Identification and Prediction of Differentially Expressed MicroRNAs Associated with Detoxification Pathways in Larvae of Spodoptera frugiperda
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Collection and Rearing
2.2. Small RNA Library Preparation and Sequencing
2.3. Unigene Assembly, Analysis, and Annotation
2.4. Differential Expression Analysis of miRNAs
2.5. Quantitative Reverse Transcription PCR
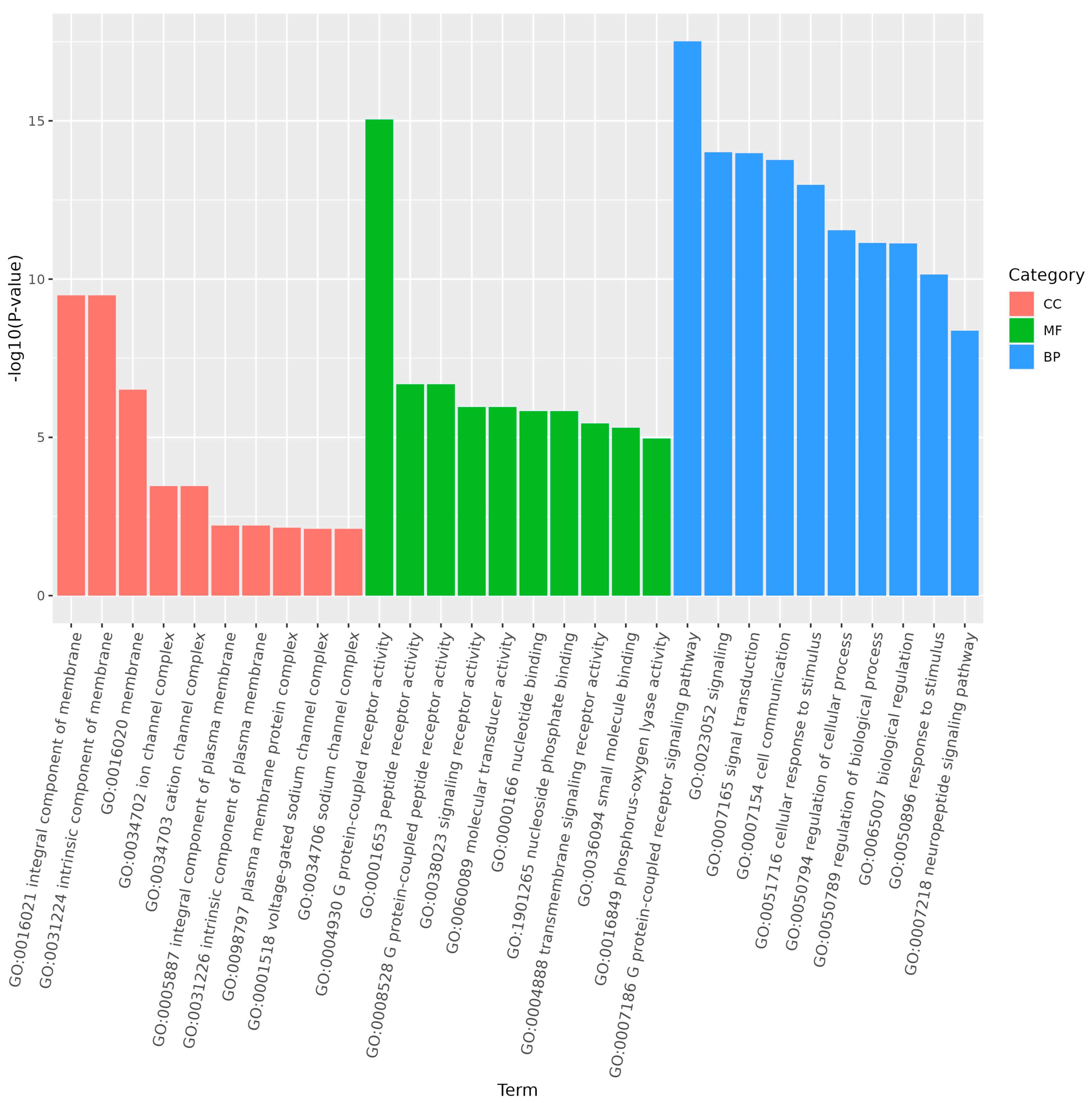
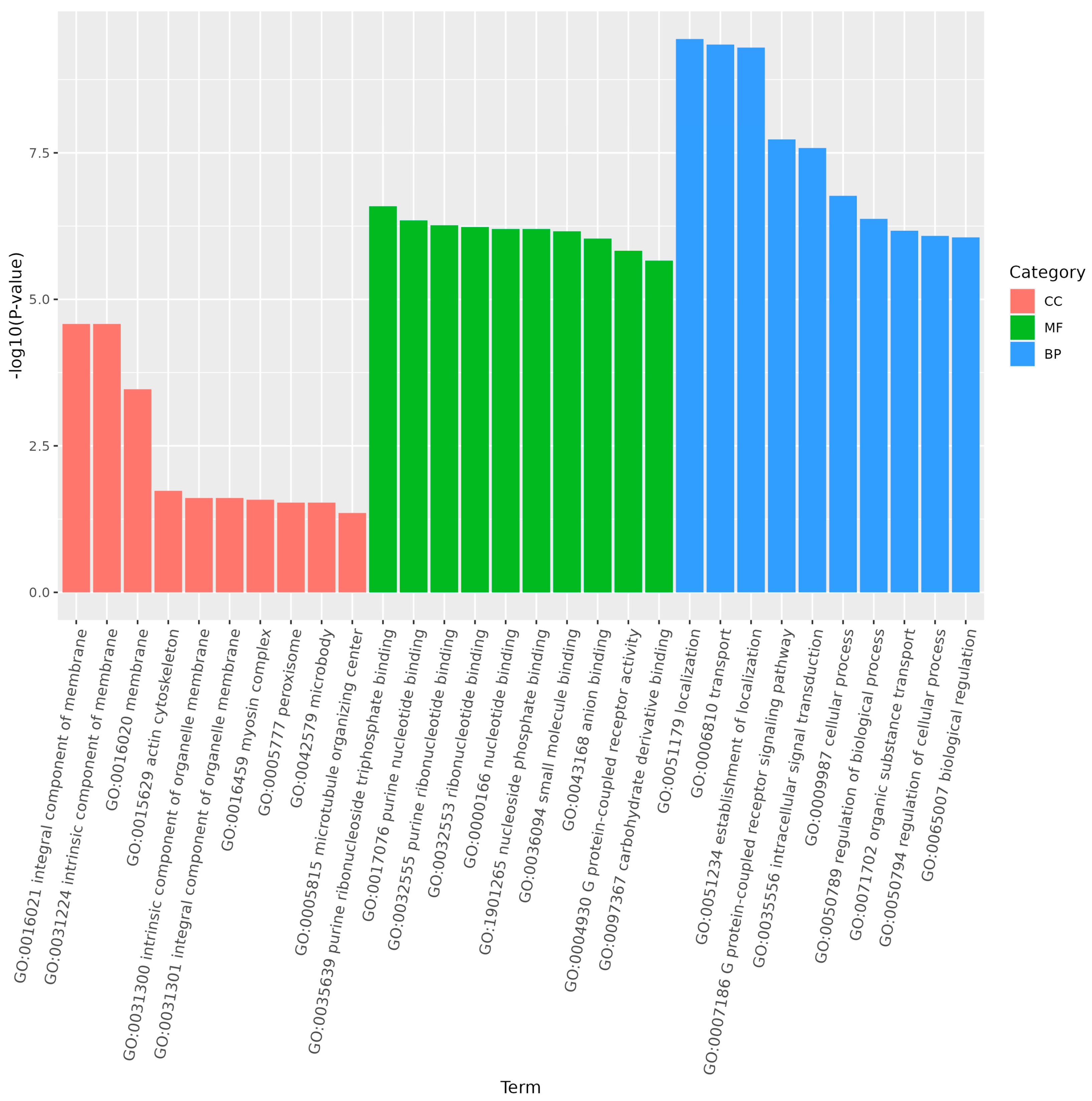
2.6. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Enrichment Analysis
2.7. Statistical and Data Analysis
3. Results
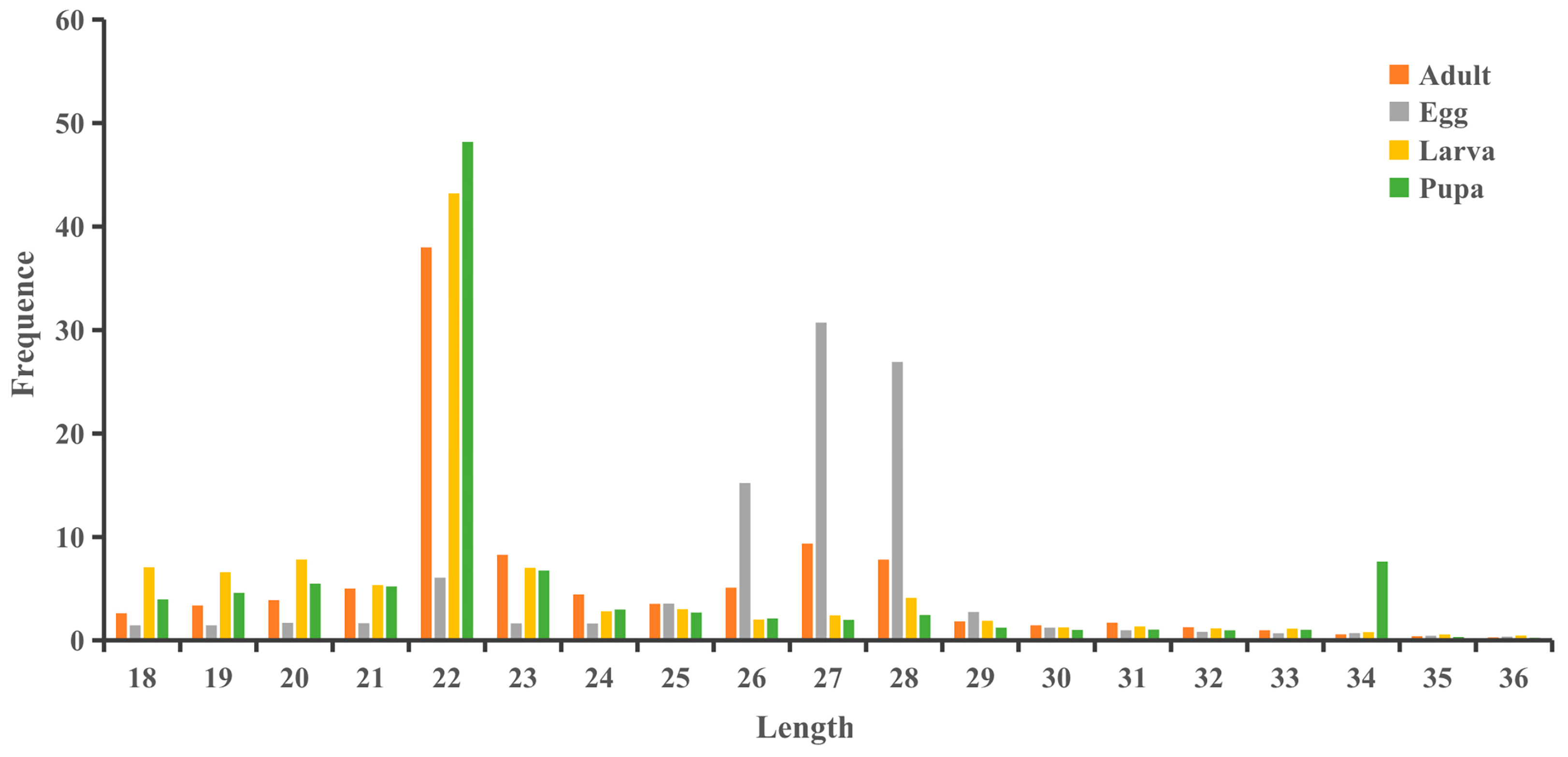
3.1. Small RNA Sequencing Data in S. frugiperda
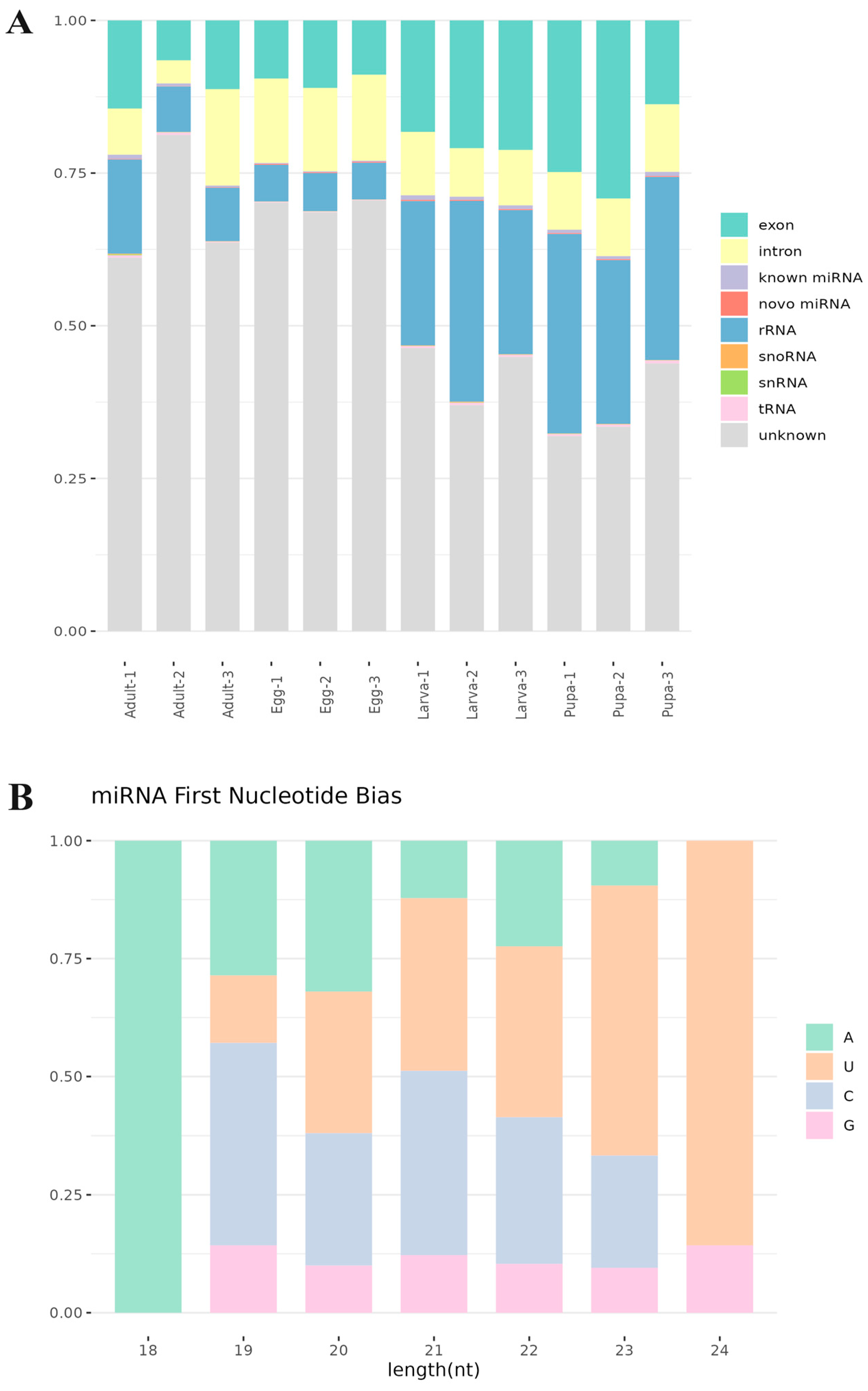
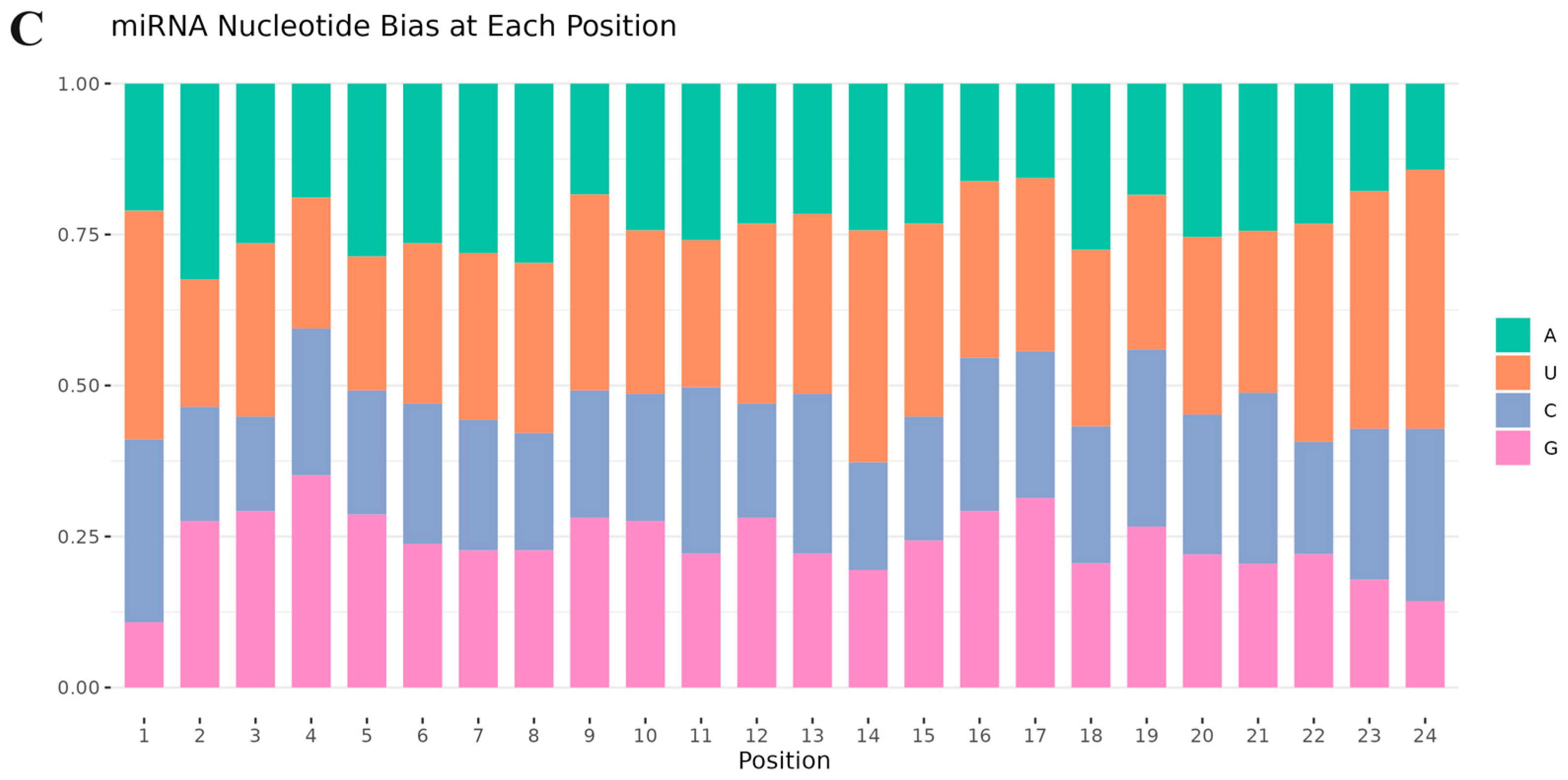
3.2. Small RNA Classification and Annotation
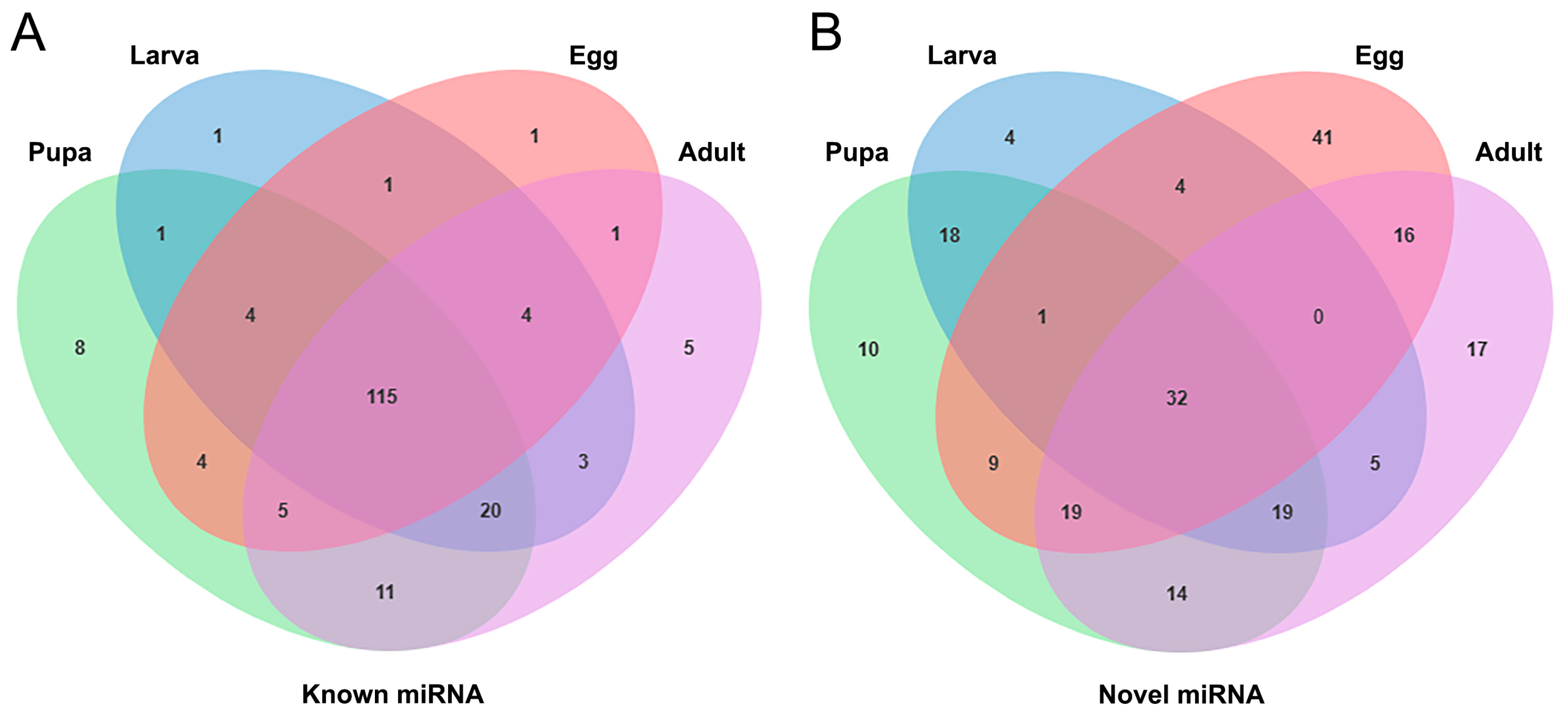
3.3. Identification of Known and Novel miRNAs in S. frugiperda
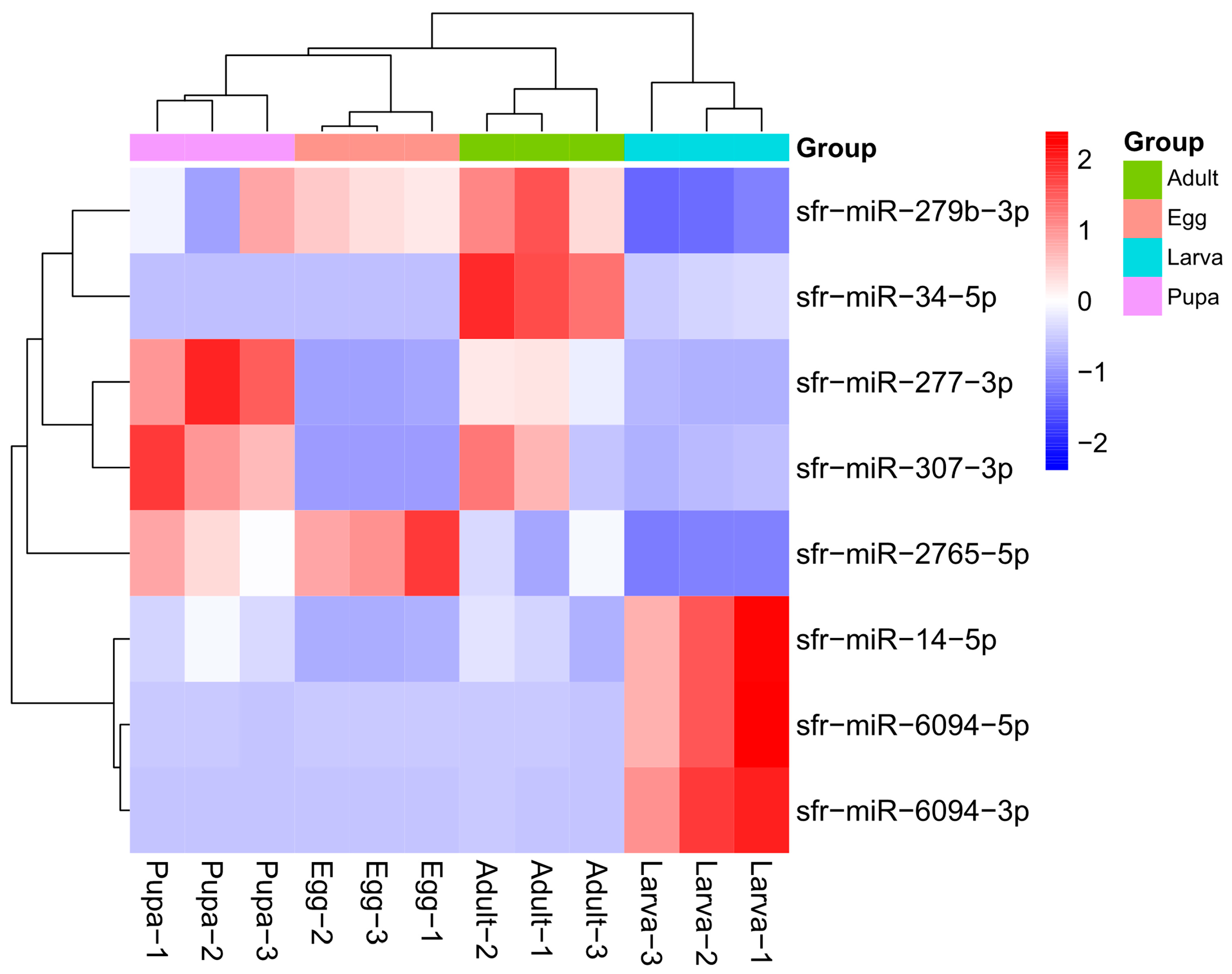
3.4. Identification of Differentially Expressed miRNAs in Larvae
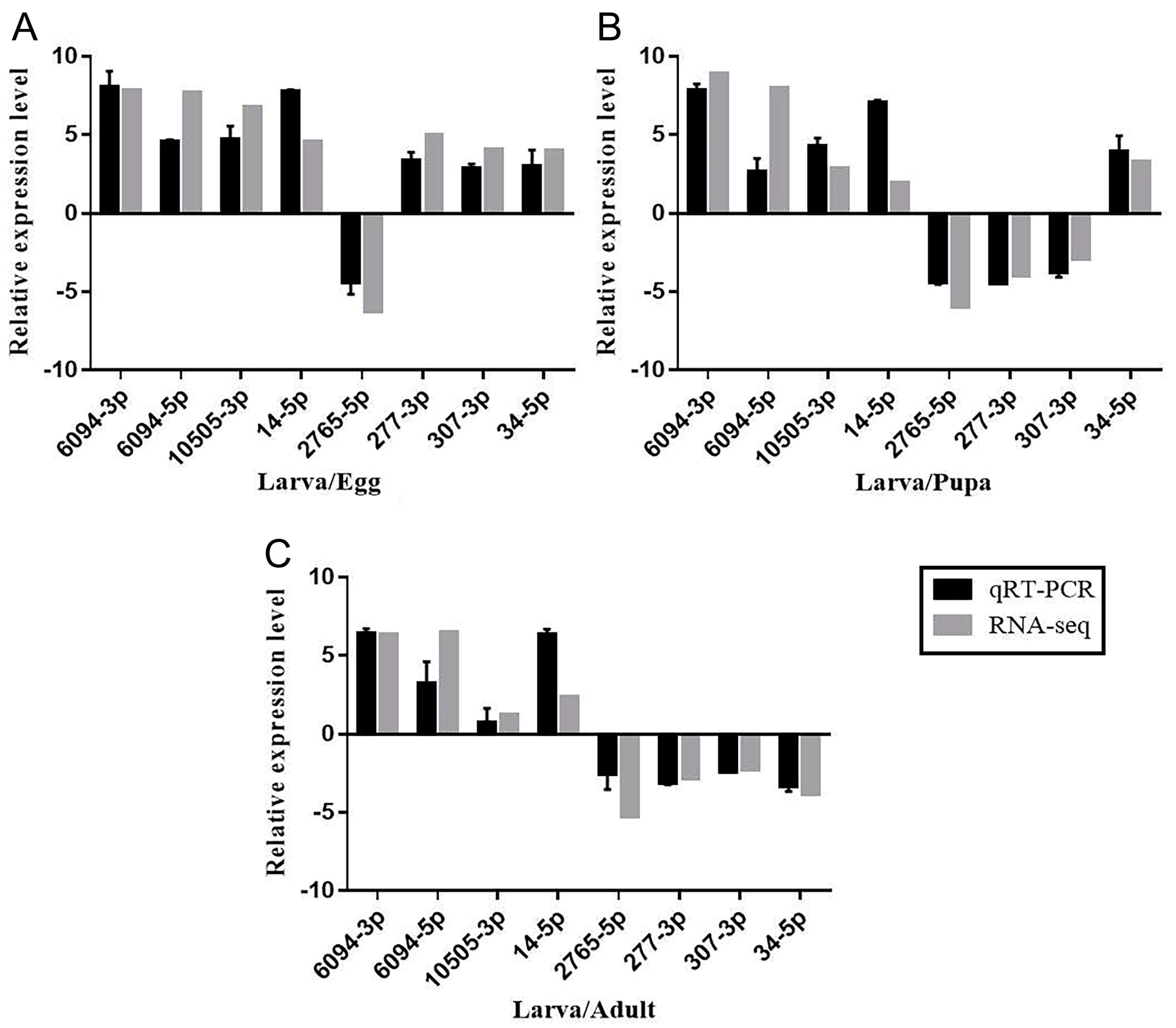
3.5. qRT-PCR Validation of Differentially Expressed miRNAs in Larvae
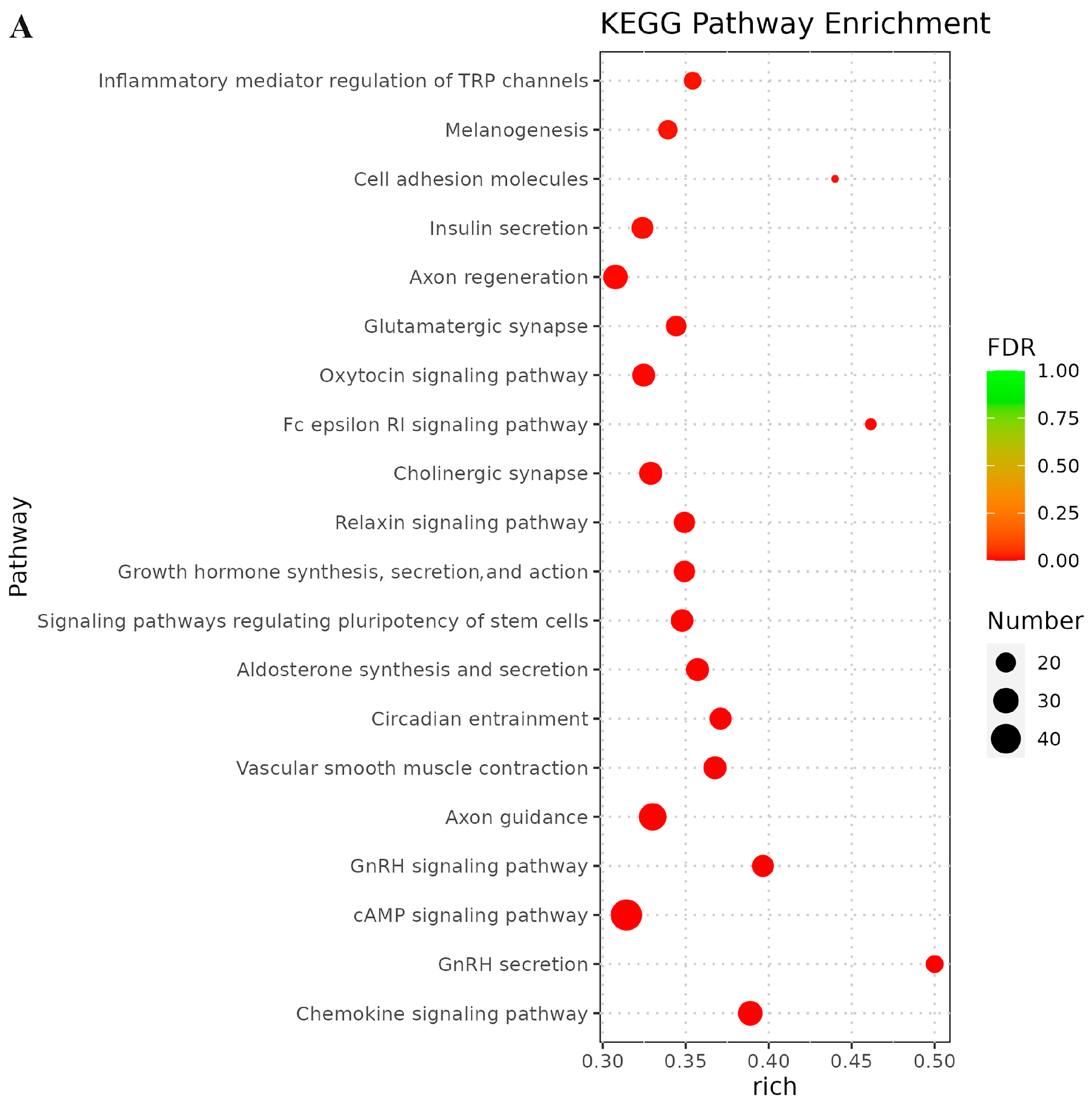
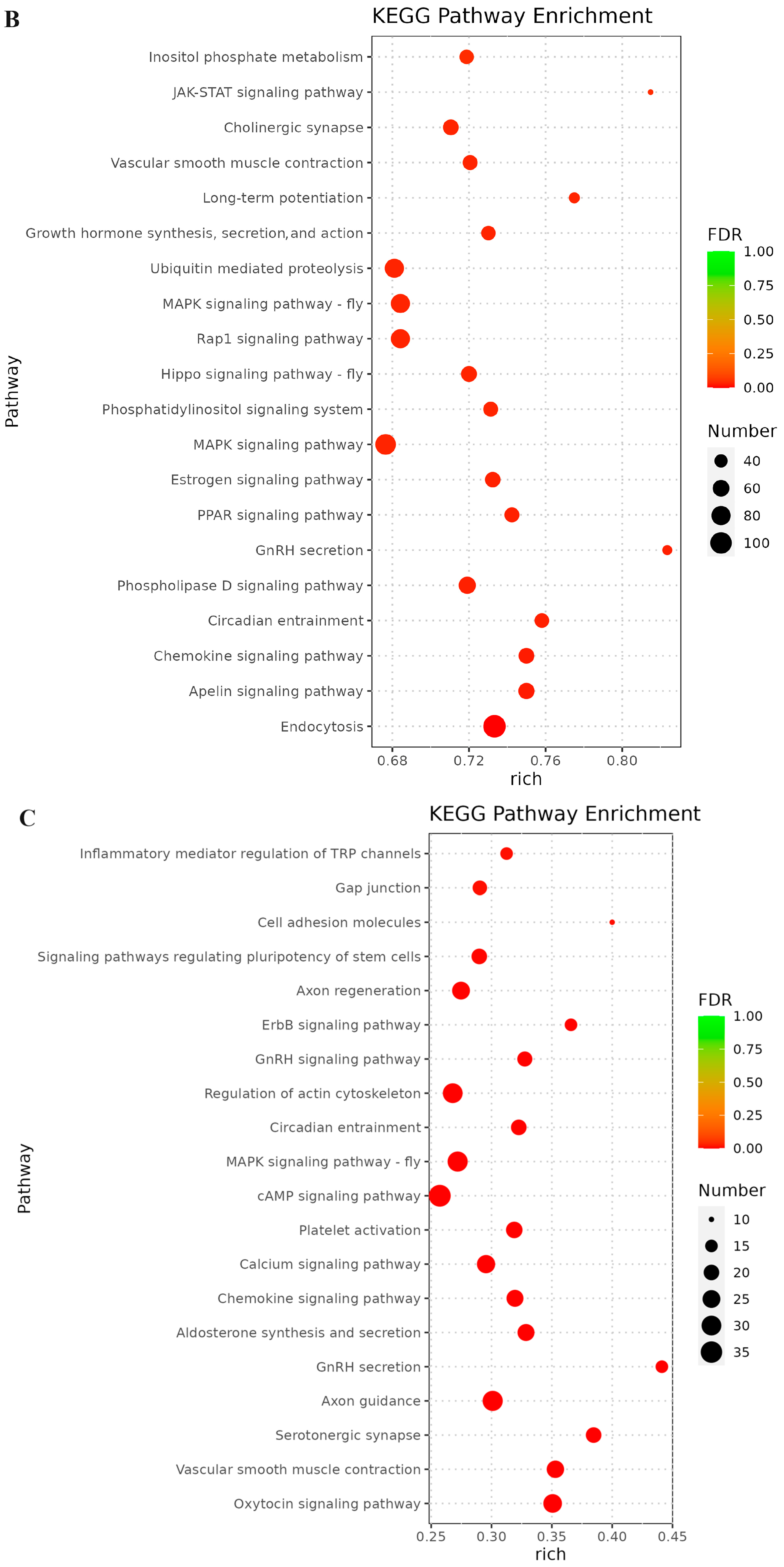
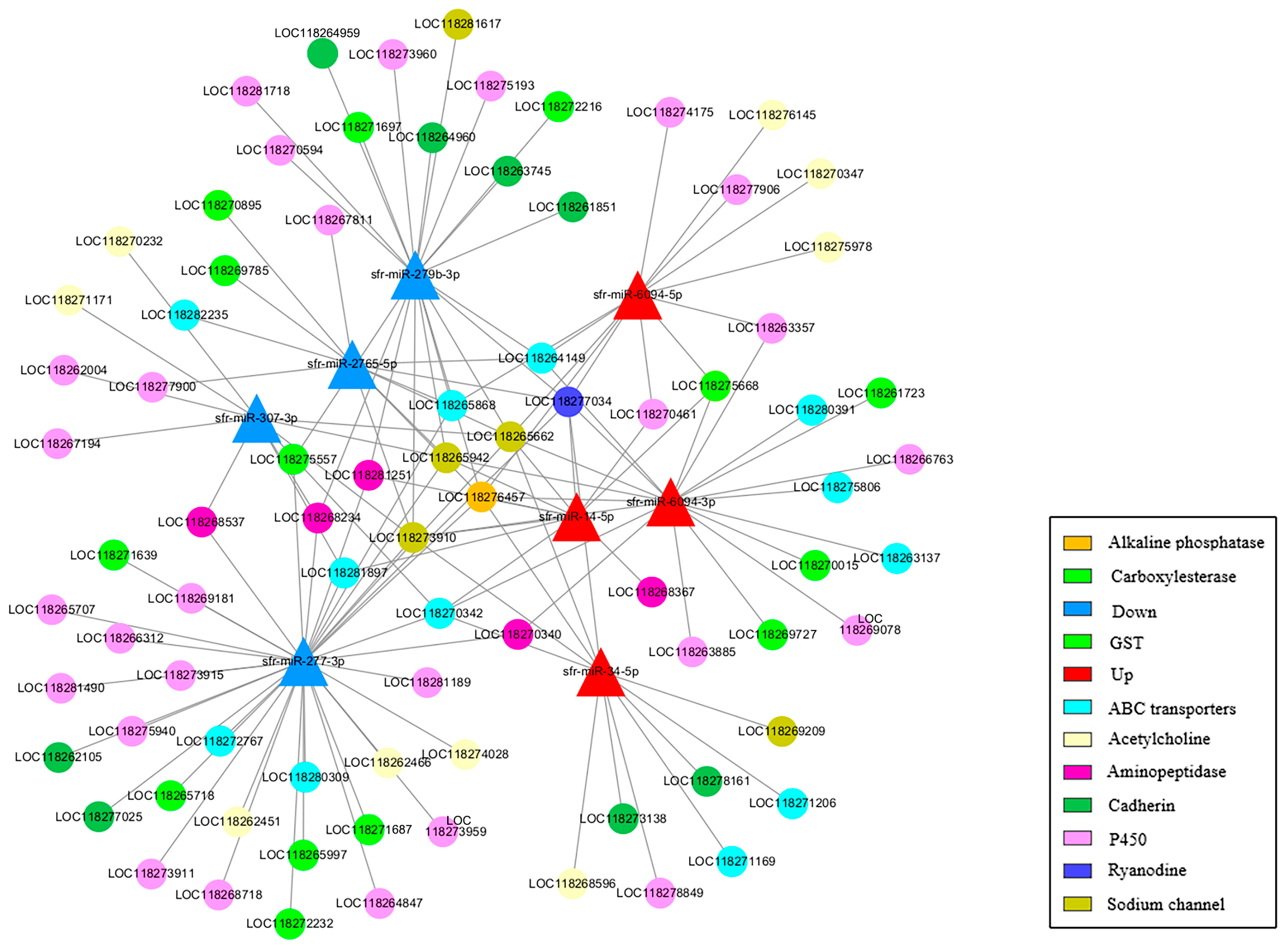
3.6. Prediction of Targeted Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Asgari, S. Venom proteins from polydnavirus-producing endoparasitoids: Their role in host-parasite interactions. Arch. Insect Biochem. 2006, 61, 146–156. [Google Scholar] [CrossRef]
- Ibanez-Ventoso, C.; Vora, M.; Driscoll, M. Sequence relationships among C. elegans, D. melanogaster and human microRNAs highlight the extensive conservation of microRNAs in biology. PLoS ONE 2008, 3, e2818. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Orte, E.; Belles, X. MicroRNA-dependent metamorphosis in hemimetabolan insects. Proc. Natl. Acad. Sci. USA 2009, 106, 21678–21682. [Google Scholar] [CrossRef]
- Asgari, S. Role of MicroRNAs in insect host-microorganism interactions. Front. Physiol. 2011, 2, 48. [Google Scholar] [CrossRef]
- Freitak, D.; Knorr, E.; Vogel, H.; Vilcinskas, A. Gender- and stressor-specific microRNA expression in Tribolium castaneum. Biol. Lett. 2012, 8, 860–863. [Google Scholar] [CrossRef]
- Fullaondo, A.; Lee, S.Y. Identification of putative miRNA involved in Drosophila melanogaster immune response. Dev. Comp. Immunol. 2012, 36, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Zhou, Q.; Cai, Y.; Luo, Q.; Lin, H.; Hu, S.; Yu, J. A discovery of novel microRNAs in the silkworm (Bombyx mori) genome. Genomics 2009, 94, 438–444. [Google Scholar] [CrossRef]
- Legeai, F.; Rizk, G.; Walsh, T.; Edwards, O.; Gordon, K.; Lavenier, D.; Leterme, N.; Mereau, A.; Nicolas, J.; Tagu, D.; et al. Bioinformatic prediction, deep sequencing of microRNAs and expression analysis during phenotypic plasticity in the pea aphid, Acyrthosiphon pisum. BMC Genom. 2010, 11, 281. [Google Scholar] [CrossRef]
