mesenchymal cell commitment and differentiation through Sox9. Cell Metab. 2009, 9, 287–302. [Google Scholar] [CrossRef]
- Hudak, C.S.; Gulyaeva, O.; Wang, Y.; Park, S.M.; Lee, L.; Kang, C.; Sul, H.S. Pref-1 marks very early mesenchymal precursors required for adipose tissue development and expansion. Cell Rep. 2014, 8, 678–687. [Google Scholar] [CrossRef] [PubMed]
- Kilroy, G.; Burk, D.H.; Floyd, Z.E. Siah2 Protein Mediates Early Events in Commitment to an Adipogenic Pathway*. J. Biol. Chem. 2016, 291, 27289–27297. [Google Scholar] [CrossRef] [PubMed]
- Dang, T.N.; Taylor, J.L.; Kilroy, G.; Yu, Y.; Burk, D.H.; Floyd, Z.E. SIAH2 is Expressed in Adipocyte Precursor Cells and Interacts with EBF1 and ZFP521 to Promote Adipogenesis. Obesity 2021, 29, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Cristancho, A.G.; Schupp, M.; Lefterova, M.I.; Cao, S.; Cohen, D.M.; Chen, C.S.; Steger, D.J.; Lazar, M.A. Repressor transcription factor 7-like 1 promotes adipogenic competency in precursor cells. Proc. Natl. Acad. Sci. USA 2011, 108, 16271–16276. [Google Scholar] [CrossRef] [PubMed]
- Scimè, A.; Grenier, G.; Huh, M.S.; Gillespie, M.A.; Bevilacqua, L.; Harper, M.E.; Rudnicki, M.A. Rb and p107 regulate preadipocyte differentiation into white versus brown fat through repression of PGC-1alpha. Cell Metab. 2005, 2, 283–295. [Google Scholar] [CrossRef] [PubMed]
- Calo, E.; Quintero-Estades, J.A.; Danielian, P.S.; Nedelcu, S.; Berman, S.D.; Lees, J.A. Rb regulates fate choice and lineage commitment in vivo. Nature 2010, 466, 1110–1114. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Zhou, Y.; Yang, Y.; Peng, J.; Song, T.; Xu, T.; Wei, H.; Jiang, S.; Peng, J. Identification of zinc finger protein Bcl6 as a novel regulator of early adipose commitment. Open Biol. 2016, 6, 160065. [Google Scholar] [CrossRef] [PubMed]
- Ireland, M.; Al-Hasan, M.; Craft, J.; Graham, A.; Bartholomew, C. EVI1 Mediated Stimulation of 3T3-L1 Preadipocyte Differentiation Is CtBP Dependent. Yangtze Med. 2017, 1, 133–147. [Google Scholar] [CrossRef][Green Version]
- An, Q.; Wu, D.; Ma, Y.; Zhou, B.; Liu, Q. Suppression of Evi1 promotes the osteogenic differentiation and inhibits the adipogenic differentiation of bone marrow-derived mesenchymal stem cells in vitro. Int. J. Mol. Med. 2015, 36, 1615–1622. [Google Scholar] [CrossRef]
- Ishibashi, J.; Firtina, Z.; Rajakumari, S.; Wood, K.H.; Conroe, H.M.; Steger, D.J.; Seale, P. An Evi1-C/EBPβ complex controls peroxisome proliferator-activated receptor γ2 gene expression to initiate white fat cell differentiation. Mol. Cell. Biol. 2012, 32, 2289–2299. [Google Scholar] [CrossRef] [PubMed]
- Xiang, H.; Zhong, Z.-X.; Peng, Y.-D.; Jiang, S.-W. The Emerging Role of Zfp217 in Adipogenesis. Int. J. Mol. Sci. 2017, 18, 1367. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Zhao, Y.; Wu, R.; Jiang, Q.; Cai, M.; Bi, Z.; Liu, Y.; Yao, Y.; Feng, J.; Wang, Y.; et al. ZFP217 regulates adipogenesis by controlling mitotic clonal expansion in a METTL3-m(6)A dependent manner. RNA Biol. 2019, 16, 1785–1793. [Google Scholar] [CrossRef] [PubMed]
- Song, T.; Yang, Y.; Wei, H.; Xie, X.; Lu, J.; Zeng, Q.; Peng, J.; Zhou, Y.; Jiang, S.; Peng, J. Zfp217 mediates m6A mRNA methylation to orchestrate transcriptional and post-transcriptional regulation to promote adipogenic differentiation. Nucleic Acids Res. 2019, 47, 6130–6144. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, R.; Tomaru, Y.; de Hoon, M.; Suzuki, H.; Hayashizaki, Y.; Shin, J.W. Identification of ZNF395 as a novel modulator of adipogenesis. Exp. Cell Res. 2013, 319, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.K.; Arany, Z.; Seale, P.; Mepani, R.J.; Ye, L.; Conroe, H.M.; Roby, Y.A.; Kulaga, H.; Reed, R.R.; Spiegelman, B.M. Transcriptional control of preadipocyte determination by Zfp423. Nature 2010, 464, 619–623. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.K.; Mepani, R.J.; Kleiner, S.; Lo, J.C.; Khandekar, M.J.; Cohen, P.; Frontini, A.; Bhowmick, D.C.; Ye, L.; Cinti, S.; et al. Zfp423 expression identifies committed preadipocytes and localizes to adipose endothelial and perivascular cells. Cell Metab. 2012, 15, 230–239. [Google Scholar] [CrossRef]
- Quach, J.M.; Walker, E.C.; Allan, E.; Solano, M.; Yokoyama, A.; Kato, S.; Sims, N.A.; Gillespie, M.T.; Martin, T.J. Zinc finger protein 467 is a novel regulator of osteoblast and adipocyte commitment. J. Biol. Chem. 2011, 286, 4186–4198. [Google Scholar] [CrossRef] [PubMed]
- Le, P.T.; Liu, H.; Alabdulaaly, L.; Vegting, Y.; Calle, I.L.; Gori, F.; Lanske, B.; Baron, R.; Rosen, C.J. The role of Zfp467 in mediating the pro-osteogenic and anti-adipogenic effects on bone and bone marrow niche. Bone 2021, 144, 115832. [Google Scholar] [CrossRef] [PubMed]
- Addison, W.N.; Fu, M.M.; Yang, H.X.; Lin, Z.; Nagano, K.; Gori, F.; Baron, R. Direct transcriptional repression of Zfp423 by Zfp521 mediates a bone morphogenic protein-dependent osteoblast versus adipocyte lineage commitment switch. Mol. Cell. Biol. 2014, 34, 3076–3085. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Akerblad, P.; Kiviranta, R.; Gupta, R.K.; Kajimura, S.; Griffin, M.J.; Min, J.; Baron, R.; Rosen, E.D. Regulation of early adipose commitment by Zfp521. PLoS Biol. 2012, 10, e1001433. [Google Scholar] [CrossRef] [PubMed]
- Gubelmann, C.; Schwalie, P.C.; Raghav, S.K.; Röder, E.; Delessa, T.; Kiehlmann, E.; Waszak, S.M.; Corsinotti, A.; Udin, G.; Holcombe, W.; et al. Identification of the transcription factor ZEB1 as a central component of the adipogenic gene regulatory network. eLife 2014, 3, e03346. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Schill, R.L.; O’Donnell, M.; Xu, K.; Bagchi, D.P.; MacDougald, O.A.; Koenig, R.J.; Xu, B. The transcription factor NKX1-2 promotes adipogenesis and may contribute to a balance between adipocyte and osteoblast differentiation. J. Biol. Chem. 2019, 294, 18408–18420. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Zhang, L.; Zhou, X.; Jiang, Z.; Hausman, G.; Zan, L.; Dodson, M. Emerging roles of zinc finger proteins in regulating adipogenesis. Cell. Mol. Life Sci. 2013, 70, 4569–4584. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Liu, S.; He, Z.; Luo, E.; Liu, H. The role of zinc finger proteins in the fate determination of mesenchymal stem cells during osteogenic and adipogenic differentiation. Int. J. Biochem. Cell Biol. 2024, 167, 106507. [Google Scholar] [CrossRef] [PubMed]
- Cristancho, A.G.; Lazar, M.A. Forming functional fat: A growing understanding of adipocyte differentiation. Nat. Rev. Mol. Cell Biol. 2011, 12, 722–734. [Google Scholar] [CrossRef] [PubMed]
- Saykally, J.N.; Dogan, S.; Cleary, M.P.; Sanders, M.M. The ZEB1 transcription factor is a novel repressor of adiposity in female mice. PLoS ONE 2009, 4, e8460. [Google Scholar] [CrossRef] [PubMed]