- Kloosterman, W.P.; Plasterk, R.H. The diverse functions of microRNAs in animal development and disease. Dev. Cell 2006, 11, 441–450. [Google Scholar] [CrossRef]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, X.; Ge, X.; Jiang, J.; Li, M.; Jia, S.; Yang, X.; Kan, Y.; Miao, X.; Zhao, G.; et al. Insect-Specific microRNA involved in the development of the silkworm Bombyx mori. PLoS ONE 2009, 4, e4677. [Google Scholar] [CrossRef]
- Etebari, K.; Asgari, S. Conserved microRNA miR-8 blocks activation of the Toll pathway by upregulating Serpin 27 transcripts. RNA Biol. 2013, 10, 1356–1364. [Google Scholar] [CrossRef]
- Lucas, K.J.; Zhao, B.; Liu, S.; Raikhel, A.S. Regulation of physiological processes by microRNAs in insects. Curr. Opin. Insect Sci. 2015, 11, 1–7. [Google Scholar] [CrossRef]
- Ling, L.; Kokoza, V.A.; Zhang, C.; Aksoy, E.; Raikhel, A.S. MicroRNA-277 targets insulin-like peptides 7 and 8 to control lipid metabolism and reproduction in Aedes aegypti mosquitoes. Proc. Natl. Acad. Sci. USA 2017, 114, E8017–E8024. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, X.; Shiotsuki, T.; Wang, Z.; Xu, X.; Huang, Y.; Li, M.; Li, K.; Tan, A. Depletion of juvenile hormone esterase extends larval growth in Bombyx mori. Insect Biochem. Mol. Biol. 2017, 81, 72–79. [Google Scholar] [CrossRef]
- Song, J.; Li, W.; Zhao, H.; Gao, L.; Fan, Y.; Zhou, S. The microRNAs let-7 and miR-278 regulate insect metamorphosis and oogenesis by targeting the juvenile hormone early-response gene Kruppel-homolog 1. Development 2018, 145, 170670. [Google Scholar] [CrossRef]
- Shang, F.; Niu, J.; Ding, B.Y.; Zhang, W.; Wei, D.D.; Wei, D.; Jiang, H.B.; Wang, J.J. The miR-9b microRNA mediates dimorphism and development of wing in aphids. Proc. Natl. Acad. Sci. USA 2020, 117, 8404–8409. [Google Scholar] [CrossRef]
- Lin, L. Functional Analysis of microRNA let-7 and miR-2 Regulating Development in Bombyx mori. Ph.D. Thesis, Northwestern Polytechnical University, Xi’an, China, 2017. [Google Scholar]
- Wang, Y.L.; Yang, M.L.; Jiang, F.; Zhang, J.Z.; Kang, L. MicroRNA-dependent development revealed by RNA interference-mediated gene silencing of LmDicer1 in the migratory locust. Insect Sci. 2013, 20, 53–60. [Google Scholar] [CrossRef]
- Lozano, J.; Montanez, R.; Belles, X. MiR-2 family regulates insect metamorphosis by controlling the juvenile hormone signaling pathway. Proc. Natl. Acad. Sci. USA 2015, 112, 3740–3745. [Google Scholar] [CrossRef]
- Chen, X.; Fu, J. The microRNA miR-14 regulates egg-laying by targeting EcR in honeybees (Apis mellifera). Insects 2021, 12, 351. [Google Scholar] [CrossRef]
- He, K.; Xiao, H.; Sun, Y.; Ding, S.; Situ, G.; Li, F. Transgenic microRNA-14 rice shows high resistance to rice stem borer. Plant Biotechnol. J. 2019, 17, 461–471. [Google Scholar] [CrossRef]