- Higashi, Y.; Moribe, H.; Takagi, T.; Sekido, R.; Kawakami, K.; Kikutani, H.; Kondoh, H. Impairment of T cell development in deltaEF1 mutant mice. J. Exp. Med. 1997, 185, 1467–1479. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.Q.; Otto, T.C.; Lane, M.D. Commitment of C3H10T1/2 pluripotent stem cells to the adipocyte lineage. Proc. Natl. Acad. Sci. USA 2004, 101, 9607–9611. [Google Scholar] [CrossRef] [PubMed]
- Hammarstedt, A.; Hedjazifar, S.; Jenndahl, L.; Gogg, S.; Grunberg, J.; Gustafson, B.; Klimcakova, E.; Stich, V.; Langin, D.; Laakso, M.; et al. WISP2 regulates preadipocyte commitment and PPARgamma activation by BMP4. Proc. Natl. Acad. Sci. USA 2013, 110, 2563–2568. [Google Scholar] [CrossRef]
- You, L.; Chen, L.; Pan, L.; Gu, W.S.; Chen, J.Y. Zinc finger protein 467 regulates Wnt signaling by modulating the expression of sclerostin in adipose derived stem cells. Biochem. Biophys. Res. Commun. 2015, 456, 598–604. [Google Scholar] [CrossRef] [PubMed]
- You, L.; Pan, L.; Chen, L.; Chen, J.Y.; Zhang, X.; Lv, Z.; Fu, D. Suppression of zinc finger protein 467 alleviates osteoporosis through promoting differentiation of adipose derived stem cells to osteoblasts. J. Transl. Med. 2012, 10, 11. [Google Scholar] [CrossRef] [PubMed]
- Suo, J.; Zou, S.; Wang, J.; Han, Y.; Zhang, L.; Lv, C.; Jiang, B.; Ren, Q.; Chen, L.; Yang, L.; et al. The RNA-binding protein Musashi2 governs osteoblast-adipocyte lineage commitment by suppressing PPARgamma signaling. Bone Res. 2022, 10, 31. [Google Scholar] [CrossRef] [PubMed]
- Poissonnet, C.M.; Burdi, A.R.; Garn, S.M. The chronology of adipose tissue appearance and distribution in the human fetus. Early Hum. Dev. 1984, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Orsso, C.E.; Colin-Ramirez, E.; Field, C.J.; Madsen, K.L.; Prado, C.M.; Haqq, A.M. Adipose Tissue Development and Expansion from the Womb to Adolescence: An Overview. Nutrients 2020, 12, 2735. [Google Scholar] [CrossRef] [PubMed]
- Spalding, K.L.; Arner, E.; Westermark, P.O.; Bernard, S.; Buchholz, B.A.; Bergmann, O.; Blomqvist, L.; Hoffstedt, J.; Naslund, E.; Britton, T.; et al. Dynamics of fat cell turnover in humans. Nature 2008, 453, 783–787. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Berry, D.C.; Tang, W.; Graff, J.M. Independent stem cell lineages regulate adipose organogenesis and adipose homeostasis. Cell Rep. 2014, 9, 1007–1022. [Google Scholar] [CrossRef] [PubMed]
- Hong, K.Y.; Bae, H.; Park, I.; Park, D.Y.; Kim, K.H.; Kubota, Y.; Cho, E.S.; Kim, H.; Adams, R.H.; Yoo, O.J.; et al. Perilipin+ embryonic preadipocytes actively proliferate along growing vasculatures for adipose expansion. Development 2015, 142, 2623–2632. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.; Pang, Y.; Park, J.; Liu, L.; Lukas, B.E.; Kim, S.H.; Kim, K.W.; Xu, P.; Berry, D.C.; Jiang, Y. Dynamic control of adipose tissue development and adult tissue homeostasis by platelet-derived growth factor receptor alpha. eLife 2020, 9, e56189. [Google Scholar] [CrossRef] [PubMed]
- Vishvanath, L.; MacPherson, K.A.; Hepler, C.; Wang, Q.A.; Shao, M.; Spurgin, S.B.; Wang, M.Y.; Kusminski, C.M.; Morley, T.S.; Gupta, R.K. Pdgfrbeta+ Mural Preadipocytes Contribute to Adipocyte Hyperplasia Induced by High-Fat-Diet Feeding and Prolonged Cold Exposure in Adult Mice. Cell Metab. 2016, 23, 350–359. [Google Scholar] [CrossRef] [PubMed]
- Ambrosi, T.H.; Scialdone, A.; Graja, A.; Gohlke, S.; Jank, A.M.; Bocian, C.; Woelk, L.; Fan, H.; Logan, D.W.; Schurmann, A.; et al. Adipocyte Accumulation in the Bone Marrow during Obesity and Aging Impairs Stem Cell-Based Hematopoietic and Bone Regeneration. Cell Stem Cell 2017, 20, 771–784.e6. [Google Scholar] [CrossRef] [PubMed]
- Uezumi, A.; Fukada, S.; Yamamoto, N.; Takeda, S.; Tsuchida, K. Mesenchymal progenitors distinct from satellite cells contribute to ectopic fat cell formation in skeletal muscle. Nat. Cell Biol. 2010, 12, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Joe, A.W.; Yi, L.; Natarajan, A.; Le Grand, F.; So, L.; Wang, J.; Rudnicki, M.A.; Rossi, F.M. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat. Cell Biol. 2010, 12, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Reggio, A.; Rosina, M.; Palma, A.; Cerquone Perpetuini, A.; Petrilli, L.L.; Gargioli, C.; Fuoco, C.; Micarelli, E.; Giuliani, G.; Cerretani, M.; et al. Adipogenesis of skeletal muscle fibro/adipogenic progenitors is affected by the WNT5a/GSK3/beta-catenin axis. Cell Death Differ. 2020, 27, 2921–2941. [Google Scholar] [CrossRef] [PubMed]
- Laharrague, P.; Casteilla, L. The emergence of adipocytes. Endocr. Dev. 2010, 19, 21–30. [Google Scholar] [CrossRef]
- Ruge, T.; Hodson, L.; Cheeseman, J.; Dennis, A.L.; Fielding, B.A.; Humphreys, S.M.; Frayn, K.N.; Karpe, F. Fasted to fed trafficking of Fatty acids in human adipose tissue reveals a novel regulatory step for enhanced fat storage. J. Clin. Endocrinol. Metab. 2009, 94, 1781–1788. [Google Scholar] [CrossRef] [PubMed]
- Todorcevic, M.; Manuel, A.R.; Austen, L.; Michailidou, Z.; Hazlehurst, J.M.; Neville, M.; Stradling, J.R.; Karpe, F. Markers of adipose tissue hypoxia are elevated in subcutaneous adipose tissue of severely obese patients with obesity hypoventilation syndrome but not in the moderately obese. Int. J. Obes. 2021, 45, 1618–1622. [Google Scholar] [CrossRef] [PubMed]
- Muir, L.A.; Neeley, C.K.; Meyer, K.A.; Baker, N.A.; Brosius, A.M.; Washabaugh, A.R.; Varban, O.A.; Finks, J.F.; Zamarron, B.F.; Flesher, C.G.; et al. Adipose tissue fibrosis, hypertrophy, and hyperplasia: Correlations with diabetes in human obesity. Obesity 2016, 24, 597–605. [Google Scholar] [CrossRef]
- Chait, A.; den Hartigh, L.J. Adipose Tissue Distribution, Inflammation and Its Metabolic Consequences, Including Diabetes and Cardiovascular Disease. Front. Cardiovasc. Med. 2020, 7, 22. [Google Scholar] [CrossRef] [PubMed]
- Mittal, B. Subcutaneous adipose tissue & visceral adipose tissue. Indian J. Med. Res. 2019, 149, 571–573. [Google Scholar] [CrossRef] [PubMed]
- Arner, E.; Westermark, P.O.; Spalding, K.L.; Britton, T.; Ryden, M.; Frisen, J.; Bernard, S.; Arner, P. Adipocyte turnover: Relevance to human adipose tissue morphology. Diabetes 2010, 59, 105–109. [Google Scholar] [CrossRef] [PubMed]
- Storz, C.; Heber, S.D.; Rospleszcz, S.; Machann, J.; Sellner, S.; Nikolaou, K.; Lorbeer, R.; Gatidis, S.; Elser, S.; Peters, A.; et al. The role of visceral and subcutaneous adipose tissue measurements and their ratio by magnetic resonance imaging in subjects with prediabetes, diabetes and healthy controls from a general population without cardiovascular disease. Br. J. Radiol. 2018, 91, 20170808. [Google Scholar] [CrossRef] [PubMed]
- Spalding, K.L.; Bernard, S.; Naslund, E.; Salehpour, M.; Possnert, G.; Appelsved, L.; Fu, K.Y.; Alkass, K.; Druid, H.; Thorell, A.; et al. Impact of fat mass and distribution on lipid turnover in human adipose tissue. Nat. Commun. 2017, 8, 15253. [Google Scholar] [CrossRef] [PubMed]
- Goossens, G.H. The Metabolic Phenotype in Obesity: Fat Mass, Body Fat Distribution, and Adipose Tissue Function. Obes. Facts. 2017, 10, 207–215. [Google Scholar] [CrossRef] [PubMed]
- April-Sanders, A.K.; Rodriguez, C.J. Metabolically Healthy Obesity Redefined. JAMA Netw. Open 2021, 4, e218860. [Google Scholar] [CrossRef] [PubMed]
- Wildman, R.P.; Muntner, P.; Reynolds, K.; McGinn, A.P.; Rajpathak, S.; Wylie-Rosett, J.; Sowers, M.R. The obese without cardiometabolic risk factor clustering and the normal weight with cardiometabolic risk factor clustering: Prevalence and correlates of 2 phenotypes among the US population (NHANES 1999–2004). Arch. Intern. Med. 2008, 168, 1617–1624. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.C.; Arthur, R.; Iyengar, N.M.; Kamensky, V.; Xue, X.; Wassertheil-Smoller, S.; Allison, M.A.; Shadyab, A.H.; Wild, R.A.; Sun, Y.; et al. Association between regional body fat and cardiovascular disease risk among postmenopausal women with normal body mass index. Eur. Heart J. 2019, 40, 2849–2855. [Google Scholar] [CrossRef] [PubMed]
- Longo, M.; Zatterale, F.; Naderi, J.; Parrillo, L.; Formisano, P.; Raciti, G.A.; Beguinot, F.; Miele, C. Adipose Tissue Dysfunction as Determinant of Obesity-Associated Metabolic Complications. Int. J. Mol. Sci. 2019, 20, 2358. [Google Scholar] [CrossRef]
- Weyer, C.; Foley, J.E.; Bogardus, C.; Tataranni, P.A.; Pratley, R.E. Enlarged subcutaneous abdominal adipocyte size, but not obesity itself, predicts type II diabetes independent of insulin resistance. Diabetologia 2000, 43, 1498–1506. [Google Scholar] [CrossRef]
- Naryzhnaya, N.V.; Koshelskaya, O.A.; Kologrivova, I.V.; Kharitonova, O.A.; Evtushenko, V.V.; Boshchenko, A.A. Hypertrophy and Insulin Resistance of Epicardial Adipose Tissue Adipocytes: Association with the Coronary Artery Disease Severity. Biomedicines 2021, 9, 64. [Google Scholar] [CrossRef] [PubMed]
- Honecker, J.; Ruschke, S.; Seeliger, C.; Laber, S.; Strobel, S.; Proll, P.; Nellaker, C.; Lindgren, C.M.; Kulozik, U.; Ecker, J.; et al. Transcriptome and fatty-acid signatures of adipocyte hypertrophy and its non-invasive MR-based characterization in human adipose tissue. EBioMedicine 2022, 79, 104020. [Google Scholar] [CrossRef] [PubMed]
- Hammarstedt, A.; Gogg, S.; Hedjazifar, S.; Nerstedt, A.; Smith, U. Impaired Adipogenesis and Dysfunctional Adipose Tissue in Human Hypertrophic Obesity. Physiol. Rev. 2018, 98, 1911–1941. [Google Scholar] [CrossRef] [PubMed]
- Ghaben, A.L.; Scherer, P.E. Adipogenesis and metabolic health. Nat. Rev. Mol. Cell Biol. 2019, 20, 242–258. [Google Scholar] [CrossRef]
- Muller, C.; Zidek, L.M.; Eichwald, S.; Kortman, G.; Koster, M.H.; Calkhoven, C.F. Enhanced C/EBPbeta function promotes hypertrophic versus hyperplastic fat tissue growth and prevents steatosis in response to high-fat diet feeding. eLife 2022, 11, e62625. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Li, X.; Tang, Q.Q. Transcriptional regulation of adipocyte differentiation: A central role for CCAAT/enhancer-binding protein (C/EBP) beta. J. Biol. Chem. 2015, 290, 755–761. [Google Scholar] [CrossRef] [PubMed]
- Gustafson, B.; Nerstedt, A.; Smith, U. Reduced subcutaneous adipogenesis in human hypertrophic obesity is linked to senescent precursor cells. Nat. Commun. 2019, 10, 2757. [Google Scholar] [CrossRef] [PubMed]
- Krotkiewski, M.; Bjorntorp, P.; Sjostrom, L.; Smith, U. Impact of obesity on metabolism in men and women. Importance of regional adipose tissue distribution. J. Clin. Investig. 1983, 72, 1150–1162. [Google Scholar] [CrossRef] [PubMed]
- Faust, I.M.; Johnson, P.R.; Stern, J.S.; Hirsch, J. Diet-induced adipocyte number increase in adult rats: A new model of obesity. Am. J. Physiol. 1978, 235, E279–E286. [Google Scholar] [CrossRef] [PubMed]
- Jackman, M.R.; Steig, A.; Higgins, J.A.; Johnson, G.C.; Fleming-Elder, B.K.; Bessesen, D.H.; MacLean, P.S. Weight regain after sustained weight reduction is accompanied by suppressed oxidation of dietary fat and adipocyte hyperplasia. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2008, 294, R1117–R1129. [Google Scholar] [CrossRef] [PubMed]
- MacLean, P.S.; Higgins, J.A.; Jackman, M.R.; Johnson, G.C.; Fleming-Elder, B.K.; Wyatt, H.R.; Melanson, E.L.; Hill, J.O. Peripheral metabolic responses to prolonged weight reduction that promote rapid, efficient regain in obesity-prone rats. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2006, 290, R1577–R1588. [Google Scholar] [CrossRef] [PubMed]
- MacLean, P.S.; Higgins, J.A.; Giles, E.D.; Sherk, V.D.; Jackman, M.R. The role for adipose tissue in weight regain after weight loss. Obes. Rev. 2015, 16 (Suppl. S1), 45–54. [Google Scholar] [CrossRef] [PubMed]
- Puhl, R.M.; Heuer, C.A. Obesity stigma: Important considerations for public health. Am. J. Public Health 2010, 100, 1019–1028. [Google Scholar] [CrossRef] [PubMed]
- Sarma, S.; Sockalingam, S.; Dash, S. Obesity as a multisystem disease: Trends in obesity rates and obesity-related complications. Diabetes Obes. Metab. 2021, 23 (Suppl. S1), 3–16. [Google Scholar] [CrossRef] [PubMed]
- Gao, D.; Madi, M.; Ding, C.; Fok, M.; Steele, T.; Ford, C.; Hunter, L.; Bing, C. Interleukin-1beta mediates macrophage-induced impairment of insulin signaling in human primary adipocytes. Am. J. Physiol. Endocrinol. Metab. 2014, 307, E289–E304. [Google Scholar] [CrossRef] [PubMed]
- Lagathu, C.; Yvan-Charvet, L.; Bastard, J.P.; Maachi, M.; Quignard-Boulange, A.; Capeau, J.; Caron, M. Long-term treatment with interleukin-1beta induces insulin resistance in murine and human adipocytes. Diabetologia 2006, 49, 2162–2173. [Google Scholar] [CrossRef] [PubMed]
- van Asseldonk, E.J.; Stienstra, R.; Koenen, T.B.; van Tits, L.J.; Joosten, L.A.; Tack, C.J.; Netea, M.G. The effect of the interleukin-1 cytokine family members IL-1F6 and IL-1F8 on adipocyte differentiation. Obesity 2010, 18, 2234–2236. [Google Scholar] [CrossRef] [PubMed]
- Sabaratnam, R.; Svenningsen, P. Adipocyte-Endothelium Crosstalk in Obesity. Front. Endocrinol. 2021, 12, 681290. [Google Scholar] [CrossRef] [PubMed]
- Hammel, J.H.; Bellas, E. Endothelial cell crosstalk improves browning but hinders white adipocyte maturation in 3D engineered adipose tissue. Integr. Biol. 2020, 12, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Schwalie, P.C.; Dong, H.; Zachara, M.; Russeil, J.; Alpern, D.; Akchiche, N.; Caprara, C.; Sun, W.; Schlaudraff, K.U.; Soldati, G.; et al. A stromal cell population that inhibits adipogenesis in mammalian fat depots. Nature 2018, 559, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.Y.; Hu, L.L.; Song, T.J.; Li, X.; He, Q.; Sun, X.; Li, Y.M.; Lu, H.J.; Yang, P.Y.; Tang, Q.Q. Involvement of cytoskeleton-associated proteins in the commitment of C3H10T1/2 pluripotent stem cells to adipocyte lineage induced by BMP2/4. Mol. Cell. Proteom. 2011, 10, M110.002691. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Sobue, T.; Esliger, A.; Kronenberg, M.S.; Coffin, J.D.; Doetschman, T.; Hurley, M.M. Disruption of the Fgf2 gene activates the adipogenic and suppresses the osteogenic program in mesenchymal marrow stromal stem cells. Bone 2010, 47, 360–370. [Google Scholar] [CrossRef] [PubMed]