- Puthiyakunnon, S.; Yao, Y.; Li, Y.; Gu, J.; Peng, H.; Chen, X. Functional characterization of three MicroRNAs of the Asian tiger mosquito, Aedes albopictus. Parasite Vector 2013, 6, 230. [Google Scholar] [CrossRef]
- Peng, W.; Zheng, W.W.; Tariq, K.; Yu, S.N.; Zhang, H.Y. MicroRNA Let-7 targets the ecdysone signaling pathway E75 gene to control larval-pupal development in Bactrocera dorsalis. Insect Sci. 2019, 26, 229–239. [Google Scholar] [CrossRef]
- Yang, M.; Wei, Y.; Jiang, F.; Wang, Y.; Guo, X.; He, J.; Kang, L. MicroRNA-133 inhibits behavioral aggregation by controlling dopamine synthesis in locusts. PLoS Genet. 2014, 10, e1004206. [Google Scholar] [CrossRef]
- Zha, W.; Chang, M.; You, A. Regulation of microRNA on insect growth and development. Hubei Agric. Sci. 2019, 58, 198–202. [Google Scholar] [CrossRef]
- Deng, C.; He, Q. Advanced research of microRNA in insects. J. Heilongjiang Bayi Agric. Univ. 2021, 33, 15–21. [Google Scholar]
- Li, X.; Guo, L.; Zhou, X.; Gao, X.; Liang, P. miRNAs regulated overexpression of ryanodine receptor is involved in chlorantraniliprole resistance in Plutella xylostella (L.). Sci. Rep. 2015, 5, 14095. [Google Scholar] [CrossRef]
- Zhu, B.; Sun, X.; Nie, X.; Liang, P.; Gao, X. MicroRNA-998-3p contributes to Cry1Ac-resistance by targeting ABCC2 in lepidopteran insects. Insect. Biochem. Mol. Biol. 2020, 117, 103283. [Google Scholar] [CrossRef]
- Mao, K.; Jin, R.; Ren, Z.; Zhang, J.; Li, Z.; He, S.; Ma, K.; Wan, H.; Li, J. miRNAs targeting CYP6ER1 and CarE1 are involved in nitenpyram resistance in Nilaparvata lugens. Insect Sci. 2022, 29, 177–187. [Google Scholar] [CrossRef]
- Ma, K.; Li, F.; Tang, Q.; Liang, P.; Liu, Y.; Zhang, B.; Gao, X. CYP4CJ1-mediated gossypol and tannic acid tolerance in Aphis gossypii Glover. Chemosphere 2019, 219, 961–970. [Google Scholar] [CrossRef]
- Van den Berg, J.; Brewer, M.J.; Reisig, D.D. A special collection: Spodoptera frugiperda (fall armyworm): Ecology and management of its world-scale invasion outside of the Americas. J. Econ. Entomol. 2022, 115, 1725–1728. [Google Scholar] [CrossRef]
- Goergen, G.; Kumar, P.L.; Sankung, S.B.; Togola, A.; Tamo, M. First report of outbreaks of the fall armyworm Spodoptera frugiperda (JE Smith) (Lepidoptera, Noctuidae), a new alien invasive pest in west and central Africa. PLoS ONE 2016, 11, e165632. [Google Scholar] [CrossRef]
- Cock, M.; Beseh, P.K.; Buddie, A.G.; Cafa, G.; Crozier, J. Molecular methods to detect Spodoptera frugiperda in Ghana, and implications for monitoring the spread of invasive species in developing countries. Sci. Rep. 2017, 7, 4103. [Google Scholar] [CrossRef]
- Belay, D.K.; Huckaba, R.M.; Foster, J.E. Susceptibility of the fall armyworm, Spodoptera frugiperda (lepidoptera: Noctuidae), at Santa Isabel, Puerto Rico, to different insecticides. Fla. Entomol. 2012, 95, 476–478. [Google Scholar] [CrossRef]