- Dali-Youcef, N.; Mataki, C.; Coste, A.; Messaddeq, N.; Giroud, S.; Blanc, S.; Koehl, C.; Champy, M.F.; Chambon, P.; Fajas, L.; et al. Adipose tissue-specific inactivation of the retinoblastoma protein protects against diabesity because of increased energy expenditure. Proc. Natl. Acad. Sci. USA 2007, 104, 10703–10708. [Google Scholar] [CrossRef] [PubMed]
- Green, H.; Kehinde, O. An established preadipose cell line and its differentiation in culture. II. Factors affecting the adipose conversion. Cell 1975, 5, 19–27. [Google Scholar] [CrossRef]
- Farmer, S.R. Transcriptional control of adipocyte formation. Cell Metab. 2006, 4, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Cowherd, R.M.; Lyle, R.E.; McGehee, R.E., Jr. Molecular regulation of adipocyte differentiation. Semin. Cell Dev. Biol. 1999, 10, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Gregoire, F.M.; Smas, C.M.; Sul, H.S. Understanding adipocyte differentiation. Physiol. Rev. 1998, 78, 783–809. [Google Scholar] [CrossRef]
- Jeon, Y.; Lee, M.S.; Cheon, Y.P. Decreased contact inhibition in mouse adipose mesenchymal stem cells. Dev. Reprod. 2012, 16, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Reichert, M.; Eick, D. Analysis of cell cycle arrest in adipocyte differentiation. Oncogene 1999, 18, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.-Q.; Otto, T.C.; Lane, M.D. Mitotic clonal expansion: A synchronous process required for adipogenesis. Proc. Natl. Acad. Sci. USA 2003, 100, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Yeh, W.-C.; Bierer, B.E.; McKnight, S.L. Rapamycin inhibits clonal expansion and adipogenic differentiation of 3T3-L1 cells. Proc. Natl. Acad. Sci. USA 1995, 92, 11086–11090. [Google Scholar] [CrossRef] [PubMed]
- Patel, Y.M.; Lane, M.D. Mitotic clonal expansion during preadipocyte differentiation: Calpain-mediated turnover of p27. J. Biol. Chem. 2000, 275, 17653–17660. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Z.; Wei, Y.; Chen, N.; Jiang, M.; Wu, J.; Liao, K. DNA synthesis and mitotic clonal expansion is not a required step for 3T3-L1 preadipocyte differentiation into adipocytes. J. Biol. Chem. 2001, 276, 11988–11995. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Guan, Y.; MacNicol, M.C.; MacNicol, A.M.; McGehee, R.E. Early expression of p107 is associated with 3T3-L1 adipocyte differentiation. Mol. Cell. Endocrinol. 2002, 194, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Kreis, N.N.; Friemel, A.; Zimmer, B.; Roth, S.; Rieger, M.A.; Rolle, U.; Louwen, F.; Yuan, J. Mitotic p21Cip1/CDKN1A is regulated by cyclin-dependent kinase 1 phosphorylation. Oncotarget 2016, 7, 50215–50228. [Google Scholar] [CrossRef] [PubMed]
- Morrison, R.F.; Farmer, S.R. Role of PPARgamma in regulating a cascade expression of cyclin-dependent kinase inhibitors, p18(INK4c) and p21(Waf1/Cip1), during adipogenesis. J. Biol. Chem. 1999, 274, 17088–17097. [Google Scholar] [CrossRef] [PubMed]
- Classon, M.; Kennedy, B.K.; Mulloy, R.; Harlow, E. Opposing roles of pRB and p107 in adipocyte differentiation. Proc. Natl. Acad. Sci. USA 2000, 97, 10826–10831. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Mejia, I.C.; Castillo-Armengol, J.; Lagarrigue, S.; Fajas, L. Role of cell cycle regulators in adipose tissue and whole body energy homeostasis. Cell Mol. Life Sci. 2018, 75, 975–987. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.; Kaldis, P. Cdks, cyclins and CKIs: Roles beyond cell cycle regulation. Development 2013, 140, 3079–3093. [Google Scholar] [CrossRef] [PubMed]
- Fajas, L.; Landsberg, R.L.; Huss-Garcia, Y.; Sardet, C.; Lees, J.A.; Auwerx, J. E2Fs regulate adipocyte differentiation. Dev. Cell 2002, 3, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Hao, W.; Xiao, C.; Wang, R.; Xu, X.; Lu, H.; Chen, W.; Deng, C.X. SIRT6 Is Essential for Adipocyte Differentiation by Regulating Mitotic Clonal Expansion. Cell Rep. 2017, 18, 3155–3166. [Google Scholar] [CrossRef] [PubMed]
- Molonia, M.S.; Salamone, F.L.; Muscara, C.; Costa, G.; Vento, G.; Saija, A.; Speciale, A.; Cimino, F. Regulation of mitotic clonal expansion and thermogenic pathway are involved in the antiadipogenic effects of cyanidin-3-O-glucoside. Front. Pharmacol. 2023, 14, 1225586. [Google Scholar] [CrossRef] [PubMed]
- Lefterova, M.I.; Zhang, Y.; Steger, D.J.; Schupp, M.; Schug, J.; Cristancho, A.; Feng, D.; Zhuo, D.; Stoeckert, C.J., Jr.; Liu, X.S.; et al. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 2008, 22, 2941–2952. [Google Scholar] [CrossRef]
- Tanaka, T.; Yoshida, N.; Kishimoto, T.; Akira, S. Defective adipocyte differentiation in mice lacking the C/EBPbeta and/or C/EBPdelta gene. EMBO J. 1997, 16, 7432–7443. [Google Scholar] [CrossRef] [PubMed]
- Hishida, T.; Nishizuka, M.; Osada, S.; Imagawa, M. The role of C/EBPdelta in the early stages of adipogenesis. Biochimie 2009, 91, 654–657. [Google Scholar] [CrossRef] [PubMed]
- Hishida, T.; Eguchi, T.; Osada, S.; Nishizuka, M.; Imagawa, M. A novel gene, fad49, plays a crucial role in the immediate early stage of adipocyte differentiation via involvement in mitotic clonal expansion. FEBS J. 2008, 275, 5576–5588. [Google Scholar] [CrossRef] [PubMed]
- Merkestein, M.; Laber, S.; McMurray, F.; Andrew, D.; Sachse, G.; Sanderson, J.; Li, M.; Usher, S.; Sellayah, D.; Ashcroft, F.M.; et al. FTO influences adipogenesis by regulating mitotic clonal expansion. Nat. Commun. 2015, 6, 6792. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Guo, L.; Xie, L.Q.; Zhang, Y.Y.; Liu, X.H.; Zhang, Y.; Zhu, H.; Yang, P.Y.; Lu, H.J.; Tang, Q.Q. Proteome profiling of mitotic clonal expansion during 3T3-L1 adipocyte differentiation using iTRAQ-2DLC-MS/MS. J. Proteome Res. 2014, 13, 1307–1314. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Zhang, J.; Liang, F.; Zhu, Q.; Cai, S.; Tong, X.; He, Z.; Liu, X.; Chen, Y.; Mo, D. HMGB2 orchestrates mitotic clonal expansion by binding to the promoter of C/EBPβ to facilitate adipogenesis. Cell Death Dis. 2021, 12, 666. [Google Scholar] [CrossRef] [PubMed]
- Akerblad, P.; Lind, U.; Liberg, D.; Bamberg, K.; Sigvardsson, M. Early B-cell factor (O/E-1) is a promoter of adipogenesis and involved in control of genes important for terminal adipocyte differentiation. Mol. Cell. Biol. 2002, 22, 8015–8025. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, M.A.; Akerblad, P.; Sigvardsson, M.; Rosen, E.D. Critical role for Ebf1 and Ebf2 in the adipogenic transcriptional cascade. Mol. Cell. Biol. 2007, 27, 743–757. [Google Scholar] [CrossRef]