- Yu, S.J.; McCord, E.J. Lack of cross-resistance to indoxacarb in insecticide-resistant Spodoptera frugiperda (Lepidoptera: Noctuidae) and Plutella xylostella (Lepidoptera: Yponomeutidae). Pest Manag. Sci. 2007, 63, 63–67. [Google Scholar] [CrossRef] [PubMed]
- Sharon, G.; Segal, D.; Ringo, J.M.; Hefetz, A.; Zilber-Rosenberg, I.; Rosenberg, E. Commensal bacteria play a role in mating preference of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2010, 107, 20051–20056. [Google Scholar] [CrossRef]
- Carvalho, R.A.; Omoto, C.; Field, L.M.; Williamson, M.S.; Bass, C. Investigating the molecular mechanisms of organophosphate and pyrethroid resistance in the fall armyworm Spodoptera frugiperda. PLoS ONE 2013, 8, e62268. [Google Scholar] [CrossRef]
- Nascimento, A.R.B.D.; Farias, J.R.; Bernardi, D.; Horikoshi, R.J.; Omoto, C. Genetic basis of Spodoptera frugiperda (Lepidoptera: Noctuidae) resistance to the chitin synthesis inhibitor lufenuron. Pest Manag. Sci. 2016, 72, 810–815. [Google Scholar] [CrossRef]
- Bolzan, A.; Padovez, F.E.; Nascimento, A.R.; Kaiser, I.S.; Lira, E.C.; Amaral, F.S.; Kanno, R.H.; Malaquias, J.B.; Omoto, C. Selection and characterization of the inheritance of resistance of Spodoptera frugiperda (Lepidoptera: Noctuidae) to chlorantraniliprole and cross-resistance to other diamide insecticides. Pest Manag. Sci. 2019, 75, 2682–2689. [Google Scholar] [CrossRef]
- Lira, E.C.; Bolzan, A.; Nascimento, A.R.; Amaral, F.S.; Kanno, R.H.; Kaiser, I.S.; Omoto, C. Resistance of Spodoptera frugiperda (Lepidoptera: Noctuidae) to spinetoram: Inheritance and cross-resistance to spinosad. Pest Manag. Sci. 2020, 76, 2674–2680. [Google Scholar] [CrossRef]
- Mone, Y.; Nhim, S.; Gimenez, S.; Legeai, F.; Seninet, I.; Parrinello, H.; Negre, N.; D’Alencon, E. Characterization and expression profiling of microRNAs in response to plant feeding in two host-plant strains of the lepidopteran pest Spodoptera frugiperda. BMC Genom. 2018, 19, 804. [Google Scholar] [CrossRef]
- Karamipour, N.; Fathipour, Y.; Talebi, A.A.; Asgari, S.; Mehrabadi, M. The microRNA pathway is involved in Spodoptera frugiperda (Sf9) cells antiviral immune defense against Autographa californica multiple nucleopolyhedrovirus infection. Insect Biochem. Mol. Biol. 2019, 112, 103202. [Google Scholar] [CrossRef]
- Mahalle, R.M.; Sun, W.; Posos-Parra, O.A.; Jung, S.; Mota-Sanchez, D.; Pittendrigh, B.R.; Seong, K.M. Identification of differentially expressed miRNAs associated with diamide detoxification pathways in Spodoptera frugiperda. Sci. Rep. 2024, 14, 4308. [Google Scholar] [CrossRef]
- He, L.; Wang, T.; Chen, Y.; Ge, S.; Wyckhuys, K.; Wu, K. Larval diet affects development and reproduction of East Asian strain of the fall armyworm, Spodoptera frugiperda. J. Integr. Agric. 2021, 20, 736–744. [Google Scholar] [CrossRef]
- Andrews, S.A. Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/ (accessed on 7 December 2016).
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Friedlander, M.R.; Chen, W.; Adamidi, C.; Maaskola, J.; Einspanier, R.; Knespel, S.; Rajewsky, N. Discovering microRNAs from deep sequencing data using miRDeep. Nat. Biotechnol. 2008, 26, 407–415. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef]
- Hu, K. Become competent in generating RNA-seq heat maps in one day for novices without prior R experience. Methods Mol. Biol. 2021, 2239, 269–303. [Google Scholar] [CrossRef]
- Peng, X.; Nguyen, A.; Ghosh, D. Quantification of M13 and T7 bacteriophages by Taqman and SYBR green qPCR. J. Virol. Methods 2018, 252, 100–107. [Google Scholar] [CrossRef]
- Sriram, K.; Wiley, S.Z.; Moyung, K.; Gorr, M.W.; Salmerón, C.; Marucut, J.; French, R.P.; Lowy, A.M.; Insel, P.A. Detection and quantification of GPCR mRNA: An assessment and implications of data from high-content methods. ACS Omega 2019, 4, 17048–17059. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Yang, K.; Sablok, G.; Qiao, G.; Nie, Q.; Wen, X. IsomiR2Function: An integrated workflow for identifying microRNA variants in plants. Front. Plant Sci. 2017, 8, 322. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. Clusterprofiler: An R package for comparing biological themes among gene clusters. Omics 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Asgari, S. MicroRNA functions in insects. Insect Biochem. Mol. Biol. 2013, 43, 388–397. [Google Scholar] [CrossRef]
- Belles, X. MicroRNAs and the evolution of insect metamorphosis. Annu. Rev. Entomol. 2017, 62, 111–125. [Google Scholar] [CrossRef] [PubMed]
- Lucas, K.; Raikhel, A.S. Insect microRNAs: Biogenesis, expression profiling and biological functions. Insect Biochem. Mol. Biol. 2013, 43, 24–38. [Google Scholar] [CrossRef]
- Azzam, G.; Smibert, P.; Lai, E.C.; Liu, J.L. Drosophila Argonaute 1 and its miRNA biogenesis partners are required for oocyte formation and germline cell division. Dev. Biol. 2012, 365, 384–394. [Google Scholar] [CrossRef]
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993, 75, 855–862. [Google Scholar] [CrossRef]
- Gouin, A.; Bretaudeau, A.; Nam, K.; Gimenez, S.; Aury, J.M.; Duvic, B.; Hilliou, F.; Durand, N.; Montagne, N.; Darboux, I.; et al. Two genomes of highly polyphagous lepidopteran pests (Spodoptera frugiperda, Noctuidae) with different host-plant ranges. Sci. Rep. 2017, 7, 11816. [Google Scholar] [CrossRef]
- Rios-Diez, J.D.; Saldamando-Benjumea, C. Susceptibility of Spodoptera frugiperda (lepidoptera: Noctuidae) strains from central Colombia to two insecticides, methomyl and lambda-cyhalothrin: A study of the genetic basis of resistance. J. Econ. Entomol. 2011, 104, 1698–1705. [Google Scholar] [CrossRef] [PubMed]
- Mccord, E.; Yus, J. The mechanisms of carbaryl resistance in the fall armyworm, Spodoptera frugiperda (J.E. Smith). Pestic. Biochem. Physiol. 1987, 27, 114–122. [Google Scholar] [CrossRef]