- Amri, E.Z.; Bertrand, B.; Ailhaud, G.; Grimaldi, P. Regulation of adipose cell differentiation. I. Fatty acids are inducers of the aP2 gene expression. J. Lipid Res. 1991, 32, 1449–1456. [Google Scholar] [CrossRef]
- Herrera, R.; Ro, H.S.; Robinson, G.S.; Xanthopoulos, K.G.; Spiegelman, B.M. A direct role for C/EBP and the AP-I-binding site in gene expression linked to adipocyte differentiation. Mol. Cell. Biol. 1989, 9, 5331–5339. [Google Scholar] [CrossRef] [PubMed]
- Marion-Letellier, R.; Savoye, G.; Ghosh, S. Fatty acids, eicosanoids and PPAR gamma. Eur. J. Pharmacol. 2016, 785, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Tontonoz, P.; Hu, E.; Spiegelman, B.M. Stimulation of adipogenesis in fibroblasts by PPARγ2, a lipid-activated transcription factor. Cell 1994, 79, 1147–1156. [Google Scholar] [CrossRef] [PubMed]
- Hu, E.; Tontonoz, P.; Spiegelman, B.M. Transdifferentiation of myoblasts by the adipogenic transcription factors PPAR gamma and C/EBP alpha. Proc. Natl. Acad. Sci. USA 1995, 92, 9856–9860. [Google Scholar] [CrossRef] [PubMed]
- Ren, D.; Collingwood, T.N.; Rebar, E.J.; Wolffe, A.P.; Camp, H.S. PPARγ knockdown by engineered transcription factors: Exogenous PPARγ2 but not PPARγ1 reactivates adipogenesis. Genes Dev. 2002, 16, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Luo, H.; Xu, Z.; Yang, Z.; Du, G.; Zhang, Y.; Yu, L.; Hu, K.; Zhu, W.; Tong, Q.; et al. Bavachinin, as a novel natural pan-PPAR agonist, exhibits unique synergistic effects with synthetic PPAR-γ and PPAR-α agonists on carbohydrate and lipid metabolism in db/db and diet-induced obese mice. Diabetologia 2016, 59, 1276–1286. [Google Scholar] [CrossRef] [PubMed]
- Mueller, E.; Drori, S.; Aiyer, A.; Yie, J.; Sarraf, P.; Chen, H.; Hauser, S.; Rosen, E.D.; Ge, K.; Roeder, R.G.; et al. Genetic analysis of adipogenesis through peroxisome proliferator-activated receptor gamma isoforms. J. Biol. Chem. 2002, 277, 41925–41930. [Google Scholar] [CrossRef] [PubMed]
- Ge, K.; Guermah, M.; Yuan, C.X.; Ito, M.; Wallberg, A.E.; Spiegelman, B.M.; Roeder, R.G. Transcription coactivator TRAP220 is required for PPARγ2-stimulated adipogenesis. Nature 2002, 417, 563–567. [Google Scholar] [CrossRef] [PubMed]
- Barak, Y.; Nelson, M.C.; Ong, E.S.; Jones, Y.Z.; Ruiz-Lozano, P.; Chien, K.R.; Koder, A.; Evans, R.M. PPARγ is required for placental, cardiac, and adipose tissue development. Mol. Cell 1999, 4, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Rosen, E.D.; Sarraf, P.; Troy, A.E.; Bradwin, G.; Moore, K.; Milstone, D.S.; Spiegelman, B.M.; Mortensen, R.M. PPARγ is required for the differentiation of adipose tissue in vivo and in vitro. Mol. Cell 1999, 4, 611–617. [Google Scholar] [CrossRef] [PubMed]
- Miles, P.D.; Barak, Y.; Evans, R.M. Metabolic characterization of mice heterozygous for PPARγ deficiency. Diabetes 1999, 48, A68. [Google Scholar]
- Ma, X.; Wang, D.; Zhao, W.; Xu, L. Deciphering the Roles of PPARγ in Adipocytes via Dynamic Change of Transcription Complex. Front. Endocrinol. 2018, 9, 473. [Google Scholar] [CrossRef] [PubMed]
- Miles, P.D.; Barak, Y.; He, W.; Evans, R.M.; Olefsky, J.M. Improved insulin-sensitivity in mice heterozygous for PPARγ deficiency. J. Clin. Investig. 2000, 105, 287–292. [Google Scholar] [CrossRef] [PubMed]
- Miles, P.D.; Barak, Y.; Evans, R.M.; Olefsky, J.M. Effect of heterozygous PPARγdeficiency and TZD treatment on insulin resistance associated with age and high-fat feeding. Am. J. Physiol. Endocrinol. Metab. 2003, 284, E618–E626. [Google Scholar] [CrossRef]
- Koutnikova, H.; Cock, T.A.; Watanabe, M.; Houten, S.M.; Champy, M.F.; Dierich, A.; Auwerx, J. Compensation by the muscle limits the metabolic consequences of lipodystrophy in PPARγ hypomorphic mice. Proc. Natl. Acad. Sci. USA 2003, 100, 14457–14462. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Barak, Y.; Hevener, A.; Olson, P.; Liao, D.; Le, J.; Nelson, M.; Ong, E.; Olefsky, J.M.; Evans, R.M. Adipose-specific peroxisome proliferator-activated receptor γ knockout causes insulin resistance in fat and liver but not in muscle. Proc. Natl. Acad. Sci. USA 2003, 100, 15712–15717. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.R.; Barrick, C.; Kim, K.A.; Lindner, J.; Blondeau, B.; Fujimoto, Y.; Shiota, M.; Kesterson, R.A.; Kahn, B.B.; Magnuson, M.A. Deletion of PPARγ in adipose tissues of mice protects against high fat diet-induced obesity and insulin resistance. Proc. Natl. Acad. Sci. USA 2005, 102, 6207–6212. [Google Scholar] [CrossRef] [PubMed]
- Duan, S.Z.; Ivashchenko, C.Y.; Whitesall, S.E.; D’Alecy, L.G.; Duquaine, D.C.; Brosius, F.C., 3rd; Gonzalez, F.J.; Vinson, C.; Pierre, M.A.; Milstone, D.S.; et al. Hypotension, lipodystrophy, and insulin resistance in generalized PPARγ-deficient mice rescued from embryonic lethality. J. Clin. Investig. 2007, 117, 812–822. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Mullican, S.E.; DiSpirito, J.R.; Peed, L.C.; Lazar, M.A. Lipoatrophy and severe metabolic disturbance in mice with fat-specific deletion of PPARγ. Proc. Natl. Acad. Sci. USA 2013, 110, 18656–18661. [Google Scholar] [CrossRef] [PubMed]
- Barroso, I.; Gurnell, M.; Crowley, V.E.; Agostini, M.; Schwabe, J.W.; Soos, M.A.; Maslen, G.L.; Williams, T.D.; Lewis, H.; Schafer, A.J.; et al. Dominant negative mutations in human PPARγ associated with severe insulin resistance, diabetes mellitus and hypertension. Nature 1999, 402, 880–883. [Google Scholar] [CrossRef] [PubMed]
- Monajemi, H.; Zhang, L.; Li, G.; Jeninga, E.H.; Cao, H.; Maas, M.; Brouwer, C.B.; Kalkhoven, E.; Stroes, E.; Hegele, R.A.; et al. Familial partial lipodystrophy phenotype resulting from a single-base mutation in deoxyribonucleic acid-binding domain of peroxisome proliferator-activated receptor-gamma. J. Clin. Endocrinol. Metab. 2007, 92, 1606–1612. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Umek, R.M.; McKnight, S.L. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev. 1991, 5, 1538–1552. [Google Scholar] [CrossRef]
- Freytag, S.O.; Geddes, T.J. Reciprocal regulation of adipogenesis by Myc and C/EBP alpha. Science 1992, 256, 379–382. [Google Scholar] [CrossRef] [PubMed]
- Heath, V.J.; Gillespie, D.A.; Crouch, D.H. Inhibition of the terminal stages of adipocyte differentiation by cMyc. Exp. Cell Res. 2000, 254, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.T.; Lane, M.D. CCAAT/enhancer binding protein alpha is sufficient to initiate the 3T3-L1 adipocyte differentiation program. Proc. Natl. Acad. Sci. USA 1994, 91, 8757–8761. [Google Scholar] [CrossRef] [PubMed]
- Freytag, S.O.; Paielli, D.L.; Gilbert, J.D. Ectopic expression of the CCAAT/enhancer-binding protein alpha promotes the adipogenic program in a variety of mouse fibroblastic cells. Genes Dev. 1994, 8, 1654–1663. [Google Scholar] [CrossRef] [PubMed]
- Lane, M.D.; Lin, F.T.; MacDougald, O.A.; Vasseur-Cognet, M. Control of adipocyte differentiation by CCAAT/enhancer binding protein alpha (C/EBP alpha). Int. J. Obes. Relat. Metab. Disord. 1996, 20 (Suppl. S3), S91–S96. [Google Scholar] [PubMed]
- Wu, Z.; Rosen, E.D.; Brun, R.; Hauser, S.; Adelmant, G.; Troy, A.E.; McKeon, C.; Darlington, G.J.; Spiegelman, B.M. Cross-regulation of C/EBP alpha and PPARγ controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol. Cell 1999, 3, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Rosen, E.D.; Hsu, C.H.; Wang, X.; Sakai, S.; Freeman, M.W.; Gonzalez, F.J.; Spiegelman, B.M. C/EBPalpha induces adipogenesis through PPARγ: A unified pathway. Genes Dev. 2002, 16, 22–26. [Google Scholar] [CrossRef] [PubMed]
- Madsen, M.S.; Siersbaek, R.; Boergesen, M.; Nielsen, R.; Mandrup, S. Peroxisome proliferator-activated receptor gamma and C/EBPalpha synergistically activate key metabolic adipocyte genes by assisted loading. Mol. Cell. Biol. 2014, 34, 939–954. [Google Scholar] [CrossRef] [PubMed]
- Picard, F.; Kurtev, M.; Chung, N.; Topark-Ngarm, A.; Senawong, T.; Machado De Oliveira, R.; Leid, M.; McBurney, M.W.; Guarente, L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-γ. Nature 2004, 429, 771–776. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Zhang, F.; Ge, X.; Yan, T.; Chen, X.; Shi, X.; Zhai, Q. SIRT1 improves insulin sensitivity under insulin-resistant conditions by repressing PTP1B. Cell Metab. 2007, 6, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Jin, Q.; Zhang, F.; Yan, T.; Liu, Z.; Wang, C.; Ge, X.; Zhai, Q. C/EBPalpha regulates SIRT1 expression during adipogenesis. Cell Res. 2010, 20, 470–479. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.D.; Finegold, M.J.; Bradley, A.; Ou, C.N.; Abdelsayed, S.V.; Wilde, M.D.; Taylor, L.R.; Wilson, D.R.; Darlington, G.J. Impaired energy homeostasis in C/EBP alpha knockout mice. Science 1995, 269, 1108–1112. [Google Scholar] [CrossRef] [PubMed]
- Darlington, G.J.; Ross, S.E.; MacDougald, O.A. The role of C/EBP genes in adipocyte differentiation. J. Biol. Chem. 1998, 273, 30057–30060. [Google Scholar] [CrossRef] [PubMed]
- Linhart, H.G.; Ishimura-Oka, K.; DeMayo, F.; Kibe, T.; Repka, D.; Poindexter, B.; Bick, R.J.; Darlington, G.J. C/EBPalpha is required for differentiation of white, but not brown, adipose tissue. Proc. Natl. Acad. Sci. USA 2001, 98, 12532–12537. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Croniger, C.M.; Lekstrom-Himes, J.; Zhang, P.; Fenyus, M.; Tenen, D.G.; Darlington, G.J.; Hanson, R.W. Metabolic response of mice to a postnatal ablation of CCAAT/enhancer-binding protein alpha. J. Biol. Chem. 2005, 280, 38689–38699. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, R.; Bhattacharya, P.; Gavrilova, O.; Glass, K.; Moitra, J.; Myakishev, M.; Pack, S.; Jou, W.; Feigenbaum, L.; Eckhaus, M.; et al. Suppression of the C/EBP family of transcription factors in adipose tissue causes lipodystrophy. J. Mol. Endocrinol. 2011, 46, 175–192. [Google Scholar] [CrossRef]
- Mori, T.; Sakaue, H.; Iguchi, H.; Gomi, H.; Okada, Y.; Takashima, Y.; Nakamura, K.; Nakamura, T.; Yamauchi, T.; Kubota, N.; et al. Role of Kruppel-like factor 15 (KLF15) in transcriptional regulation of adipogenesis. J. Biol. Chem. 2005, 280, 12867–12875. [Google Scholar] [CrossRef] [PubMed]
- Asada, M.; Rauch, A.; Shimizu, H.; Maruyama, H.; Miyaki, S.; Shibamori, M.; Kawasome, H.; Ishiyama, H.; Tuckermann, J.; Asahara, H. DNA binding-dependent glucocorticoid receptor activity promotes adipogenesis via Kruppel-like factor 15 gene expression. Lab. Investig. J. Tech. Methods Pathol. 2011, 91, 203–215. [Google Scholar] [CrossRef]
- Lee, D.S.; Choi, H.; Han, B.S.; Kim, W.K.; Lee, S.C.; Oh, K.J.; Bae, K.H. c-Jun regulates adipocyte differentiation via the KLF15-mediated mode. Biochem. Biophys. Res. Commun. 2016, 469, 552–558. [Google Scholar] [CrossRef] [PubMed]
- Pollak, N.M.; Hoffman, M.; Goldberg, I.J.; Drosatos, K. Kruppel-like factors: Crippling and un-crippling metabolic pathways. JACC Basic Transl. Sci. 2018, 3, 132–156. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Nino, W.R.; Zazueta, C. New insights of Kruppel-like transcription factors in adipogenesis and the role of their regulatory neighbors. Life Sci. 2021, 265, 118763. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.S.; Feinberg, M.W.; Watanabe, M.; Gray, S.; Haspel, R.L.; Denkinger, D.J.; Kawahara, R.; Hauner, H.; Jain, M.K. The Kruppel-like factor KLF2 inhibits peroxisome proliferator-activated receptor-γ expression and adipogenesis. J. Biol. Chem. 2003, 278, 2581–2584. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Srinivasan, S.V.; Neumann, J.C.; Lingrel, J.B. The KLF2 transcription factor does not affect the formation of preadipocytes but inhibits their differentiation into adipocytes. Biochemistry 2005, 44, 11098–11105. [Google Scholar] [CrossRef] [PubMed]
- Sue, N.; Jack, B.H.; Eaton, S.A.; Pearson, R.C.; Funnell, A.P.; Turner, J.; Czolij, R.; Denyer, G.; Bao, S.; Molero-Navajas, J.C.; et al. Targeted disruption of the basic Kruppel-like factor gene (Klf3) reveals a role in adipogenesis. Mol. Cell. Biol. 2008, 28, 3967–3978. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, Y.; Tanaka, Y.; Kawamori, R.; Maeda, S. Overexpression of Kruppel-like factor 7 regulates adipocytokine gene expressions in human adipocytes and inhibits glucose-induced insulin secretion in pancreatic beta-cell line. Mol. Endocrinol. 2006, 20, 844–856. [Google Scholar] [CrossRef] [PubMed]
- Bagchi, D.P.; MacDougald, O.A. Wnt Signaling: From Mesenchymal Cell Fate to Lipogenesis and Other Mature Adipocyte Functions. Diabetes 2021, 70, 1419–1430. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Wang, J. Wnt/β-Catenin Signaling and Obesity. Front. Physiol. 2018, 9, 792. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.Y.; Jang, H.J.; Muthamil, S.; Shin, U.C.; Lyu, J.H.; Kim, S.W.; Go, Y.; Park, S.H.; Lee, H.G.; Park, J.H. Novel insights into regulators and functional modulators of adipogenesis. Biomed. Pharmacother. 2024, 177, 117073. [Google Scholar] [CrossRef] [PubMed]
- Rosen, E.D.; MacDougald, O.A. Adipocyte differentiation from the inside out. Nat. Rev. Mol. Cell Biol. 2006, 7, 885–896. [Google Scholar] [CrossRef] [PubMed]
- Christodoulides, C.; Lagathu, C.; Sethi, J.K.; Vidal-Puig, A. Adipogenesis and WNT signalling. Trends Endocrinol. Metab. 2009, 20, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Wang, J.; Zhu, T.; Shen, Y.; Tang, X.; Fang, L.; Xu, Y. Cross-Talking Between PPAR and WNT Signaling and its Regulation in Mesenchymal Stem Cell Differentiation. Curr. Stem Cell Res. Ther. 2016, 11, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Li, Q.; Luo, S.; Liu, Z.; Luo, D.; Zhang, B.; Zhang, D.; Rao, P.; Xiao, J. PPARγ and Wnt Signaling in Adipogenic and Osteogenic Differentiation of Mesenchymal Stem Cells. Curr Stem Cell Res. Ther. 2016, 11, 216–225. [Google Scholar] [CrossRef] [PubMed]