- Gutiérrez-Moreno, R.; Mota-Sanchez, D.; A Blanco, C.; E Whalon, M.; Terán-Santofimio, H.; Rodriguez-Maciel, J.C.; DiFonzo, C. Field-evolved resistance of the fall armyworm (Lepidoptera: Noctuidae) to synthetic insecticides in Puerto Rico and Mexico. J. Econ. Entomol. 2019, 112, 792–802. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhang, Y.; Wang, A.; Duan, A.; Xue, C.; Wang, K.; Zhao, M.; Zhang, J. Four microRNAs, miR-13b-3p, miR-278-5p, miR-10483-5p, and miR-10485-5p, mediate insecticide tolerance in Spodoptera frugiperda. Front. Genet. 2021, 12, 820778. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Li, X.; Liu, Y.; Gao, X.; Liang, P. Global identification of microRNAs associated with chlorantraniliprole resistance in diamondback moth Plutella xylostella (L.). Sci. Rep. 2017, 7, 40713. [Google Scholar] [CrossRef]
- Li, J.M.; Zhou, Y.R.; Sun, Z.T.; Wang, X.; Xie, L.; Chen, J.P. Identification and profiling of conserved and novel microRNAs in Laodelphax striatellus in response to rice black-streaked dwarf virus (RBSDV) infection. Genom. Data 2015, 3, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.Z.; Hu, G.L.; Lu, L.Y.; Hu, S.F.; Li, Y.S.; Su, X.; Dong, W.Y.; Zhen, C.A.; Liu, R.Q.; Kong, F.B.; et al. Identification of differentially expressed microRNAs under imidacloprid exposure in Sitobion miscanthi. Pestic. Biochem. Phys. 2021, 177, 104885. [Google Scholar] [CrossRef]
- Shen, Z.J.; Zhu, F.; Liu, Y.J.; Li, Z.; Moural, T.W.; Liu, X.M.; Liu, X. MicroRNAs miR-14 and miR-2766 regulate tyrosine hydroxylase to control larval-pupal metamorphosis in Helicoverpa armigera. Pest Manag. Sci. 2022, 78, 3540–3550. [Google Scholar] [CrossRef]
- Li, X.; Hu, S.; Zhang, H.; Yin, H.; Wang, H.; Zhou, D.; Sun, Y.; Ma, L.; Shen, B.; Zhu, C. MiR-279-3p regulates deltamethrin resistance through CYP325BB1 in Culex pipiens pallens. Parasite Vector 2021, 14, 528. [Google Scholar] [CrossRef]
- Etebari, K.; Hussain, M.; Asgari, S. Identification of microRNAs from Plutella xylostella larvae associated with parasitization by Diadegma semiclausum. Insect Biochem. Mol. Biol. 2013, 43, 309–318. [Google Scholar] [CrossRef]
- Bai, J.; Lin, G.; Wang, Y.; Zhao, X.; Peng, L.; Xie, M.; Yang, J.; He, W.; You, M. Identification and functional characterization of larval-based microRNAs in the diamondback moth, Plutella xylostella. J. Biosaf. 2022, 31, 64–74. [Google Scholar] [CrossRef]
- Yang, S. Function Analysis of Protein Coding Gene and miRNA in Pupation and Pupae Development of in Chilo suppressalis. Ph.D. Thesis, Nanjing Agricultural University, Nanjing, China, 2015. [Google Scholar]