- Nishizuka, M.; Koyanagi, A.; Osada, S.; Imagawa, M. Wnt4 and Wnt5a promote adipocyte differentiation. FEBS Lett. 2008, 582, 3201–3205. [Google Scholar] [CrossRef]
- Tang, Q.; Chen, C.; Zhang, Y.; Dai, M.; Jiang, Y.; Wang, H.; Yu, M.; Jing, W.; Tian, W. Wnt5a regulates the cell proliferation and adipogenesis via MAPK-independent pathway in early stage of obesity. Cell Biol. Int. 2018, 42, 63–74. [Google Scholar] [CrossRef] [PubMed]
- de Winter, T.J.J.; Nusse, R. Running Against the Wnt: How Wnt/beta-Catenin Suppresses Adipogenesis. Front. Cell Dev. Biol. 2021, 9, 627429. [Google Scholar] [CrossRef] [PubMed]
- Prestwich, T.C.; Macdougald, O.A. Wnt/beta-catenin signaling in adipogenesis and metabolism. Curr. Opin. Cell Biol. 2007, 19, 612–617. [Google Scholar] [CrossRef] [PubMed]
- Kanazawa, A.; Tsukada, S.; Kamiyama, M.; Yanagimoto, T.; Nakajima, M.; Maeda, S. Wnt5b partially inhibits canonical Wnt/beta-catenin signaling pathway and promotes adipogenesis in 3T3-L1 preadipocytes. Biochem. Biophys. Res. Commun. 2005, 330, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Bilkovski, R.; Schulte, D.M.; Oberhauser, F.; Gomolka, M.; Udelhoven, M.; Hettich, M.M.; Roth, B.; Heidenreich, A.; Gutschow, C.; Krone, W.; et al. Role of WNT-5a in the determination of human mesenchymal stem cells into preadipocytes. J. Biol. Chem. 2010, 285, 6170–6178. [Google Scholar] [CrossRef]
- Guo, L.; Huang, J.X.; Liu, Y.; Li, X.; Zhou, S.R.; Qian, S.W.; Liu, Y.; Zhu, H.; Huang, H.Y.; Dang, Y.J.; et al. Transactivation of Atg4b by C/EBPbeta promotes autophagy to facilitate adipogenesis. Mol. Cell. Biol. 2013, 33, 3180–3190. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.Q.; Lane, M.D. Role of C/EBP homologous protein (CHOP-10) in the programmed activation of CCAAT/enhancer-binding protein-beta during adipogenesis. Proc. Natl. Acad. Sci. USA 2000, 97, 12446–12450. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.W.; Klemm, D.J.; Vinson, C.; Lane, M.D. Role of CREB in transcriptional regulation of CCAAT/enhancer-binding protein beta gene during adipogenesis. J. Biol. Chem. 2004, 279, 4471–4478. [Google Scholar] [CrossRef] [PubMed]
- Tong, Q.; Dalgin, G.; Xu, H.; Ting, C.N.; Leiden, J.M.; Hotamisligil, G.S. Function of GATA transcription factors in preadipocyte-adipocyte transition. Science 2000, 290, 134–138. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Li, N.; Lin, Y.; Wang, M.; Peng, Y.; Lewi, K.; Wang, Q. Glucagon Like Peptide-1 Promotes Adipocyte Differentiation via the Wnt4 Mediated Sequestering of Beta-Catenin. PLoS ONE 2016, 11, e0160212. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Torrens, J.I.; Anand, A.; Spiegelman, B.M.; Friedman, J.M. Krox20 stimulates adipogenesis via C/EBPbeta-dependent and -independent mechanisms. Cell Metab. 2005, 1, 93–106. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.B.; Moon, H.M.; Kim, W.S.; Lee, Y.S.; Jeong, H.W.; Yoo, E.J.; Ham, J.; Kang, H.; Park, M.G.; Steffensen, K.R.; et al. Activated liver X receptors stimulate adipocyte differentiation through induction of peroxisome proliferator-activated receptor γ expression. Mol. Cell. Biol. 2004, 24, 3430–3444. [Google Scholar] [CrossRef] [PubMed]
- Mayoral, R.; Osborn, O.; McNelis, J.; Johnson, A.M.; Oh, D.Y.; Izquierdo, C.L.; Chung, H.; Li, P.; Traves, P.G.; Bandyopadhyay, G.; et al. Adipocyte SIRT1 knockout promotes PPARγ activity, adipogenesis and insulin sensitivity in chronic-HFD and obesity. Mol. Metab. 2015, 4, 378–391. [Google Scholar] [CrossRef] [PubMed]
- Leow, S.C.; Poschmann, J.; Too, P.G.; Yin, J.; Joseph, R.; McFarlane, C.; Dogra, S.; Shabbir, A.; Ingham, P.W.; Prabhakar, S.; et al. The transcription factor SOX6 contributes to the developmental origins of obesity by promoting adipogenesis. Development 2016, 143, 950–961. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.B.; Spiegelman, B.M. ADD1/SREBP1 promotes adipocyte differentiation and gene expression linked to fatty acid metabolism. Genes Dev. 1996, 10, 1096–1107. [Google Scholar] [CrossRef] [PubMed]
- Choy, L.; Derynck, R. Transforming growth factor-beta inhibits adipocyte differentiation by Smad3 interacting with CCAAT/enhancer-binding protein (C/EBP) and repressing C/EBP transactivation function. J. Biol. Chem. 2003, 278, 9609–9619. [Google Scholar] [CrossRef] [PubMed]
- Ross, S.E.; Hemati, N.; Longo, K.A.; Bennett, C.N.; Lucas, P.C.; Erickson, R.L.; MacDougald, O.A. Inhibition of adipogenesis by Wnt signaling. Science 2000, 289, 950–953. [Google Scholar] [CrossRef] [PubMed]
- Chirumbolo, S.; Bjørklund, G. Can Wnt5a and Wnt non-canonical pathways really mediate adipocyte de-differentiation in a tumour microenvironment? Eur. J. Cancer 2016, 64, 96–100. [Google Scholar] [CrossRef] [PubMed]
- van Tienen, F.H.; Laeremans, H.; van der Kallen, C.J.; Smeets, H.J. Wnt5b stimulates adipogenesis by activating PPARgamma, and inhibiting the beta-catenin dependent Wnt signaling pathway together with Wnt5a. Biochem. Biophys. Res. Commun. 2009, 387, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Cawthorn, W.P.; Bree, A.J.; Yao, Y.; Du, B.; Hemati, N.; Martinez-Santibañez, G.; MacDougald, O.A. Wnt6, Wnt10a and Wnt10b inhibit adipogenesis and stimulate osteoblastogenesis through a β-catenin-dependent mechanism. Bone 2012, 50, 477–489. [Google Scholar] [CrossRef] [PubMed]
- Kennell, J.A.; MacDougald, O.A. Wnt signaling inhibits adipogenesis through beta-catenin-dependent and -independent mechanisms. J. Biol. Chem. 2005, 280, 24004–24010. [Google Scholar] [CrossRef] [PubMed]
- Longo, K.A.; Wright, W.S.; Kang, S.; Gerin, I.; Chiang, S.-H.; Lucas, P.C.; Opp, M.R.; MacDougald, O.A. Wnt10b Inhibits Development of White and Brown Adipose Tissues. J. Biol. Chem. 2004, 279, 35503–35509. [Google Scholar] [CrossRef] [PubMed]
- Sha, H.; He, Y.; Chen, H.; Wang, C.; Zenno, A.; Shi, H.; Yang, X.; Zhang, X.; Qi, L. The IRE1alpha-XBP1 pathway of the unfolded protein response is required for adipogenesis. Cell Metab. 2009, 9, 556–564. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.M.; Kwak, S.N.; Joo, N.S.; Kim, D.H.; Lee, A.H.; Kim, K.S.; Seo, J.B.; Jeong, S.W.; Kwon, O.J. X-box binding protein 1 is a novel key regulator of peroxisome proliferator-activated receptor γ2. FEBS J. 2014, 281, 5132–5146. [Google Scholar] [CrossRef] [PubMed]
- Meruvu, S.; Hugendubler, L.; Mueller, E. Regulation of adipocyte differentiation by the zinc finger protein ZNF638. J. Biol. Chem. 2011, 286, 26516–26523. [Google Scholar] [CrossRef] [PubMed]
- Scheja, L.; Heeren, J. The endocrine function of adipose tissues in health and cardiometabolic disease. Nat. Rev. Endocrinol. 2019, 15, 507–524. [Google Scholar] [CrossRef] [PubMed]
- Church, C.; Horowitz, M.; Rodeheffer, M. WAT is a functional adipocyte? Adipocyte 2012, 1, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Moitra, J.; Mason, M.M.; Olive, M.; Krylov, D.; Gavrilova, O.; Marcus-Samuels, B.; Feigenbaum, L.; Lee, E.; Aoyama, T.; Eckhaus, M.; et al. Life without white fat: A transgenic mouse. Genes Dev. 1998, 12, 3168–3181. [Google Scholar] [CrossRef] [PubMed]
- Temple, N.J. The Origins of the Obesity Epidemic in the USA–Lessons for Today. Nutrients 2022, 14, 4253. [Google Scholar] [CrossRef] [PubMed]
- Setiawan, A.M.; Kamarudin, T.A.; Abd Ghafar, N. The role of BMP4 in adipose-derived stem cell differentiation: A minireview. Front. Cell Dev. Biol. 2022, 10, 1045103. [Google Scholar] [CrossRef] [PubMed]
- Gustafson, B.; Hammarstedt, A.; Hedjazifar, S.; Hoffmann, J.M.; Svensson, P.A.; Grimsby, J.; Rondinone, C.; Smith, U. BMP4 and BMP Antagonists Regulate Human White and Beige Adipogenesis. Diabetes 2015, 64, 1670–1681. [Google Scholar] [CrossRef] [PubMed]
- Modica, S.; Wolfrum, C. The dual role of BMP4 in adipogenesis and metabolism. Adipocyte 2017, 6, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, J.M.; Grunberg, J.R.; Church, C.; Elias, I.; Palsdottir, V.; Jansson, J.O.; Bosch, F.; Hammarstedt, A.; Hedjazifar, S.; Smith, U. BMP4 Gene Therapy in Mature Mice Reduces BAT Activation but Protects from Obesity by Browning Subcutaneous Adipose Tissue. Cell Rep. 2017, 20, 1038–1049. [Google Scholar] [CrossRef] [PubMed]
- van Baak, M.A.; Mariman, E.C.M. Mechanisms of weight regain after weight loss—The role of adipose tissue. Nat. Rev. Endocrinol. 2019, 15, 274–287. [Google Scholar] [CrossRef] [PubMed]
- Busetto, L.; Bettini, S.; Makaronidis, J.; Roberts, C.A.; Halford, J.C.G.; Batterham, R.L. Mechanisms of weight regain. Eur. J. Intern. Med. 2021, 93, 3–7. [Google Scholar] [CrossRef] [PubMed]
- Qian, S.W.; Tang, Y.; Li, X.; Liu, Y.; Zhang, Y.Y.; Huang, H.Y.; Xue, R.D.; Yu, H.Y.; Guo, L.; Gao, H.D.; et al. BMP4-mediated brown fat-like changes in white adipose tissue alter glucose and energy homeostasis. Proc. Natl. Acad. Sci. USA 2013, 110, E798–E807. [Google Scholar] [CrossRef] [PubMed]
- Sugihara, H.; Yonemitsu, N.; Miyabara, S.; Yun, K. Primary cultures of unilocular fat cells: Characteristics of growth in vitro and changes in differentiation properties. Differentiation 1986, 31, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Sugihara, H.; Yonemitsu, N.; Miyabara, S.; Toda, S. Proliferation of unilocular fat cells in the primary culture. J. Lipid Res. 1987, 28, 1038–1045. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Song, T.; Hu, X.; Zhou, Y.; Wei, H.; Peng, J.; Jiang, S. Phenotypic and Functional Properties of Porcine Dedifferentiated Fat Cells during the Long-Term Culture In Vitro. BioMed Res. Int. 2015, 2015, 673651. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Jeong, S.; Kwon, H.R.; Olson, L.E. Regulation of adipocyte dedifferentiation at the skin wound edge. bioRxiv 2023. [Google Scholar]
- Song, T.; Kuang, S. Adipocyte dedifferentiation in health and diseases. Clin. Sci. 2019, 133, 2107–2119. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; He, Y.; Tang, H.; Li, J.; Cai, J.; Liao, Y. Dedifferentiated fat cells: Current applications and future directions in regenerative medicine. Stem Cell Res. Ther. 2023, 14, 207. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Duarte, M.S.; Zan, L.; Du, M.; Jiang, Z.; Guan, L.; Chen, J.; Hausman, G.J.; Dodson, M.V. Cellular and molecular implications of mature adipocyte dedifferentiation. J. Genom. 2013, 1, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Gustafson, B.; Smith, U. Activation of canonical wingless-type MMTV integration site family (Wnt) signaling in mature adipocytes increases beta-catenin levels and leads to cell dedifferentiation and insulin resistance. J. Biol. Chem. 2010, 285, 14031–14041. [Google Scholar] [CrossRef] [PubMed]
- Cote, J.A.; Lessard, J.; Pelletier, M.; Marceau, S.; Lescelleur, O.; Fradette, J.; Tchernof, A. Role of the TGF-beta pathway in dedifferentiation of human mature adipocytes. FEBS Open Bio 2017, 7, 1092–1101. [Google Scholar] [CrossRef] [PubMed]
- Taylor, B.; Shah, A.; Bielczyk-Maczynska, E. TGF-beta is insufficient to induce adipocyte state loss without concurrent PPARgamma downregulation. Sci. Rep. 2020, 10, 14084. [Google Scholar] [CrossRef] [PubMed]
- Chartoumpekis, D.V.; Palliyaguru, D.L.; Wakabayashi, N.; Khoo, N.K.; Schoiswohl, G.; O’Doherty, R.M.; Kensler, T.W. Notch intracellular domain overexpression in adipocytes confers lipodystrophy in mice. Mol. Metab. 2015, 4, 543–550. [Google Scholar] [CrossRef] [PubMed]
- Bi, P.; Yue, F.; Karki, A.; Castro, B.; Wirbisky, S.E.; Wang, C.; Durkes, A.; Elzey, B.D.; Andrisani, O.M.; Bidwell, C.A.; et al. Notch activation drives adipocyte dedifferentiation and tumorigenic transformation in mice. J. Exp. Med. 2016, 213, 2019–2037. [Google Scholar] [CrossRef] [PubMed]
- Siouti, E.; Salagianni, M.; Manioudaki, M.; Pavlos, E.; Klinakis, A.; Galani, I.E.; Andreakos, E. Notch signaling in adipose tissue macrophages prevents diet-induced inflammation and metabolic dysregulation. Eur. J. Immunol. 2024, 54, e2350669. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Bergen, W.G.; Hausman, G.J.; Zan, L.; Dodson, M.V. Cell culture purity issues and DFAT cells. Biochem. Biophys. Res. Commun. 2013, 433, 273–275. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dowker-Key, P.D.; Jadi, P.K.; Gill, N.B.; Hubbard, K.N.; Elshaarrawi, A.; Alfatlawy, N.D.; Bettaieb, A. A Closer Look into White Adipose Tissue Biology and the Molecular Regulation of Stem Cell Commitment and Differentiation. Genes 2024, 15, 1017. https://doi.org/10.3390/genes15081017
Dowker-Key PD, Jadi PK, Gill NB, Hubbard KN, Elshaarrawi A, Alfatlawy ND, Bettaieb A. A Closer Look into White Adipose Tissue Biology and the Molecular Regulation of Stem Cell Commitment and Differentiation. Genes. 2024; 15(8):1017. https://doi.org/10.3390/genes15081017
Chicago/Turabian StyleDowker-Key, Presley D., Praveen Kumar Jadi, Nicholas B. Gill, Katelin N. Hubbard, Ahmed Elshaarrawi, Naba D. Alfatlawy, and Ahmed Bettaieb. 2024. "A Closer Look into White Adipose Tissue Biology and the Molecular Regulation of Stem Cell Commitment and Differentiation" Genes 15, no. 8: 1017. https://doi.org/10.3390/genes15081017
APA StyleDowker-Key, P. D., Jadi, P. K., Gill, N. B., Hubbard, K. N., Elshaarrawi, A., Alfatlawy, N. D., & Bettaieb, A. (2024). A Closer Look into White Adipose Tissue Biology and the Molecular Regulation of Stem Cell Commitment and Differentiation. Genes, 15(8), 1017. https://doi.org/10.3390/genes15081017