- Liang, Z.; Yang, Y.; Sun, X.; Du, J.; Wang, Q.; Zhang, G.; Zhang, J.; Yin, X.; Singh, D.; Su, P.; et al. Integrated analysis of microRNA and mRNA expression profiles in the fat bodies of MbMNPV-Infected Helicoverpa armigera. Viruses 2022, 15, 19. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Guo, X.; Li, Y.; Zhang, J.; Fu, Y. miR-34-5p, encoded by Spodoptera frugiperda, participates in anti-baculovirus by regulating innate immunity in the insect host. Int. J. Biol. Macro. Mol. 2022, 222, 2190–2199. [Google Scholar] [CrossRef]
- Liu, Z.; Xu, J.; Ling, L.; Luo, X.; Yang, D.; Yang, X.; Zhang, X.; Huang, Y. miR-34 regulates larval growth and wing morphogenesis by directly modulating ecdysone signaling and cuticle protein in Bombyx mori. RNA Biol. 2020, 17, 1342–1351. [Google Scholar] [CrossRef]
- Wu, Y.; Guo, Y.; Fan, X.; Zhao, H.; Zhang, Y.; Guo, S.; Jing, X.; Liu, Z.; Feng, P.; Liu, X.; et al. ame-miR-34 modulates the larval body weight and immune response of Apis mellifera Workers to Ascosphara apis Invasion. Int. J. Mol. Sci. 2023, 24, 1214. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Liu, N. Role of the G-protein-coupled receptor signaling pathway in insecticide resistance. Int. J. Mol. Sci. 2019, 20, 4300. [Google Scholar] [CrossRef] [PubMed]
- Hilliou, F.; Chertemps, T.; Maïbèche, M.; Le Goff, G. Resistance in the genus Spodoptera: Key insect detoxification genes. Insects 2021, 12, 544. [Google Scholar] [CrossRef] [PubMed]
- Etebari, K.; Afrad, M.H.; Tang, B.; Silva, R.; Furlong, M.J.; Asgari, S. Involvement of microRNA miR-2b-3p in regulation of metabolic resistance to insecticides in Plutella xylostella. Insect Mol. Biol. 2018, 27, 478–491. [Google Scholar] [CrossRef]
- Gong, P.P.; Wei, X.G.; Liu, S.N.; Yang, J.; Fu, B.L.; Liang, J.J.; Huang, M.J.; Du, T.H.; Yin, C.; Ji, Y.; et al. Novel_miR-1517 mediates CYP6CM1 to regulate imidacloprid resistance in Bemisia tabaci (Hemiptera: Gennadius). Pestic. Biochem. Phys. 2023, 194, 105469. [Google Scholar] [CrossRef]
- Giraudo, M.; Hilliou, F.; Fricaux, T.; Audant, P.; Feyereisen, R.; Le Goff, G. Cytochrome p450s from the fall armyworm (Spodoptera frugiperda): Responses to plant allelochemicals and pesticides. Insect Mol. Biol. 2015, 24, 115–128. [Google Scholar] [CrossRef]

| Primer | Sequence (from 5′ End to 3′ End) |
|---|---|
| miR-6094-5P-RT | GTCGTATCCAGTGCAGGGTCCGAGGTAT TCGCACTGGATACGACAGGTAC |
| miR-6094-5P-F | TCAGCGGTGGCCTGGG |
| miR-6094-3P-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAGGATC |
| miR-6094-3P-F | CGCGTATTCGAGACCTCTGCT |
| miR-10505-3p-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGAGCCA |
| miR-10505-3p-F | CGCGCGTAGGGTTAGAAACT |
| miR-14-5p-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCGAGT |
| miR-14-5p-F | GCGGGGGGAGGAATTG |
| miR-2765-5p-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGCCAAC |
| miR-2765-5p-F | CGCGCGTGGTAACTCCACCACC |
| miR-277-3p-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTGTCGT |
| miR-277-3p-F | CGCGCGTAAATGCACTATCTGGT |
| miR-307-3p-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGCTCAC |
| miR-307-3p-F | CGCGCGTCACAACCTCCTTGA |
| miR-34-5p-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACAACC |
| miR-34-5p-F | CGCGCGTGGCAGTGTGGTTAGCT |
| miR-R | AGTGCAGGGTCCGAGGTATT |
| Sf-actin(nei)-F | CGAGAAGATGACCCAGAT |
| Sf-actin(nei)-R | GATAGCACAGCCTGGATA |
| probe | FAM-CGCACTGGATACGAC-MGB |
| Sample | Flag | Raw Reads | Average Raw Reads | Clean Reads | Average Clean Reads |
|---|---|---|---|---|---|
| Egg-1 | E01 | 23,489,347 | 20,128,612 | 20,154,610 | 17,231,905 |
| Egg-2 | E02 | 24,772,159 | 20,128,612 | 20,586,799 | 17,231,905 |
| Egg-3 | E03 | 12,124,331 | 20,128,612 | 10,954,307 | 17,231,905 |
| Larva-1 | L01 | 10,837,072 | 16,315,920 | 1,502,965 | 3,731,980 |
| Larva-2 | L02 | 17,172,714 | 16,315,920 | 2,494,144 | 3,731,980 |
| Larva-3 | L03 | 20,937,973 | 16,315,920 | 7,198,831 | 3,731,980 |
| Pupa-1 | P01 | 20,479,951 | 23,520,239 | 14,980,746 | 17,141,575 |
| Pupa-2 | P02 | 25,798,866 | 23,520,239 | 19,742,017 | 17,141,575 |
| Pupa-3 | P03 | 24,281,901 | 23,520,239 | 16,701,963 | 17,141,575 |
| Adult-1 | A01 | 23,403,358 | 17,593,809 | 13,367,017 | 12,107,130 |
| Adult-2 | A02 | 12,435,180 | 17,593,809 | 10,254,039 | 12,107,130 |
| Adult-3 | A03 | 16,942,888 | 17,593,809 | 12,700,333 | 12,107,130 |
| Control | Case | Upregulated Genes | Downregulated Genes | Total DEGs |
|---|---|---|---|---|
| Egg | Larva | 32 | 22 | 54 |
| Adult | Larva | 9 | 9 | 18 |
| Pupa | Larva | 6 | 9 | 15 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.-P.; Chen, X.-Y.; Pu, D.-Q.; Yi, C.-Y.; Liu, C.-H.; Zhang, C.-C.; Wei, Z.-Z.; Guo, J.-W.; Yu, W.-J.; Chen, S.; et al. Identification and Prediction of Differentially Expressed MicroRNAs Associated with Detoxification Pathways in Larvae of Spodoptera frugiperda. Genes 2024, 15, 1021. https://doi.org/10.3390/genes15081021
Wang Y-P, Chen X-Y, Pu D-Q, Yi C-Y, Liu C-H, Zhang C-C, Wei Z-Z, Guo J-W, Yu W-J, Chen S, et al. Identification and Prediction of Differentially Expressed MicroRNAs Associated with Detoxification Pathways in Larvae of Spodoptera frugiperda. Genes. 2024; 15(8):1021. https://doi.org/10.3390/genes15081021
Chicago/Turabian StyleWang, Yan-Ping, Xing-Yu Chen, De-Qiang Pu, Chun-Yan Yi, Chang-Hua Liu, Cui-Cui Zhang, Zhen-Zhen Wei, Jing-Wei Guo, Wen-Juan Yu, Song Chen, and et al. 2024. "Identification and Prediction of Differentially Expressed MicroRNAs Associated with Detoxification Pathways in Larvae of Spodoptera frugiperda" Genes 15, no. 8: 1021. https://doi.org/10.3390/genes15081021
APA StyleWang, Y.-P., Chen, X.-Y., Pu, D.-Q., Yi, C.-Y., Liu, C.-H., Zhang, C.-C., Wei, Z.-Z., Guo, J.-W., Yu, W.-J., Chen, S., & Liu, H.-L. (2024). Identification and Prediction of Differentially Expressed MicroRNAs Associated with Detoxification Pathways in Larvae of Spodoptera frugiperda. Genes, 15(8), 1021. https://doi.org/10.3390/genes15081021






